Abstract
Stringent regulation of the interferon (IFN) signalling pathway is essential for maintaining the immune response to pathogens and tumours. The transcription factor STAT1 is a crucial mediator of this response. Here, we show that hCAF1/CNOT7 regulates class I and II IFN pathways at different crucial steps. In resting cells, hCAF1 can control STAT1 trafficking by interacting with the latent form of STAT1 in the cytoplasm. IFN treatment induces STAT1 release, suggesting that hCAF1 may shield cytoplasmic STAT1 from undesirable stimulation. Consistently, hCAF1 silencing enhances STAT1 basal promoter occupancy associated with increased expression of a subset of STAT1-regulated genes. Consequently, hCAF1 knockdown cells exhibit an increased protection against viral infection and reduced viral replication. Furthermore, hCAF1 participates in the extinction of the IFN signal, through its deadenylase activity, by speeding up the degradation of some STAT1-regulated mRNAs. Since abnormal and unbalanced JAK/STAT activation is associated with immune disorders and cancer, hCAF1 could play a major role in innate immunity and oncogenesis, contributing to tumour escape.
Keywords: CCR4–NOT, deadenylation, hCAF1/CNOT7, interferon signalling, STAT1
Introduction
The CCR4–NOT complex is an evolutionarily conserved multi-subunit complex that regulates several aspects of eukaryotic gene expression, including the repression and activation of mRNA translation, control of mRNA elongation, deadenylation and subsequent degradation of mRNA and even protein degradation (for review, see Collart and Panasenko, 2012 and Miller and Reese, 2012). Deadenylase (Tucker et al, 2001) and E3 ubiquitin ligase (Panasenko and Collart, 2011) are two enzymatic activities that have been described for different subunits of the CCR4–NOT complex and could mediate most of its functions. The CAF1 and CCR4 subunits were initially identified as the major cytoplasmic deadenylases in budding yeast. Homologues of these proteins have been identified in Metazoa, indicating that this mRNA degradation pathway is evolutionarily conserved (Denis and Chen, 2003; Bianchin et al, 2005; Bartlam and Yamamoto, 2010). Both proteins have been shown to localize to cytoplasmic P-bodies with translationally repressed mRNA and miRNAs (Jakymiw et al, 2005; Pillai, 2005; Eulalio et al, 2009). CAF1 deadenylase activity has been shown to be responsible, at least in part, for miRNA-mediated gene repression (Behm-Ansmant et al, 2006; Eulalio et al, 2009; Fabian et al, 2009). Furthermore, mammalian CAF1 appears to be a crucial partner of the antiproliferative BTG/TOB proteins (Bogdan et al, 1998; Rouault et al, 1998; Ikematsu et al, 1999; Prevot et al, 2001) that are also implicated in mRNA turnover (Ezzeddine et al, 2007; Mauxion et al, 2008). hCAF1 has also been described to regulates transcription of several nuclear receptors (Prevot et al, 2001; Garapaty et al, 2008; Aslam et al, 2009) and the activity of the protein arginine methyl transferase PRMT1 (Robin-Lespinasse et al, 2007).
The biological role of CAF1 has been examined in a range of eukaryotic species. In yeast, caf1-deleted strains are sensitive to high temperatures and caffeine (Hata et al, 1998). In C. elegans, CAF1 is essential for embryonic and larval development (Molin and Puisieux, 2005). Additionally, CAF1 is required for normal growth of trypanosomes (Schwede et al, 2009). Mice that lack CAF1 are infertile due to impaired maturation of spermatogenic cells resulting in multiple defects during spermatic differentiation (Berthet et al, 2004; Nakamura et al, 2004). In plants, CAF1 has been shown to be involved in both development and response to biotic stresses. The Arabidopsis CAF1-like gene regulates the deadenylation of stress responsive mRNAs and plant defence responses to pathogen infection (Liang et al, 2009). Despite considerable progress in the understanding of CAF1-mediated regulation of gene expression, the physiological targets and the specific pathways by which it exerts its functions remain unclear.
In this study, we found that hCAF1 physically interacts with STAT1 and negatively regulates interferon (IFN)/STAT1 signalling by acting at different crucial steps of this pathway. Accordingly, hCAF1 depletion resulted in a hyper-activated subset of STAT1-regulated genes that led to retarded cell growth and an enhanced response to viral infection.
Results
hCAF1 regulates IFN-inducible genes
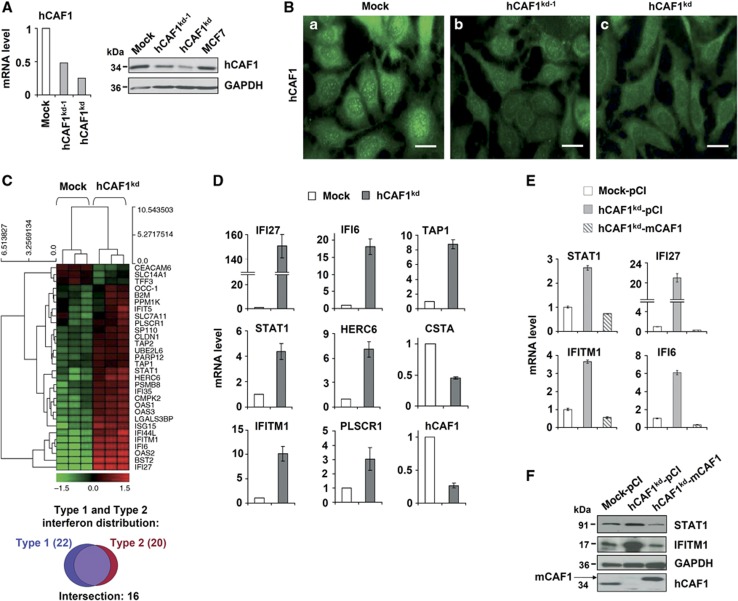
To identify novel cellular pathways regulated by hCAF1, we first ascertained its physiologically relevant targets. As a starting point, we knocked down hCAF1 in the human cell line MCF7. We produced BLOCK-iT Pol II miR RNAi vectors expressing specific miRNAs targeting hCAF1 (kd and kd-1) and we used a non-specific miRNA (mock) as a control. As shown in Figure 1A, the residual levels of hCAF1 mRNA and protein in depleted cells were about 25% in hCAF1 knockdown cells (hCAF1kd) and 50% in hCAF1 knockdown 1 cells (hCAF1kd-1) compared to control mock cells. hCAF1 depletion was also confirmed by immunofluorescence. In control cells, hCAF1 localized in both the cytoplasm and nuclear speckles as previously described (Robin-Lespinasse et al, 2007) (Figure 1Ba) and we observed decreased staining in hCAF1kd-1 and hCAF1kd cells (Figures 1Bb and c).
