Abstract
Comprehensive discovery of genetic mechanisms of drug resistance and identification of in vivo drug targets represent significant challenges. Here we present a functional variomics technology in the model organism Saccharomyces cerevisiae. This tool analyzes numerous genetic variants and effectively tackles both problems simultaneously. Using this tool, we discovered almost all genes that, due to mutations or modest overexpression, confer resistance to rapamycin, cycloheximide, and amphotericin B. Most significant among the resistance genes were drug targets, including multiple targets of a given drug. With amphotericin B, we discovered the highly conserved membrane protein Pmp3 as a potent resistance factor and a possible novel target. Widespread application of this tool should allow rapid identification of conserved resistance mechanisms and targets of many more compounds. New genes and alleles that confer resistance to other stresses can also be discovered. Similar tools in other systems such as human cell lines will also be useful.
Introduction
Genetic alterations in pathogens or cancer cells are major causes of drug resistance, with mutations and overexpression in drug target(s) or target pathways representing the major mechanisms. Discovering such resistance genes and mutations has thus also traditionally been exploited to identify drug targets (Barnes et al., 1984; Heitman et al., 1991; Kaufer et al., 1983; Rine et al., 1983). Discovering resistance genes could be done using the recently emerged genome sequencing (Albert et al., 2005) or high throughput complementation (Ho et al., 2009) methods. However, the discovery scope afforded by these methods is typically limited to the number of resistant isolates or cell lines being studied. Considering that every drug could encounter potentially multiple resistance mechanisms and that many drugs may each have multiple targets (Rask-Andersen et al.; Yildirim et al., 2007), many resistant isolates will have to be independently analyzed using these methods, and this could be costly and inconvenient.
Here we describe the systematic construction and screening of numerous genetic variants of the genes in the model organism S. cerevisiae. This technology, which we termed “functional variomics”, allows simultaneous, unbiased, and rapid identification of almost all genes that confer drug resistance due to mutations or modest overexpression. This technology centers on a set of high complexity random mutagenesis (or variomic) libraries of ~90% yeast genes expressed from low-copy centromeric plasmids. Screening these libraries as mixed populations of yeast cells against three test compounds rapidly identified most known resistance factors as well as novel genes. Most significant among these were the drug targets, including multiple targets of a given drug. Using this tool, we discovered Pmp3, a small membrane protein highly conserved in fungi and plants (Mitsuya et al., 2005; Navarre and Goffeau, 2000; Wang and Shiozaki, 2006), as an important amphotericin B (AmB) resistance factor, revealing a novel aspect of the mechanism of action of this commonly used antifungal drug. A Candida albicans homologue also caused AmB-resistance when expressed in S. cerevisiae, suggesting possibly a conserved mechanism across species.
Results
Constructing the variomic libraries
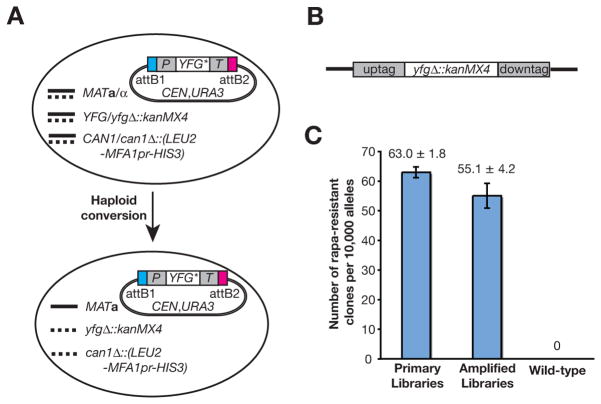
Each variant allele was expressed largely under control of the native upstream and downstream regulatory sequences from a centromeric plasmid, with URA3 as the selection marker (Figures 1A & S1). The variant alleles were flanked by attB1 and attB2 Gateway recombination sequences to facilitate their transfer to other vectors (Figure 1A). Each library was directly constructed in the corresponding heterozygous diploid deletion mutant that harbored a haploid selection reporter (can1Δ::LEU2-MFA1pr-HIS3) (Pan et al., 2004), and variant alleles could be tested both as heterozygous diploid cells and as haploid MATa cells in the absence of the chromosomal wild-type gene after haploid conversion (Huang et al., 2008) (Figures 1A & S1 and Table S1). A small subset of variomic libraries were constructed in heterozygous diploid deletion mutants of other genes due to a lack of corresponding deletion mutants (Table S1). The preexisting barcodes in each yeast deletion host strain (Giaever et al., 2002; Winzeler et al., 1999) also identify the corresponding variomic library (Figure 1A & 1B). A total of 5,847 variomic libraries were constructed, the majority (>99%) of which contained >2.0 × 105 independent primary alleles (Table S1). The relatively high genetic diversity of such a variomic library was previously demonstrated, with both singular and multiple mutations present in certain alleles. Mutations within a library likely have affected almost all amino acid residues of the encoded protein (Huang et al., 2008). However, mutational bias as a result of founder effects during error-prone PCR cannot be ruled out. Such variomic libraries have been stably amplified for at least 1,000-fold without a discernible loss in the capacity for discovering drug resistance genes and alleles, as demonstrated with two independent TOR2 variomic libraries (Figure 1C) and the genome-wide screens discussed below.