Figure 1.
Characterization of hCAF1-depleted cells. MCF7 cells were transfected with vectors expressing control miRNA (mock) or with two alternative miRNAs (called kd, and kd-1) targeting hCAF1. After vector transfection and selection, mock, hCAF1kd and hCAF1kd-1 cells were obtained. Total RNA or protein extracts were prepared to test hCAF1 knockdown efficiency. (A) Left panel, SYBR green real-time RT–PCR analysis was performed for detection of transcript levels of hCAF1. Results were normalized using 36B4 mRNA level as an internal control. Transcript levels in control cells (mock) were set to 1. Right panel, hCAF1 protein levels were analysed by western blot from 30 μg of protein extracts. (B) Endogenous hCAF1 expression and localization in hCAF1kd and hCAF1kd-1 compared to the mock control, by Immunofluorescent staining using mouse polyclonal anti-CAF1 antibody and goat anti-mouse Alexa 488-conjugated secondary antibody (green). Scale bar=20 μm. (C) Upper: summary of Human Exon 1.0 ST Array analysis results. Diagram of the hierarchical clustering of gene expression profiles of control versus hCAF1-deficient hCAF1kd cell lines. Probes are represented vertically, whereas conditions are shown horizontally. Lower: type I and II interferon distribution by ISG Database. (D) Validation of the DNA array screen. SYBR green RT-qPCR analysis of the upregulated gene products IFI27, IFI6, TAP1, STAT1, IFITM1, HERC6, PLSCR1 and the downregulated gene CSTA. Total RNA isolated from mock and hCAF1kd cells was reverse transcribed, and PCR was performed with primers specific for the transcripts of the indicated genes. Gene expression levels were normalized to internal controls 36B4 and shown as expression levels of hCAF1kd cells relative to expression levels in control cells (arbitrarily set to 1). (E, F) Rescue of hCAF1 functions in knockdown cells. hCAF1kd cells were stably transfected with a plasmid expressing mouse flag CAF1 (insensitive to miRNAs), or with the empty vector used as a control. (E) The expression of the indicated genes was analysed by real-time RT-qPCR as in (D). (F) Efficiency of flag mCAF1 overexpression and the protein expression of the indicated hCAF1-regulated genes was assessed by western blot. The experiments illustrated in (D) and (E) were performed in triplicate and expressed as mean values of three independent experiments. Standard deviations are shown.
Source data for this figure is available on the online supplementary information page.
We utilized microarray analysis to identify any hCAF1-regulated genes, comparing the gene expression profiles of hCAF1kd cells and mock cells. With a cutoff of 1.5 for fold change and 0.05 for the P-value, the analysis of expression data showed that the most differentially expressed genes were upregulated in hCAF1 knockdown cells, consistent with hCAF1 acting as a repressor of gene expression (gene lists in Supplementary Table 1). We analysed gene co-occurrence in common biological functions and found that about 40% of upregulated transcripts encoded proteins involved in IFN-mediated immunity (Supplementary Table 2). Finally, a comparison between the list of genes upregulated after hCAF1 depletion and the ISG Database (http://www.interferome.org) showed that over 50% of the upregulated genes corresponded to genes involved in both type I (α and β) and type II (γ) IFN signalling pathways (Figure 1C, lower panel). Using RNA from mock and hCAF1kd cells, we confirmed the microarray results on a subset of hCAF1-target genes by RT–qPCR (Figure 1D). We also obtained similar results in MCF7 cells transiently depleted of hCAF1 by two independent small-interfering RNAs (siRNAs) targeting different regions of hCAF1 mRNA (Supplementary Figure 1A). To rule out the possibility of off-target effects and further confirm the specificity of the silencing, we expressed a tagged form of mouse CAF1 cDNA (flag-mCAF1) (resistant to miRNA silencing) in hCAF1kd and control cells. In this rescue experiment, we found that the expression of mCAF1 restored the expression of several genes upregulated by hCAF1 knockdown, almost to wild-type levels (Figure 1E). Overexpression and rescue of STAT1 and IFITM1 were also confirmed at the protein level (Figure 1F). Altogether, these data demonstrate that the effects of hCAF1 depletion are specific and reversible.
Physiological outcome of hCAF1 knockdown
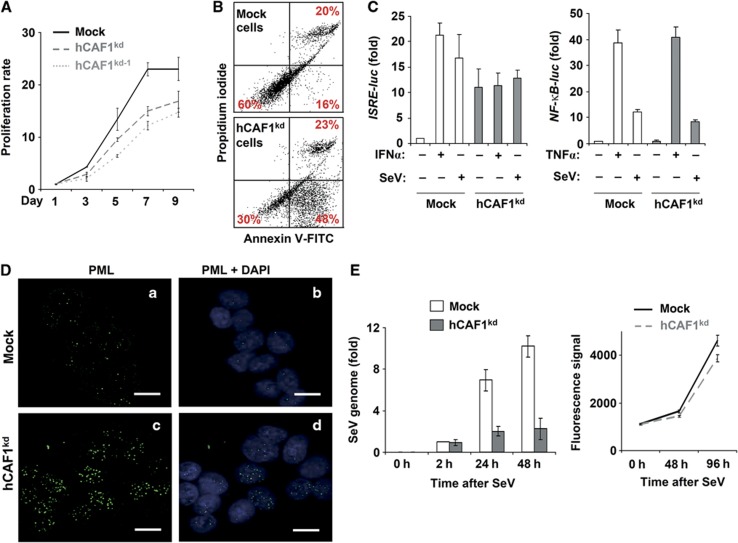
We then investigated the physiological consequence of the negative regulation on IFN signalling caused by hCAF1. hCAF1kd cells exhibited reduced growth rates, as measured by the Uptiblue assay (Figure 2A), and the number of cells in early apoptosis increased (Annexin V positive) compared to control (Figure 2B).
Figure 2.
Physiological outcome of hCAF1 knockdown cells. (A) Cell growth of mock and hCAF1kd cells was measured by the Uptiblue assay, and (B) apoptosis by using the Annexin-V-FITC assay. (C) Mock and hCAF1kd cells transfected with the reporter plasmids ISRE-Luc (left panel) or with NF-κB-Luc (right panel). ISRE- and NF-κB-dependent luciferase activities were measured after treatment with IFNα or infection with Sendai virus (SeV) (left panel) or with TNFα or SeV (right panel), respectively. Luciferase activities were normalized to the activity of the internal control Renilla luciferase and expressed as activation relative to the basal level in mock control cells (arbitrarily set to 1). (D) Confocal fluorescence microscopy experiments using anti-PML antibodies on untreated mock (a, b) and hCAF1kd cells (c, d). Scale bar=20 μm. (E) Mock and hCAF1kd cells were infected with SeV (left panel). The virus genome was quantified by RT–qPCR at the indicated times after the infection. Each set of experiments was performed in triplicate and repeated at least three times. Uptiblue Reagent was used to assess cell viability and cell proliferation of hCAFkd and mock cells after SeV infection (right panel). All the illustrated data were performed in triplicate and are expressed as mean values of three independent experiments. Standard deviations are shown.