Figure 1. A summary of the yeast variomic libraries.
A. The variomic library of YFG (or Your Favorite Gene) was constructed in a heterozygous diploid deletion mutant that contains a haploid specific reporter can1Δ::(LEU2-MFA1pr-HIS3). Each mutant allele (YFG*) is expressed under control of the endogenous promoter (P) and terminator (T) from a centromeric (CEN) plasmid. The library can be converted to haploid MATa cells that lack the chromosomal wild-type gene. B. The diagram of a deletion cassette that contains two unique barcodes (uptag and downtag). C. The frequency for isolating rapamycin (Rapa) resistance alleles from TOR2 variomic libraries both before and after an ~1,000-fold amplification. Results of two independent experiments were averaged and plotted. (See also Figures S1 and S2, and Table S1)
Interrogating the libraries for drug resistance genes
Typically, only ~0.5–2% of variant alleles of a true drug resistance gene would confer resistance phenotypes (data not shown), we therefore anticipated a need to test a relatively large number of independent alleles in order to evaluate a gene’s possible role in drug resistance. We estimated that an average of ~10,000 alleles for each gene would be sufficient and manageable on a genome-wide scale. To screen for resistance genes, we assembled and amplified a pool of all available variomic libraries, and converted an aliquot of this pool into haploid MATa G418R Ura+ cells after meiosis (Figure S2). For each drug, we screened ~6 × 107 such haploid cells at ≥IC100 to rapidly enrich resistant alleles. Cells of all resistant colonies were harvested as a pool and analyzed with barcode deep sequencing (Smith et al., 2009) to identify the responsible genes (Figure S2). With each candidate gene, we subsequently screened ~10,000 alleles from the individual variomic library for resistant alleles and re-tested them for resistance phenotypes to establish a causative relationship (Figure S2).
Identifying rapamycin- and cycloheximide-resistance genes and drug targets
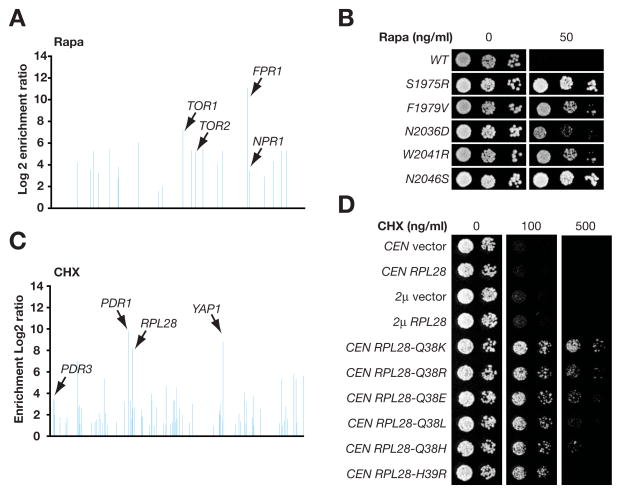
We first tested the immunosuppressant drug rapamycin, which forms a complex with the FKBP12 peptidyl-prolyl cis-trans isomerase (PPIase) to inhibit Tor kinases (Cardenas and Heitman, 1995; Chiu et al., 1994; Choi et al., 1996; Lorenz and Heitman, 1995; Sabatini et al., 1994). Recessive inactivating mutations in FKBP12 (encoded by FPR1) and dominant mutations in Tor (encoded by TOR1 and TOR2) confer high levels of resistance to rapamycin (Heitman et al., 1991). Similar mutations have also been shown to cause rapamycin-resistance in human cells and fungal pathogens (Bastidas et al., 2012; Cruz et al., 1999; Dumont et al., 1995). By screening the variomic libraries against rapamycin with barcode sequencing (Smith et al., 2009), we found that FPR1, TOR1, and TOR2 were enriched within the resistant population (all with P values < 1e-300) (Figure 2A and Table S2). We also found that inactivating mutations in NPR1 confers rapamycin-resistance (P value < 1e-300) (Figure 2A and data not shown), consistent with a previous report (Schmidt et al., 1998). Therefore, we were able to simultaneously rediscover all four known genes that confer rapamycin-resistance due to mutations. Significantly, three of these four genes represent the drug’s targets, demonstrating that screening the variomic libraries can simultaneously and accurately identify potentially multiple targets of a given drug.
Figure 2. Rapamycin (Rapa) and cycloheximide (CHX) resistance genes and alleles identified from screening the variomic libraries.
A. Rapa-resistance genes identified from screening the variomic libraries. Representation of each gene in both a drug resistant and a control population was compared. For simplicity, only genes with log2 enrichment ratios of >1.0 were plotted, with names of validated resistance genes also provided. The graphs in this and the following panels were derived from Tables S2. B. Rapamycin resistant alleles isolated from the TOR2 variomic library. Cells expressing wild-type (WT) or mutant TOR2 of tthe indicated genotypes from a centromeric plasmid were grown in the presence or absence of rapamycin (50ng/ml) at 30°C for 2 days. C. CHX-resistance genes identified from screening the variomic libraries. D. Alleles of RPL28 that confer resistance to CHX. Cells of a wild-type strain BY4743a/α carrying plasmids of indicated genotypes were grown in the presence or absence of cycloheximide at 30°C for 3 days. CEN is a centromeric low copy plasmid and 2μ is a high copy plasmid. (See also Figure S3 and Tables S2 and S3)
The variomic libraries have also provided an excellent opportunity for discovering key mutations that are responsible for drug resistance, some of which may help to define drug-binding sites on a target protein. For example, mutations residing within the FKBP12-rapamycin-binding (FRB) domain of Tor (S1972 of Tor1, S1975, W2041, and F2049 of Tor2) confer rapamycin-resistance (Lorenz and Heitman, 1995). In fact, sequencing analysis of 10 resistant alleles each for both TOR1 and TOR2 revealed that they all contained mutations within the FRB domain, including most of the known ones (Lorenz and Heitman, 1995) and several novel mutations (Figure 2B and Table S3). Except for tor2N2036D, all TOR1 and TOR2 alleles tested were dominant or semi-dominant and conferred resistance to rapamycin at >50ng/ml (Figure S3). Therefore, screening a variomic library allows facile discovery of resistance mutations within the drug-binding domain of a target protein.
We next tested cycloheximide, an inhibitor of protein synthesis that binds to eukaryotic ribosomes (Kaufer et al., 1983; Schneider-Poetsch et al., 2010). Cycloheximide-resistance mutations were previously found in the target protein Rpl28 and transcription factors Pdr1 and Pdr3 (Katzmann et al., 1994; Kaufer et al., 1983; Meyers et al., 1992), which regulate expression of multidrug resistance transporters. We identified all three genes by screening the variomic libraries (all with P values < 1e-8 (Figure 2C and Table S2). Mutating another transcription factor Yap1 also conferred cycloheximide-resistance (P value < 1e-300) (Figure 2C). We also explored the possibility of identifying key mutations on the target protein Rpl28 that might confer resistance. A Q38E mutation was previously shown to cause cycloheximide resistance (Kaufer et al., 1983). Sequencing 64 cycloheximide-resistance RPL28 alleles revealed that all contained mutations at either Q38 or the adjacent residue H39 (Figure 2D). The exclusive identification of mutations at Q38 and H39 of Rpl28 suggests a potential binding site for cycloheximide in this region of the protein.
Identifying amphotericin B (AmB)-resistance genes
We next studied the commonly used polyene antifungal drug AmB, which binds to ergosterol and forms transmembrane pores on fungal cell membrane that leads to leakage of cellular contents and cell death (Bolard, 1986; De Kruijff and Demel, 1974). It was recently suggested that the mechanism of action by AmB and its analogs might be multifaceted (Baginski and Czub, 2009; Gray et al., 2012; te Welscher et al., 2008). In addition, clinical resistance to this drug has been rare except that deleting ERG6 was previously shown to confer resistance in vitro, likely due to reduced levels of ergosterol as the putative drug target or receptor (Broughton et al., 1991; Jensen-Pergakes et al., 1998; Kelly et al., 1994; Young et al., 2003).
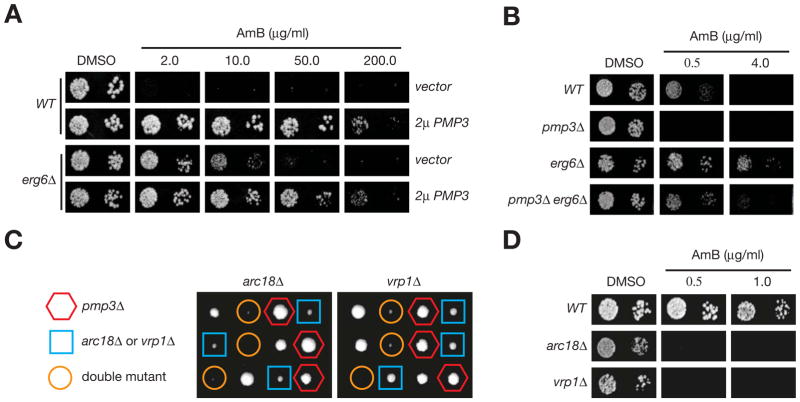
By screening the variomic libraries, we identified six AmB resistance genes (all with P values < 1e-8), including ERG6 and ERG11 of the ergosterol biosynthesis pathway, PMP3, PDR1, RPS7, and RSM24 (Figure 3A). Identification of ERG6 and ERG11 was expected and consistent with the model that membrane ergosterol is required for the toxic effect of AmB (De Kruijff and Demel, 1974). In contrast, the discovery of PMP3, which encodes a highly conserved hydrophobic small membrane protein (of 55 amino acids) involved in regulating ion homeostasis (Navarre and Goffeau, 2000), was unexpected. Pmp3 contains two predicted transmembrane helical domains and is likely mostly embedded in the plasma membrane. We found that barcodes of pmp3Δ and bsc2Δ were the most enriched, accounting for 11.23% and 83.85%, respectively, of all barcodes sequenced in the resistant cell population (Figure 3B). They were both present in the PMP3 variomic library because the host cells were accidentally a preexisting mixture of both pmp3Δ and bsc2Δ deletion mutants. We subsequently found that most alleles within the PMP3 variomic libraries were AmB-resistant, likely due to modest overexpression from the plasmids. Consistent with this idea, expressing wild-type PMP3 from the same centromeric vector also conferred AmB-resistance (Figure S4). This explained the massive overrepresentation of PMP3 barcodes in the AmB-resistant cell population (Figure 3B) and consequently possible suppression of enrichment ratios of other AmB-resistance genes (Figure 3A). As expected, expressing PMP3 from a high copy plasmid conferred AmB-resistance (Figure 3C). Overexpressing a Candida albicans Pmp3 homolog in S. cerevisiae had a similar effect (Figure 3C), indicating a conserved mechanism across species. It will be interesting to explore whether PMP3 might play a role in the rare cases of AmB resistant isolates of fungal pathogens.