Since hCAF1 depletion resulted in a strong upregulation of IFN-activated genes, we measured JAK/STAT1 signalling in hCAF1 knockdown cells using an IFN-responsive luciferase reporter gene (ISRE-Luc). As shown in Figure 2C (left panel), the reporter gene was markedly activated in these cells without IFN treatment compared to control cells. Interestingly, there was no increase in basal activation of ISRE-Luc in these cells, either after IFN treatment or infection by the Sendai virus (SeV). We confirmed the specificity of the response using the NF-κB Luc reporter gene as a control (Figure 2C, right panel). Thus, the effect of hCAF1 knockdown is reminiscent of the phenomena induced by IFN treatment. This was further substantiated by the changes in the nuclear organization observed after hCAF1 knockdown. It has been largely described that IFN treatment induces a re-organization of PML nuclear bodies (PML NBs) (Everett and Chelbi-Alix, 2007), associated with transcriptionally active parts of the genome, which persist after transcriptional shutoff (Gialitakis et al, 2010). Therefore, we analysed PML NBs in hCAF1kd cells by confocal immunofluorescence. In these cells, the PML NBs appeared increased in both number and size compared to the control cells (Figures 2Da and c).
Finally, to directly analyse whether hCAF1 is involved in IFN-dependent antiviral responses, we infected both hCAF1 knockdown and control cells with SeV. Viral replication was analysed by RT-qPCR of the viral genome at various time periods after infection. As shown in the left panel of Figure 2E, viral replication was strongly inhibited in hCAF1 knockdown cells compared to mock cells. As viral infection did not affect growth and viability of hCAF1kd either control cells (Figure 2E, right panel), hCAF1kd cells exhibited an increased protection against viral infection.
Altogether, these results indicate that knockdown of hCAF1 mimics the IFN response and inhibits viral replication.
Differential effects of hCAF1 on the decay rates of IFN-responsive genes
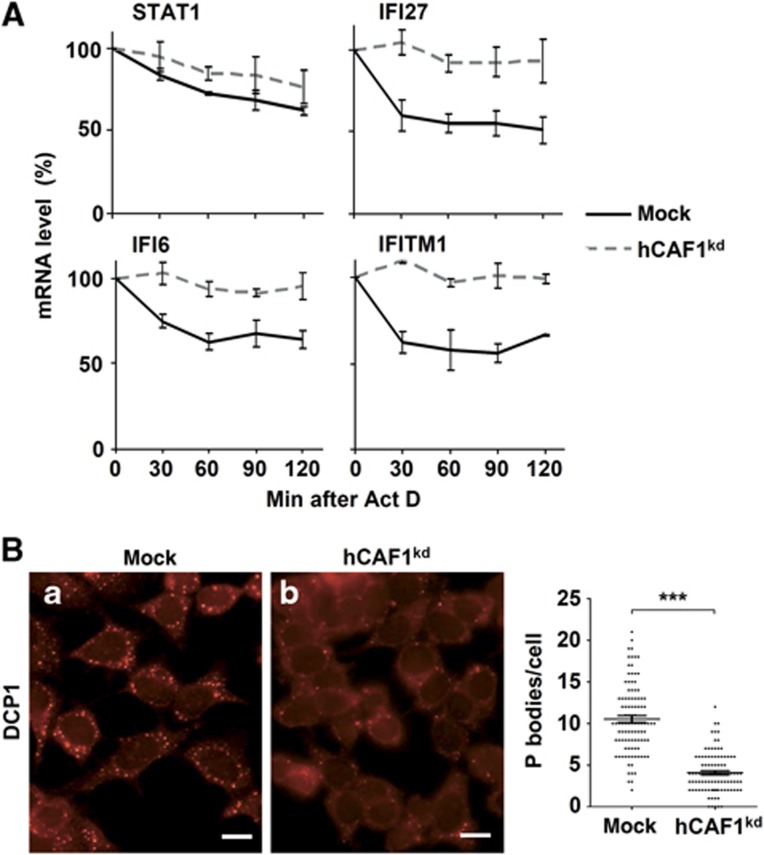
Accumulating evidence indicates that hCAF1 can regulate gene expression at both transcriptional and post-transcriptional levels. Therefore to determine the molecular mechanism of how hCAF1 regulates IFN signalling, we tested whether the increased mRNA levels we observed by microarray analysis resulted, at least in part, from the loss of mRNA deadenylation, which led to the stabilization of transcripts. Thus, we measured the mRNA levels of several hCAF1-regulated genes before and after the treatment of hCAF1kd and control cells with actinomycin D. We focused our analysis on STAT1 and the STAT1-regulated genes IFI27, IFITM1 and IFI6, involved in both type I and type II IFN signalling pathways. Figure 3A shows the results obtained after short times of treatment, (between 0 and 2 h) indicating that the stability of IFI27, IFITM1 and IFI6 mRNAs was clearly enhanced in hCAF1 knockdown cells compared to control cells. By contrast, the stability of STAT1 mRNA was not significantly different from that of control cells (Figure 3A). These results, confirmed at longer treatment times (Supplementary Figure S2), imply that hCAF1 regulates IFN-induced genes by different mechanisms besides the control of mRNA turnover.
Figure 3.
Determination of the stability of hCAF1-regulated genes. (A) Mock and hCAF1kd cells were treated with Actinomycin D and total mRNA was isolated at the indicated times after treatment. mRNA levels of the indicated genes were determined by SYBR green real-time RT–PCR as described in Figure 1. Results were plotted as a function of time from drug addiction. Results are expressed as mean values of at least three independent experiments. Standard deviations are shown. (B) Effect of hCAF1 knockdown on P-body formation, analysed using a rabbit antibody directed against Dcp1a. Fluorescence microscopy is representative of at least three independent experiments. Scale bar=20 μm. Quantification of the number of signals per cell (n=100) was performed (mean±s.e.m.) using Image J software. Student’s t-test. ***P<0.001.
Furthermore, hCAF1kd cells show a reduced amount of P-bodies, which are specific cytoplasmic foci enriched in proteins involved in mRNA metabolism (Eulalio et al, 2007; Parker and Sheth, 2007) (Figure 3B compare a to b). This strongly indicates that deadenylation is impaired in hCAF1kd cells as this function has been linked to the presence of P-bodies (Zheng et al, 2008).
hCAF1 affects the activation of the JAK/STAT pathway
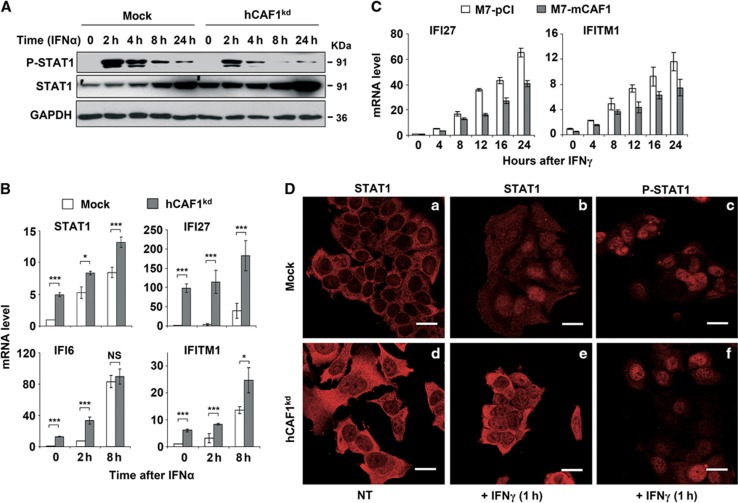
STAT1 plays a pivotal role in both type I (α/β) and type II (γ) IFN signalling pathways (Bromberg and Darnell, Jr., 2000; Ramana et al, 2002). IFN binding to type I (α/β) and type II (γ) IFN receptors results in the dimerization of receptor complexes and the activation of the Janus family of protein tyrosine kinases (JAKs), followed by phosphorylation of latent, cytosolic STAT monomers. When phosphorylated, STATs form homodimers or heterodimers, move to the nucleus and activate the transcription of target genes (for reviews, see Bromberg et al, 2000; Ramana et al, 2000 and Levy and Darnell, Jr, 2002). Therefore, we examined whether activation of the JAK/STAT pathway was affected in hCAF1 knockdown cells. As shown in Figure 4A, in the absence of IFN stimulation, the level of STAT1 protein was consistently greater in hCAF1kd cells than in control cells. Phosphorylation of STAT1 was not observed in either untreated cell line. After IFNα stimulation for the indicated times, the level of tyrosine 701 phosphorylation of STAT1 was monitored in hCAF1kd cells with respect to control cells. p-STAT1 was detected after 2 h of treatment and then decreased over several hours in both hCAF1kd and control cells. Notably, although the basal level of STAT1 protein was consistently greater in hCAF1kd cells than in control cells, we observed a reduced phosphorylation of STAT1 in hCAF1kd cells compared to control cells, indicating that only a fraction of STAT1 was activated by IFN treatment. This reduced amount of activated STAT1 is likely to be responsible for the transcriptional response of these cells to IFN. Indeed, concomitant to the decrease in p-STAT1, newly synthesized STAT1 appeared, beginning after about 8 h of IFNα treatment and accumulating at 24 h, in both hCAF1-depleted and control cells. The reduced phosphorylation of STAT1 was confirmed after IFNγ treatment and was more evident within a shorter time after activation (Supplementary Figure 3A). We next investigated the activation of STAT1-target genes in hCAF1kd and mock cells treated with IFNα at various times. qPCR profiles confirmed that in the absence of IFN stimulation, the levels of STAT1, IFI27, IFI6 and IFITM1 transcripts were consistently greater in hCAF1kd cells than in control cells (Figure 4B). In addition, these transcripts were not only efficiently induced in response to IFN stimulation, their activation appeared earlier in hCAF1 knockdown cells compared to control cells (Figure 4B). Consistently, ectopic expression of hCAF1 in MCF7 cells delayed the expression of the STAT1-target genes IFI27 and IFITM1 in response to IFN stimulation (Figure 4C).
Figure 4.
STAT1 activation in hCAF1 knockdown cells. (A) After IFN stimulation for the indicated times, the level of tyrosine 701 phosphorylation of STAT1 was measured in hCAF1kd cells and control cells by western blot. (B, C) Kinetic induction of STAT1 and STAT1-target genes upon interferon stimulation for the indicated times in (B) hCAF1 knockdown and control cells and in (C) MCF7 cells stably expressing mCAF1 and control cells was analysed using quantitative PCR as described in Figure 1. The illustrated experiments were performed in triplicate, expressed as mean values of three independent experiments. Standard deviations are shown and the P-value was determined by Student’s t-test: *P<0.05; ***P<0.001. (D) Confocal fluorescence microscopy experiments showing the subcellular distribution of endogenous STAT1 and p-STAT1 in hCAF1-depleted cells. Mock and hCAF1kd cells were grown on coverslips into 12-well plates and treated with 5 ng/ml of IFNγ for 1 h. STAT1 and p-STAT1 were analysed by immunofluorescence. Scale bar=20 μm.
Source data for this figure is available on the online supplementary information page.
The subcellular localization of both endogenous STAT1 and p-STAT1 in hCAF1-depleted cells, compared with control cells, induced or not with IFN, was analysed by confocal fluorescence microscopy experiments. Figure 4Da shows that in the absence of IFN stimulation, STAT1 localized to the cytoplasm in control cells, as expected. IFN treatment induced a rapid nuclear translocation and phosphorylation of STAT1 as detected by anti-STAT1 (Figure 4Db) and anti-p-STAT1 (Figure 4Dc) antibodies. Interestingly, in untreated hCAF1kd cells we observed that depletion of hCAF1 resulted in high expression of the protein STAT1, consistent with microarray results (Figure 4Dd). Even though a major amount of protein localized in the cytoplasm, STAT1 nuclear staining was still visible in these cells (Figure 4Dd). After IFN activation, only a fraction of the protein pool migrated to the nucleus (Figure 4De), and was phosphorylated (Figure 4Df), confirming the results shown in Figures 4A and B. We did not detect any p-STAT1 in either unstimulated control or hCAF1kd cells (unpublished results). Taken together, these findings suggest that the effects of hCAF1 on IFN signalling are, at least in part, mediated by STAT1.