Figure 3. AmB resistance genes identified from screening the variomic libraries.
A. AmB resistance genes identified from screening the variomic libraries. B. Barcode percentage of verified resistance genes in the AmB resistant cell population derived from screening the variomic libraries. C. Overexpression of both Saccharomyces cerevisiae (S.c.) and Candida albicans (C.a.) PMP3 confer resistance to AmB. Cells shown in this and the next panel were grown in the presence or absence of AmB at 30°C for 2 days. D. A pmp3Δ mutation confers hypersensitivity to AmB. (See also Figure S4 and Table S2)
Pmp3 antagonizes the action of AmB
Consistent with an active role of Pmp3 in AmB-resistance, we also found that a pmp3Δ mutant was hypersensitive to the drug (Figure 3D). Given that Pmp3 is required for salt resistance (Navarre and Goffeau, 2000) (Figure 4A), we initially thought that it might regulate the flux of ions through pores formed by the AmB-ergosterol complex on the membrane. However, overexpression of PMP3 did not confer resistance to a high concentration of NaCl in a wild-type strain or suppress salt-hypersensitivity of two mutants (crz1Δ and mck1Δ) (Figure 4A). In contrast, AmB-hypersensitivity of both mutants was suppressed by PMP3 overexpression (Figure 4A). In addition, a sky1Δ mutation that suppresses the salt-sensitivity of a pmp3Δ mutant (Erez and Kahana, 2002) failed to suppress its hypersensitivity to AmB (data not shown). These results together suggest that the role of PMP3 in AmB-resistance does not involve its role in ion homeostasis.
Figure 4. Pmp3 specifically antagonizes AmB but not its analogs.
A. PMP3 overexpression confers resistance to AmB but not to high salt. Isogenic strains of indicated genotypes were grown in the presence or absence of AmB or NaCl at 30°C for 2 (DMSO) or 3 days (NaCl and AmB). Note that growth of the wild-type (WT) strain was similarly impaired although not completely blocked by NaCl both in the presence and absence of PMP3 overexpression. B. PMP3 overexpression specifically antagonizes the effect of AmB (6.0 μg/ml) but not that of nystatin (6.0 μg/ml) on Sytox uptake. An erg6Δ mutant was used as a control in this and the following panel. The result under each condition was the average of two independent experiments and the error bar represents the standard error of the mean. The percentage of permeated cells under all condition were: 4.43±0.63 (WT, DMSO); 3.60±0.95 (PMP3, DMSO); 11.55±2.45 (erg6Δ, DMSO); 98.60±0.60 (WT, AmB); 23.35±0.73 (PMP3, AmB); 16.14±1.55 (erg6Δ, AmB); 61.47±7.52 (WT, nystatin); 65.45±3.78 (PMP3, nystatin); 26.90±8.90 (erg6Δ, nystatin). C. PMP3 overexpression confers resistance specifically to AmB but not nystatin, filipin, and natamycin. Cells were grown at 30°C for 3 days.