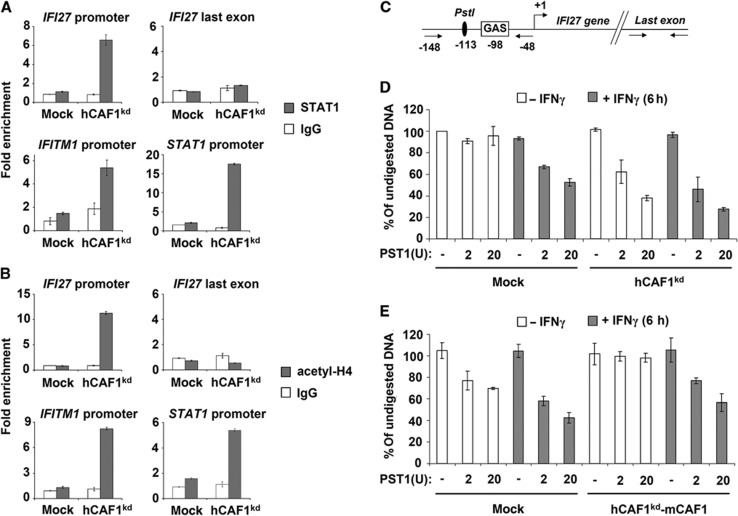
hCAF1 depletion affects STAT1 basal promoter occupancy
Previous work from several laboratories has shown that STAT1 can drive gene expression even in the absence of tyrosine phosphorylation (Chatterjee-Kishore et al, 2000; Yang and Stark, 2008). In particular, the group of G.R. Stark demonstrated that high expression of exogenous unphosphorylated STAT1 (u-STAT1) increases the expression of many immune regulatory genes (Cheon and Stark, 2009) reported to be upregulated in chemo- or radiation-resistant cancer cells and designated as the IFN-related DNA damage signature (Weichselbaum et al, 2008). Interestingly, many of these genes are also upregulated in hCAF1-depleted cells (see Supplementary Table 1). We postulated that the high amount of STAT1 protein could be responsible for the constitutive expression of its target genes, in the absence of IFN induction. Indeed, in resting hCAF1kd cells STAT1 was not phosphorylated (Figure 4A) and the expression of two transcriptional targets of p-STAT1, IRF1 and SOCS1, was not affected (Supplementary Figure 1B). To directly test this hypothesis, we performed chromatin immunoprecipitation (ChIP) assays. We found a significant increase in STAT1 recruitment at both the STAT1 promoter and promoters of its target genes, IFI27 and IFITM1, in hCAF1 knockdown cells relative to control cells, in the absence of IFN induction (Figure 5A). Interestingly, knockdown of hCAF1 also increased H4 total acetylation, a recognized hallmark of transcriptionally active chromatin (Kouzarides, 2007; Figure 5B). We did not observe any significant H4 acetylation in control cells at any of the promoters tested in the same conditions. STAT1 recruitment and H4 acetylation after IFN stimulation are shown in Supplementary Figures 4A and B. These results suggest that in hCAF1kd cells the chromatin architecture of some STAT1-target promoters is permissive for transcription, in the absence of IFN activation. To test this hypothesis, we used a restriction enzyme hypersensitivity assay (REHA) to analyse changes in the chromatin architecture of the IFI27 gene promoter, whose expression was strongly upregulated in hCAF1kd cells. REHA allows a high-resolution analysis of changes in the chromatin architecture by assaying nucleosome remodelling, which is often a prerequisite for transcriptional activation (Sproul et al, 2005). hCAF1kd and control cells were either exposed to IFNγ, or not, for 6 h. Isolated nuclei were then treated with a limiting concentration of PST1 restriction enzyme, which cuts near the STAT1-binding element in the IFI27 promoter (Ni et al, 2005; Figure 5C). DNA was then purified and the level of intact DNA was determined by qPCR using oligos flanking the PST1 restriction site or control region (Figure 5C). As shown in Figure 5D, right panel, the RE accessibility was largely increased in untreated hCAF1kd cells compared with control cells (left panel). Remarkably, the re-expression of mCAF1 in these cells (see Figure 5E) was sufficient to totally rescue the RE sensitivity phenotype (Figure 5E, right panel). These data indicate that STAT1 is recruited to the promoter of some of its target genes in unstimulated hCAF1kd cells. This basal promoter occupancy is associated with a decondensation of chromatin on these promoters.
Figure 5.
Constitutive recruitment of STAT1 at a subset of STAT1-target promoters in hCAF1 knockdown cells. ChIP assays of untreated hCAF1kd and control cells were performed using antibodies anti-STAT1 (A) and anti-acetyl H4 (B). Enriched DNA fragments were quantified by qPCR using specific primers for the indicated promoters with respect to the input DNA and normalized to a reference locus (3′ downstream region of the GAPDH gene). Rabbit IgGs were used as a negative control. (C–E) hCAF1 affects chromatin accessibility. (C) Schematic representation of GAS, PST1 site and primer positions on IFI27 promoter. (D) hCAF1kd and control cells and (E) hCAF1kd transfected with empty pCIflag (mock) or with pCIflag-mCAF1 (hCAF1kd-mCAF1, rescued cells expressing mCAF1) were exposed or not to IFNγ for 6 h. Isolated nuclei were then treated with a limiting concentration of PST1 restriction enzyme, which cut GAS containing region in IFI27 promoter. DNA was then purified and the level of intact DNA was determined by qPCR using oligos flanking the GAS element or control region illustrated in (C). The experiments were performed in triplicate, expressed as mean values and are representative of at least three independent experiments. Standard deviations are shown.
hCAF1 physically interacts with STAT1 in the cytoplasm of unstimulated cells
These results prompted us to investigate a possible physical interaction between hCAF1 and STAT1. Pull-down assays, using either GST-tagged hCAF1 or CCR4 (the preferential partner of CAF1), revealed a strong direct interaction of STAT1 with hCAF1 (Figure 6A). We did not detect any interactions between STAT1 and either CCR4 or GST. The interaction between endogenous hCAF1 and STAT1 was confirmed in both MCF7 and U937 cell lines. We incubated cellular lysates from MCF7 (Figure 6B) and U937 cells (Supplementary Figure 5) with anti-CAF1 polyclonal antibodies, resulting in co-immunoprecipitation of STAT1. The interaction between hCAF1 and STAT1 was strongly decreased when STAT1 was transiently depleted by siRNAs, compared to transfection with control siRNA (Figure 6B; Supplementary Figure 6B). Finally, the interaction between hCAF1 and STAT1 is most likely direct and not mediated by RNA because RNase treatment of MCF7 cell lysates did not affect their co-precipitation (Figure 6C).