We next tested the possibility that Pmp3 directly antagonizes AmB’s known membrane permeating effect (De Kruijff and Demel, 1974). When treated with AmB at ≥1.0 μg/ml in liquid cultures, wild-type yeast cells were mostly dead as reflected by Sytox uptake and staining (Figure 4B and data not shown). At an AmB concentration of 1.0 μg/ml, Sytox uptake mainly reflects membrane permeation because the antifungal effect of the drug under this condition is mediated by membrane permeation rather than other mechanisms such as ergosterol binding (Gray et al., 2012). In contrast, cells overexpressing PMP3 were resistant to membrane permeation under similar conditions and most of them were alive (Figure 4B and data not shown). Therefore, Pmp3 counteracts the membrane permeation effect of AmB. We subsequently investigated whether Pmp3 makes the plasma membrane more refractory to general chemical perturbations or specifically to the action(s) of AmB. In support of the latter possibility, PMP3 overexpression had no effect on the membrane permeating activity of nystatin (Figure 4B), a structural analogue of AmB. Consistently, PMP3 overexpression failed to confer resistance to nystatin (Figure 4C) and two other AmB analogues, filipin and natamycin (Figure 4C). A pmp3Δ mutation also did not confer hypersensitivity to these AmB analogs (Data not shown). The specificity in Pmp3’s effect toward AmB was also demonstrated in comparison with erg6Δ, which confers resistance to AmB and all three AmB analogues in both membrane permeation and cell growth assays (Figures 4B & 4C).
Pmp3 is a possible target of AmB
To further investigate the relationship between Pmp3 and AmB, we compared the effects of Pmp3 and ergosterol synthesis on the toxicity of AmB. PMP3 overexpression and erg6Δ were two genetic factors that confer the highest levels of AmB resistance among all factors tested (Data not shown). We found that PMP3 overexpression confers much higher levels of AmB-resistance even when compared to erg6Δ and there was little additive effect between these two (Figure 5A). We also found that pmp3Δ partly suppressed AmB-resistance conferred by erg6Δ (Figure 5B). These results together suggested that Pmp3 likely antagonizes AmB in a manner that is independent of ergosterol synthesis or functions. The fact that PMP3 overexpression confers the highest levels of AmB-resistance among all factors tested also suggests that its effects on AmB-resistance are unlikely mediated by other genes. These results, together with the observation that Pmp3 specifically antagonizes the effect of AmB but not its structural analogs (Figures 4B & 4C), suggest that Pmp3 might directly binds to AmB. A prediction of this model is that AmB might also inhibit certain functions of Pmp3. To test this possibility, we took advantage of synthetically lethal or sick interactions between pmp3Δ and mutations affecting actin polarization (arc18Δ and vrp1Δ) (Costanzo et al., 2010) (Figure 5C) and found that both the arc18Δ and vrp1Δ mutants are hypersensitivity to AmB (Figure 5D). Based on these genetic results, we speculate that Pmp3 might be a target of AmB in yeast.
Figure 5. Pmp3 is a possible target of AmB.
A. Different effects of PMP3 overexpression and erg6Δ on high levels of AmB resistance. Cells were grown in the presence or absence of AmB at 30°C for 3 days. B. A pmp3Δ mutation partly abrogates AmB-resistance conferred by erg6Δ. Cells were incubated at 30°C for 2 days. C. The arc18Δ and vrp1Δ mutations are synthetically lethal or sick with a pmp3Δ mutation as revealed by tetrad analysis. D. The arc18Δ and vrp1Δ mutations are hypersensitive to AmB. Cells of isogenic strains of indicated genotypes were incubated in the presence or absence of AmB at 30°C for 3 days.
Taken together, there is likely a bidirectional relationship between Pmp3 and AmB. In one direction, Pmp3 directly antagonizes the membrane permeation effect of AmB, likely in an ergosterol-independent manner. It likely also antagonize the erogsterol-binding effect of this drug given that PMP3 overexpression has prevented cell death at AmB concentrations that are much higher than needed for killing through the ergosterol-binding mechanism (Figure 5A) (Gray et al., 2012). In the other direction, AmB might inhibit Pmp3. However, it is also possible that Pmp3 acts simply by preventing insertion of AmB into the plasma membrane. Such possibilities will need to be further investigated in the future using biochemical or biophysical assays.
Discussion
Here we described a powerful functional variomics tool in the yeast S. cerevisiae for systematic discovery of drug resistance genes and alleles and drug targets. It shares some similarity with a previously described genome sequencing approach (Albert et al., 2005; Wacker et al., 2012) but offers several advantages. First, it is more comprehensive, with most, if not all, genes of a genome being simultaneously evaluated without bias. As a result, it discovers multiple resistance genes to a drug simultaneously and, consequently, enables simultaneous identification of potentially multiple targets of a given drug (e.g., Tor1, Tor2, and FKBP12 for rapamycin). In contrast, the scope of discovery afforded by the sequencing approach is limited to the particular mutations harbored within typically a few drug-resistant cell lines or isolates being sequenced. Even if a large number of such cell lines or isolates are sequenced regardless of the experimental cost, there is still a possible “hot-spot” issue, where the same gene(s) are discovered over and over again while others are completely overlooked. Second, the presence of numerous pre-constructed variant alleles within the variomic libraries makes it much easier to obtain drug-resistant isolates for subsequent gene identification than with genome sequencing, which typically relies on spontaneous mutations that occur at much lower mutational rates. In many cases, even isolating drug resistant cell lines is a significant challenge with the sequencing approach. As a result, functional variomics has the potential to provide much higher experimental throughput because many different drugs can be simultaneously screened with the same pre-constructed libraries. In fact, all three screens described in this study were performed in parallel. Third, identifying drug resistance genes with quantitative barcode sequencing analysis (in the context of functional variomics) is much simpler than discovering single base substitutions within a whole genome with the sequencing approach. The huge potential in sample multiplexing with barcode sequencing analysis (Smith et al., 2009) could also dramatically reduce the experimental costs with the functional variomics approach. Fourth, the pre-constructed variomic library of a target gene permits convenient isolation of many distinct resistant alleles that could help to define amino acid residues critical for drug binding or regulation of the target’s activity. On the other hand, the functional variomics technology does have limitations—it is not applicable in organisms that do not have convenient genetic tools and does not allow discovery of resistance mechanisms that simultaneously involve multiple genes. However, we are optimistic that a similar tool could be applicable in human cell culture systems, where other high throughput functional genomic tools have already been successfully implemented.
With the three drugs studied, this functional variomics tool is also advantageous compared to other existing tools such as genome-wide haploinsufficiency profiling (Giaever et al., 1999) and dosage suppression (Butcher et al., 2006) screens in identifying targets. We rediscovered all known or expected targets or target pathways of the three drugs, including Tor1, Tor2, and FKBP12 of rapamycin, Rpl28 of cycloheximide, and Erg6 and Erg11 of AmB. We also discovered Pmp3 as a possible novel target of AmB by taking advantage of potentially modest gene overexpression associated with the variomic libraries. In comparison, haploinsufficiency profiling would have been successful with only Tor1 and Tor2 and the dosage suppression would have been successful with Tor1, Tor2 and Pmp3 based on individually testing all target genes (data not shown). Both methods would have failed to identify FKBP12, Rpl28, Erg6, and Erg11. However, mutating a drug target might not always confer drug resistance. For example, mutations in a target protein that would have prevented drug binding, and thus caused drug resistance, might also inactivate the protein. In such a case, haploinsufficiency profiling and dosage suppression could be more successful.
In this study, we deliberately chose drug concentrations at or slightly higher than IC100 in order to rapidly enrich resistant colonies. This has allowed the discovery of most of the significant resistance genes and alleles but might have precluded those with marginal effects. Regarding drug target identification, there is a tendency or maybe a need to further narrow down the drug-resistance gene list, possibly by using an even higher drug concentration in the initial screen. However, as with any positive selection screen, resistant colonies could arise from spontaneous mutations that are unnecessarily associated with the barcoded genes and these could lead to false positive discoveries. Depending on the particular genes affected by such spontaneous mutations, these false positive discoveries may or may not persist when a higher drug concentration is used in the screen. In addition, as discussed with PMP3 and BSC2, pre-existing cross-contamination of strains used to host the libraries could also contribute to false positive discoveries. Given these considerations, we prefer using IC100 of a drug in an initial screen and subsequently validating the hits with higher drug concentrations both to weed out false positives and to further distinguish resistance levels among the true resistance genes and alleles. This will allow discovery of most of the drug resistance genes, which may or may not confer the same level of drug resistance, allowing the identification of possibly multiple targets of a given drug.
Here we mainly focused on the application of the functional variomics tool in systematic and rapid discovery of drug resistance genes and drug targets. This technology can also be used to rapidly identify genes and alleles that confer resistance to other types of stresses such as high temperature, high salt, high levels of ethanol, and extreme pH, etc. The variant alleles could also be combined with en masse mating to create strains with novel phenotypes. In addition, the individual variomic libraries could be screened to identify conditional or hypomorphic alleles for studying gene functions. That said, this yeast functional variomics tool is a very valuable resource to the research community. Such tools in other genetically tractable organisms (e.g., cultured mammalian cells) could also be implemented and will be similarly useful.
Materials and Methods
Strains and plasmids
Yeast strains used in this study include the wild-type diploid strain BY4743a/α (MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 lys2Δ0/LYS2 met15Δ0/MET15) (Brachmann et al., 1998) and genome-wide haploid convertible heterozygous diploid yeast deletion mutants (MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 lys2Δ0/LYS2 met15Δ0/MET15 can1Δ ::LEU2-MFA1pr::HIS3/CAN1 YFG/yfgΔ::KanMX; “YFG” stands for Your Favorite Gene) (Pan et al., 2006). Bacterial strain DH5α was used as the host for constructing the promoter/terminator clones and for recovering plasmids from yeast cells.
The variomic libraries were built on a yeast-bacteria shuttle vector pXP597 or pXP688, both derived from the centromeric plasmid pRS416 (Brachmann et al., 1998), with URA3 as the selectable marker in yeast. Both vectors contained a single SmaI site flanked by the Gateway recombination attB1 and attB2 sites (Figure S1). Their DNA sequences are available upon request. They are essentially the same except that the latter contains an additional 22 bp sequence between the attB1 and attB2 sites to allow more efficient cloning using homology-dependent methods. All plasmid-borne variant alleles used in the validation assays were derived from screening the corresponding variomic libraries. The overexpression clones were constructed on a 2μ high copy plasmid pXP684 derived from YEplac195 (Gietz and Sugino, 1988), with each gene expressed under control of the endogenous promoter and terminator sequences. A PMP3 homologue from Candida albicans was reverse translated using optimal codons of S. cereviciae, synthesized with overlapping PCR, and expressed under control of the 5′- and 3′-UTRs of the S. cerevisiae PMP3 from pXP684. The DNA sequences of these constructs are also available upon request.