Figure 6.
hCAF1 physically interacts with STAT1. (A) Direct interaction between hCAF1 and STAT1 was analysed by GST-pulldown experiments. In vitro translated STAT1 was incubated with equivalent amounts of GST, GST-CAF1 and GST-CCR4 (Supplementary Figure 5A) bounded to glutathione-Sepharose beads. The eluted proteins were analysed by immunoblotting using anti-STAT1 antibody. (B) Endogenous hCAF1 and STAT1 interact. Extracts from MCF7 cells treated with STAT1 and hCAF1-specific siRNAs or control (siControl) siRNA were immunoprecipitated (IP) with either normal rabbit IgG or anti-hCAF1 antibody. Immunoprecipitates were then analysed by immunoblotting with anti-STAT1 antibody. (C) MCF7 protein extracts were treated with or without RNAseA before immunoprecipitation (IP) with an anti-hCAF1 antibody followed by western blot with anti STAT1 antibody. (D) STAT1 and hCAF1 co-localize in the cytoplasm of the unstimulated MCF7 cells. MCF7 were grown on coverslips in 12-well plates and then treated with 5 ng/ml of Interferon γ for 1 h. Proximity Ligation Assay (PLA) was used to detect the cellular co-localization of endogenous hCAF1 and STAT1 according to manufacturer’s instructions (Duolink, Eurogentech). (a) PLA using anti-hCAF1 and anti-STAT1 on untreated MCF7 cells and (b) MCF7 cells treated with 5 ng/ml of Interferon γ for 1 h. (E) PLA control on MCF7 cells transfected with (a) control siRNA or with (b) siRNA against STAT1. Data are representative of at least three independent experiments. Scale bar=20 μm.
Source data for this figure is available on the online supplementary information page.
To determine in which cellular compartment this interaction occurred and whether it was regulated by IFN induction, we used an in situ proximity ligation assay (PLA), a technology capable of detecting protein interactions and the localization of interactions with high specificity and sensitivity. In the absence of IFN, the discrete spot-like signals indicated the interaction between endogenous hCAF1 and STAT1, as shown in Figure 6Da. Importantly, since the image in Figure 6Da was recorded in the fixed plane, the localization of each spot inside the cell could be unclear. By imaging cells with confocal microscopy, Z-stack projections showed unambiguously that the interaction spots were exclusively localized in the cytoplasm of resting MCF7 cells (see Supplementary Figure 7; compare a reconstitution of a three-dimensional (3D) image in Supplementary Figure 7A to a single median section cutting the nucleus, in Supplementary Figure 7B).
The interaction spots were strongly decreased when STAT1 expression was knocked down using siRNA in MCF-7 cells compared to mock MCF7 cells transfected with control siRNA (Figure 6E, compare panels a and b), supporting the specificity of this interaction. STAT1 knockdown efficiency was determined by qPCR analysis (Supplementary Figure 6B). Notably, IFNγ treatment (1 h) induced a visible and significant reduction in the signal, reflecting the dissociation of the hCAF1/STAT1 complex (Figure 6Db). Since the level of both proteins did not decrease after IFN treatment, hCAF1/STAT1 dissociation could be triggered by the phosphorylation and nuclear migration of STAT1 after IFN treatment, as shown in Figures 4Db and c. Importantly, the dynamic interaction between hCAF1 and STAT1 was confirmed in two different cellular models, HeLa and U937 cells (Supplementary Figure 8). Then we asked whether STAT1 interacted with other components of CCR4–NOT complex. Using immunoprecipitation assays we did not obtain convincing results, probably due to the lack of strong antibodies to detect co-immunoprecipitation of endogenous proteins or to a weak or indirect interaction (data not shown). Conversely, taking advantage of the high sensitivity of PLA technology, we detected the interaction of STAT1 with NOT1 on the cytoplasm of MCF7 cells (Supplementary Figure 9). Consistent with GST-pull down results (Figure 6A) we did not detect any direct interaction between STAT1 and CCR4 by PLA (data not shown).
Discussion
Living organisms are constantly exposed to a variety of internal and external stimuli. The innate immune system is genetically programmed to rapidly detect and respond to infection by viruses or other pathogens and to play a key role in immune surveillance of tumours (Dunn et al, 2006). A successful innate immune response requires the action of type I or II IFNs and the activation, via phosphorylation, of latent cytosolic transcription factors, STATs, as crucial mediators of this response. When phosphorylated, STATs form homodimers or heterodimers, move to the nucleus and activate the transcription of target genes (Bromberg et al, 2000; Ramana et al, 2000; Levy et al, 2002).
Although innate immunity is essential for host defence, aberrant activation of innate immune responses results in the development of autoimmune diseases and cancer. It is therefore not unexpected that this signalling pathway is tightly regulated to avoid either deficient or excessive responses. In fact, the magnitude and degree of STAT signalling is regulated by various mechanisms, including post-translational protein modifications and interaction with regulatory proteins (Levy et al, 2002).
In this report, we have defined a novel role for hCAF1 in the negative regulation of type I or II IFN signalling. Consistently, knockdown of hCAF1 renders cells more resistant to viral infection and reduced viral replication (Figure 2E).
Notably, we have found that hCAF1 forms a complex with the latent form of STAT1 in the cytoplasm of several cell lines (Figure 6Da; Supplementary Figures 7 and 8A and C). IFN treatment of cells induced the dissociation of the complex (Figure 6Db; Supplementary Figures 8B and C) permitting the release of STAT1 and its migration to the nucleus. Since the level of hCAF1 was not affected by IFN treatment (Supplementary Figure 3B), it is reasonable to attribute the dissociation of hCAF1/STAT1 complex to the phosphorylation and consequent nuclear migration of STAT1 (Figures 4Da and c). These results suggest that the formation of this complex in the cytoplasm may prevent the inappropriate migration of STAT1 in unstimulated conditions. The phosphorylation of STAT1 on tyrosine 701, following IFN stimulation and JAK activation, is crucial for dimerization and nuclear migration. However, IFN treatment of hCAF1kd cells reduced phosphorylation of STAT1 (Figure 4A), although we observed an increased amount of some STAT1-target transcripts (Figure 4B). It will be interesting to determine whether hCAF1 plays a role in the regulation of STAT1 phosphorylation and thereby its activation and migration.