Chemicals
Rapamycin, cycloheximide, amphotericin B, nystatin, filipin, and natamycin were all purchased from Sigma. Rapamycin was dissolved in water as a 1 mg/ml stock. All others were dissolved in DMSO as 10 mg/ml stocks. Stock solutions of Amphotericin B and its analogues were always made and used afresh.
High throughput screening for resistance genes
~6.0 × 107 haploid MATa G418R Ura+ cells derived from the pool variomic libraries were plated on a 150mm plate of solid synthetic complete medium lacking uracil (SC-Ura) that either contained or lacked a test compound at ≥IC100, with an average of ~10,000 cells representing each variomic library. The compounds tested were cycloheximide (at 100 ng/ml), rapamycin (at 20 ng/ml), and amphotericin B (at 4.0 μg/ml). The plates were incubated at 30°C for 3 or 4 days until the appearance of resistant colonies. Rapamycin-resistant clones were pooled and subjected to an additional round of selection to clear the relatively high background levels. Resistant clones of the other screens were pooled and directly analyzed by barcode sequencing as previously reported (Smith et al., 2009). Within each sample, the numbers of barcode sequence read for both uptag and downtag of each host strain were averaged and normalized according to a total of 1 million reads per sample. Fold enrichment of each host strain in the drug resistance was calculated by comparing the normalized barcode count against that in a control population. An differential P value was calculated for each strain between the drug treatment and control populations based on the Poison test that is widely used on sequencing data (Chen et al., 2012; Zhang et al., 2008). The P values were further corrected based on the bonferroni method (Benjamini and Hochberg 1995; Benjamini and Yekutieli 2001). The results of all screens were presented in Table S2. Genes with enrichment log2 ratios >1.0 and by a P value < 1e-8 were individually retested. The barcode deep sequencing data has been deposited at the Sequence Read Archive (SRA) and the Accession number is SRP017440.
Validating drug resistance genes and alleles
To validate a candidate resistance gene, resistant alleles were re-isolated from the corresponding variomic library by screening ~10,000 freshly derived MATa G418R Ura+ cells. Plasmid DNA of each allele was recovered from yeast cells and transformed into competent DH5α cells. Each recovered plasmid was transformed back into the corresponding haploid-convertible heterozygous diploid yeast knockout mutants or a wild-type yeast BY4743a/α by selecting on solid SC-Ura. Transformants of the heterozygous diploid knockout mutants were sporulated and converted to haploid MATa G418R Ura+ cells by growing on SC-Ura-Leu-His-Arg+Canavanine+G418 as previously described (Huang et al., 2008). Representative haploid isolates were tested for growth on solid SC-Ura either with or without a drug of interest. Both an empty vector and a plasmid expressing the corresponding wild-type gene from a centromeric plasmid were used as negative controls in all cases. The cultures were incubated at 30°C for 2 to 3 days and photographed. Transformants of BY4743a/α were spotted as 10 x serial dilutions on the surface of SC-Ura plates that either contained or lacked a drug of interest to test whether the drug-resistant alleles are dominant or recessive.
Sytox uptake assay
Cells of wild-type yeast BY4743a/α containing a vector (pXP684) or a PMP3 overexpression plasmid and an isogenic erg6Δ/erg6Δ mutant containing the vector were each grown in 5 ml of liquid SC-Ura and incubate at 30°C for overnight. Each overnight culture was used to inoculate 5 ml of fresh SC-Ura at a starting cell density of 0.4 OD600nm/ml and incubate at 30°C for 2 hrs. AmB or Nystatin was added at a final concentration of 0 μg/ml, 1.0μg/ml, or 6.0 μg/ml. 5 mM of MgCl2 and 0.2 μM of Sytox Green (Invitrogen) were also added to each culture, which was incubated at 30°C with shaking for 6 hrs. Cells were observed under an Axioplan 2 imaging microscope and photographed under both DIC and fluorescence settings. Fluorescence staining of DNA within a cell indicates uptake of Sytox green and permeation of the plasma membrane. More than 100 cells were analyzed with each sample, with the percentage of fluorescence staining cells calculated. Each experiment was carried out as two independent repeats and the results were averaged and plotted.
Supplementary Material
Highlights.
This article describes a functional variomics tool for drug target identification.
This tool compares favorably with other target identification tools.
This tool will greatly facilitate linking phenotypes to point mutations.
This work discovered Pmp3 as a novel target of amphotericin B.