Nuclear trafficking, which involves a complex combination of active and passive mechanisms and has a significant impact on STAT functions, might constitute another important ‘check-point’ in IFN/STAT signalling. This idea is further supported by several lines of evidence. Knockdown of hCAF1 resulted in a high basal transcriptional activation of STAT1 and a subset of STAT1-regulated genes. Furthermore, our results showed a local decondensation of chromatin structure within some STAT1-target promoters associated with the recruitment of STAT1 and H4 hyperacetylation, in the absence of any induction (Figure 5). Figure 4A clearly shows that STAT1 is not phosphorylated in resting hCAF1kd cells therefore we postulate that the constitutive expression of some STAT1-target genes in the absence of IFN induction, is, at least in part, consequent to the high concentration of STAT1 protein. However, the group of G.R. Stark has demonstrated that the forced expression of exogenous u-STAT1 was linked with increased expression of a class of genes (Cheon et al, 2009) previously reported to be upregulated in chemo- or radiation-resistant cancer cells overexpressing u-STAT1 (Weichselbaum et al, 2008) and classified as ‘IFN-related DNA damage signature’ (Khodarev et al, 2012). Interestingly, many of these genes are also upregulated in hCAF1-depleted cells (see Supplementary Table 1) indicating that hCAF1 can be potentially involved in cellular resistance to genotoxic stress and in prosurvival functions of STAT1. Overall, these data indicate that high amounts of u-STAT1 may, directly or not, affect the chromatin architecture of discrete genomic regions. One intriguing possibility can be that u-STAT1, when overexpressed and in absence of stimuli, can migrate to the nucleus and associate with compacted chromatin on a class of IFN-inducible promoters, acting as a ‘pioneer’ factor. These bindings would increase accessibility for subsequent transcription factor recruitments, permitting the expression of a particular class of genes, which can confer, for example, the chemo- or radiation-resistant phenotype described for several types of tumours (Weichselbaum et al, 2008). This effect can be reminiscent of the phenomena described for the HLA locus where IFN treatment induces rapid higher order chromatin modifications generating ‘primed’ transcriptionally permissive environment which facilitates subsequent inductions (Christova et al, 2007). In line with this hypothesis, hCAF1 depletion also correlates with the re-organization of PML NBs, which have been associated with transcriptionally active ‘memory’ parts of the genome after the IFN response (Figure 2D).
On the other hand, a direct association between high expression of u-STAT1 and DNA damage-resistant phenotype has been described in Drosophila, in which u-STAT92E localizes on heterochromatin maintaining the stability of transcriptionally repressed heterochromatin (Shi et al, 2008). Strikingly, animals with high levels of u-STAT92E exhibit increased levels of heterochromatin associated with increased survival rate after genotoxic stress (Yan et al, 2011). We can imagine that u-STAT1 could also be able to silence specific domains of the genome by recruiting co-repressors, as described for several pioneer factors (Zaret and Carroll, 2011). As Jak/STAT1 signalling is one of mechanisms of the tumour cell elimination in early stages of many tumour development, we can hypothesize that tumour cells in ‘secondary prolonged’ response (Cheon et al, 2009), associated with u-STAT1 transcription and primed chromatin, are more resistant to genotoxic stress and can be responsible for the selection of aggressive phenotype tumour clones.
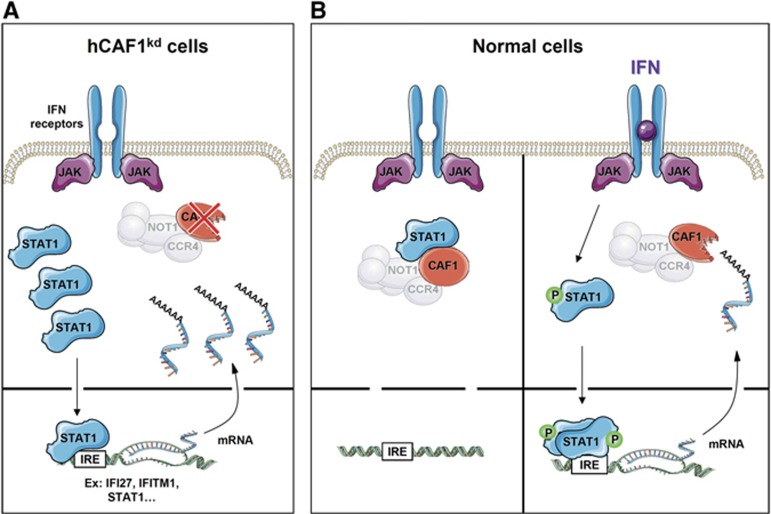
The molecular events leading to the deregulation of STAT1 observed in hCAF1kd cells are illustrated in Figure 7A. In the absence of hCAF1, a fraction of u-STAT1 migrates to the nucleus and induces the expression of STAT1 itself, thus creating a positive feedback loop that enhances its expression. In parallel, u-STAT1 stimulates the expression of several target products whose stability is prolonged by the loss of hCAF1 deadenylase activity in these cells, leading to their accumulation.
Figure 7.
Model for dual regulatory functions of hCAF1 in IFN signalling. (A) In hCAF1kd cells, hCAF1 depletion prevents the trapping of STAT1 in the cytoplasm, permitting that a fraction of u-STAT1 migrates in the nucleus and activates the expression of many target genes, including STAT1 itself. In parallel, u-STAT1 activates the expression of several target genes whose stability is prolonged and enhanced by the loss of hCAF1deadenylase activity in these cells, leading to their accumulation. (B) Under physiological conditions and in the absence of IFN induction, hCAF1 can control STAT1 trafficking by interacting with the latent form of STAT1 in the cytoplasm. IFN activation leads to the release of STAT1 and its phosphorylation. Phosphorylated STAT1 forms homodimers or heterodimers, move to the nucleus and induce the transcription of target genes to allow rapid and transient activation of immune response signalling. hCAF1 could also participate in the extinction of IFN signal via its deadenylase activity by speeding up the degradation of several STAT1-regulated mRNAs, which may constitute new physiologically relevant targets of hCAF1. The light grey colour of the CCR4–NOT complex indicates that its participation is not yet completely established.
On the basis of our results we propose a model for a previously unpredictable mechanism regulating the functions of STAT1. In Figure 7B, we illustrate how hCAF1, through its interaction with STAT1, could regulate IFN signalling at different crucial steps: (i) in resting unstimulated cells, by controlling STAT1 trafficking and (ii) in IFN-activated cells by participating to the extinction of IFN response (Figure 7B).
Despite we cannot rule out that hCAF1 interacts with and regulates STAT1 outside of the CCR4–NOT complex, increasing number of biochemical, genetic and structural studies point out that CAF1 protein, in yeast and mammalian cells, exists and acts within the CCR4–NOT multifunctional complex. Furthermore, the involvement of this complex in IFN-mediated gene activation is in agreement with its emerging role as a chaperone regulatory platform (Collart and Timmers, 2004). The CCR4–NOT complex, in addition to its housekeeping functions, can efficiently and rapidly adapt cellular gene expression in response to changes in environmental conditions and stimuli. The innate immune response to infection by viruses or other pathogens is in fact initiated by an induction phase which rapidly responds to external aggression, followed by a downregulation of the response. Since the CCR4–NOT complex acts at all levels of gene expression, from transcription to decay (mRNA or protein), it permits the rapid adaptation of cells to external stresses.