Acknowledgments
We thank Dr. Zheng Zhou for the access to the Axioplan 2 imaging microscope. H.W. was supported by postdoctoral fellowship F32EY19430. We also thank the reviewers for their constructive comments that have significantly improved the paper. This work was supported by the National Institutes of Health (NIH) grants HG004840 to X.P. and 1S10RR026550 to R.C. We would also like to acknowledge the support of the Dan L. Duncan Cancer Center (P30CA125123) and the Administrative and Genome-wide RNAi Screens Cores (IDDRC P30HD024064).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, Richmond TA, Molla M, Singh J, Green RD, Berg DE. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- Baginski M, Czub J. Amphotericin B and its new derivatives - mode of action. Curr Drug Metab. 2009;10:459–469. doi: 10.2174/138920009788898019. [DOI] [PubMed] [Google Scholar]
- Barnes G, Hansen WJ, Holcomb CL, Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol. 1984;4:2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Shertz CA, Lee SC, Heitman J, Cardenas ME. Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12-dependent inhibition of Tor. Eukaryot Cell. 2012;11:270–281. doi: 10.1128/EC.05284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Series B. 1995;57:289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta. 1986;864:257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Broughton MC, Bard M, Lees ND. Polyene resistance in ergosterol producing strains of Candida albicans. Mycoses. 1991;34:75–83. doi: 10.1111/j.1439-0507.1991.tb00623.x. [DOI] [PubMed] [Google Scholar]
- Butcher RA, Bhullar BS, Perlstein EO, Marsischky G, LaBaer J, Schreiber SL. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat Chem Biol. 2006;2:103–109. doi: 10.1038/nchembio762. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Heitman J. FKBP12-rapamycin target Tor2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, et al. DANPOS: Dynamic Analysis of Nucleosome Position and Occupancy by Sequencing. Genome Res. 2012 doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci U S A. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Cavallo LM, Gorlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kruijff B, Demel RA. Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawii cells and lecithin liposomes. 3. Molecular structure of the polyene antibiotic-cholesterol complexes. Biochim Biophys Acta. 1974;339:57–70. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]
- Dumont FJ, Staruch MJ, Grammer T, Blenis J, Kastner CA, Rupprecht KM. Dominant mutations confer resistance to the immunosuppressant, rapamycin, in variants of a T cell lymphoma. Cell Immunol. 1995;163:70–79. doi: 10.1006/cimm.1995.1100. [DOI] [PubMed] [Google Scholar]
- Erez O, Kahana C. Deletions of SKY1 or PTK2 in the Saccharomyces cerevisiae trk1Δ trk2Δ mutant cells exert dual effect on ion homeostasis. Biochem Biophys Res Commun. 2002;295:1142–1149. doi: 10.1016/s0006-291x(02)00823-9. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A. 2012;109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Sucgang RS, Lin YY, Shi X, Boeke JD, Pan X. Plasmid-chromosome shuffling for non-deletion alleles in yeast. Nat Methods. 2008;5:167–169. doi: 10.1038/nmeth.1173. [DOI] [PubMed] [Google Scholar]
- Jensen-Pergakes KL, Kennedy MA, Lees ND, Barbuch R, Koegel C, Bard M. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants. Antimicrob Agents Chemother. 1998;42:1160–1167. doi: 10.1128/aac.42.5.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Burnett PE, Golin J, Mahe Y, Moye-Rowley WS. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufer NF, Fried HM, Schwindinger WF, Jasin M, Warner JR. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983;11:3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SL, Lamb DC, Taylor M, Corran AJ, Baldwin BC, Powderly WG. Resistance to amphotericin B associated with defective sterol delta 8-->7 isomerase in a Cryptococcus neoformans strain from an AIDS patient. FEMS Microbiol Lett. 1994;122:39–42. doi: 10.1111/j.1574-6968.1994.tb07140.x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- Mitsuya S, Taniguchi M, Miyake H, Takabe T. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta. 2005;222:1001–1009. doi: 10.1007/s00425-005-0043-9. [DOI] [PubMed] [Google Scholar]
- Navarre C, Goffeau A. Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 2000;19:2515–2524. doi: 10.1093/emboj/19.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Rine J, Hansen W, Hardeman E, Davis RW. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci U S A. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, Roth FP, Giaever G, Nislow C. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher YM, ten Napel HH, Balague MM, Souza CM, Riezman H, de Kruijff B, Breukink E. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J Biol Chem. 2008;283:6393–6401. doi: 10.1074/jbc.M707821200. [DOI] [PubMed] [Google Scholar]
- Wacker SA, Houghtaling BR, Elemento O, Kapoor TM. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat Chem Biol. 2012;8:235–237. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LY, Shiozaki K. The fission yeast stress MAPK cascade regulates the pmp3+ gene that encodes a highly conserved plasma membrane protein. FEBS Lett. 2006;580:2409–2413. doi: 10.1016/j.febslet.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Young LY, Hull CM, Heitman J. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother. 2003;47:2717–2724. doi: 10.1128/AAC.47.9.2717-2724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.