To prevent excessive responses to IFNs, hCAF1 could participate in the extinction of the IFN signal via its deadenylase activity, by accelerating the degradation of some STAT1-induced mRNAs. Notably, although CCR4/CAF1 deadenylase can potentially remove poly(A) tails from any mRNA species, hCAF1 knockdown differentially affected the stability of STAT1-induced transcripts. This selectivity could be explained by differential ‘marks’ of degradation on mRNAs, as theorized by Parker (Tucker et al, 2001). The emerging idea that mRNAs are marked for degradation during transcription is supported by recent results from genome-wide gene expression, establishing that mRNA production and decay are strongly linked (Shalem et al, 2011; Bregman et al, 2011). Moreover, fast-induced responsive genes, as is the case for the STAT1-target genes, display a corresponding rapid destabilization of their mRNAs (Shalem et al, 2008). Consistently, our previous data indicated that the different regulatory roles in which the CCR4–NOT complex is involved, notably transcription and mRNA turnover, may occur via distinct complexes, whose size, composition and cellular localization changes during the cell cycle (Morel et al, 2003). Further studies may provide insight into the molecular mechanism determining and coordinating the rate of both mRNA synthesis and decay in IFN signalling. Focus on this pathway might be an appealing approach to explore how a simple complex, such as CCR4–NOT, can regulate an mRNA from ‘birth to death’. These findings identify hCAF1 as a key factor in IFN-negative regulation playing a physiologically important role in the maintenance of immune homeostasis, especially with regard to regulation of the innate immune response. Since abnormal and unbalanced JAK/STAT activation is associated with immune disorders, cancer and cellular resistance to DNA-damaging agents, hCAF1 could play a major role in oncogenesis, contributing to tumour escape. Therefore, our results give the basis to decode the molecular properties and functions of u-STAT1 and the role of its increased expression in the selection of therapy-resistant cancer clones. Finally, hCAF1 and the CCR4–NOT complex have potential useful therapeutic targets for enhancing immunity against microbial infections and inflammation-associated diseases.
Materials and methods
Cell culture
MCF7, HeLa and stable cell lines were routinely maintained in DMEM, U937 in RPMI-1640, supplemented with 10% fetal bovine serum, penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
hCAF1 knockdown
We used BLOCK-iT Pol II miR RNAi Expression Vector Kits (Invitrogen) to generate vectors containing miRNA duplexes targeting hCAF1, corresponding to the coding regions 571–591 (Kd-1), and 941–961 (kd) (hCAF1 NCBI Reference Sequence: NM_013354.5) and one non-targeting control miRNA (mock), following manufacturer’s instructions. Cells expressing the miRNA of interest (called hCAF1kd, hCAF1kd-1 and mock) were selected 48 h after vector transfections, in medium containing 5 μg/ml of blasticidin (Invitrogen) for at least 3 weeks, after which blasticidin concentration was reduced to 3 μg/ml.
Generation of stable cell lines
For the production of rescue cell lines, MCF7, and hCAF1kd cell lines were transfected with 5 μg of vectors expressing Flag-mCAF1 (pCIflag-mCAF1) or with empty pCIflag (2 × 106 cells in a 100-mm culture dish) using Exgen 500 (Euromedex). Selection was initiated 48 h after transfection in medium containing 750 μg/ml of G418. MCF7pCI and MCF7mCAF1 cell populations were maintained in medium containing 300 μg/ml of G418; mock-mCAF1, mock-pCI, hCAF1kd-mCAF1 and hCAF1kd-pCI were maintained in medium containing 300 μg/ml of G418 and 5 μg/ml of blasticidin.
Microarray analysis
Total RNA from biological triplicates of hCAF1kd and mock cells was isolated, processed and hybridized using Affymetrix HuEx arrays. HuEx arrays contain probes targeting >1 million exons from well-annotated or computationally predicted genes. In this study, we focused on genes with known cDNAs. For each gene, the average intensity of exonic probes was calculated in each sample from four independent experiments. Human Exon 1.0 ST Array data set analysis and visualization were made using EASANA (GenoSplice Technology, www.genosplice.com) based on GenoSplice’s FAST DB annotations, as described in detail in Supplementary data.
Immunoprecipitation, western blot and antibodies
We carried out immunoprecipitations and western blots as described (Robin-Lespinasse et al, 2007) except that cell lysates were treated or not with 0.5 mg/ml of RNase A for 10 min at room temperature before incubation with either normal rabbit IgG, the affinity purified rabbit polyclonal antibodies against hCAF1 or hCCR4 previously described (Morel et al, 2003). Rabbit polyclonal antibodies against CNOT1 protein were produced by Agrobio Laboratory (France) using specific peptide (KPGNLLKDKDRLKNLDC). For IF analysis, the mouse polyclonal anti-hCAF1 (AO1) from Abnova was used. The rabbit polyclonal anti-STAT1αp91 (C-24), the goat polyclonal anti-IFITM1 (P-17) and the mouse monoclonal anti-PML (PG-M3) antibodies were purchased from Santa Cruz Biotechnology. Anti-phopho Tyr701 STAT1 (#9171S) rabbit polyclonal antibody was purchased from Cell Signaling Technology, the anti-GAPDH (clone 6C5) was from Biodesign International and the anti-acetyl-Histone H4 (06-598) from Millipore. The polyclonal rabbit anti-DCP1 antibody was kindly provided by J. Lykke-Anderson (Department. of Molecular and Developmental Biology, University of Colorado, USA).
siRNA and plasmid transfection
One million cells were plated in a 10-mm culture dish and transfected with a final concentration of 50 nM of each siRNA duplex using lipofectamin 2000 (Invitrogen) according to manufacturer’s instructions. The siRNA sequences targeting hCAF1 corresponded to the coding regions 697–715 (sihCAF1 n°2) and 463–480 (sihCAF1 n°4) and the siRNA sequence targeting STAT1 corresponded to the coding region 647–669.
Real-time PCR
In all, 1 μg of total RNA purified using TRI-Reagent (Sigma) was reverse transcribed using 100 ng of random primers following the Superscript II (Invitrogen) protocol. Real-time PCR was performed with SYBR Green qPCR master mix (Applied Biosystems) in a Step One plus real-time PCR detection system (Applied Biosystems). All amplifications were performed in duplicate. Mean values of duplicate measurements were calculated according to the –ΔΔCt quantification method using 36B4 gene expression as a reference for normalization. Relative mRNA levels in control cells were equated to 1 and other values were expressed relative to this. Each PCR run also included non-template controls containing all reagents except cDNA which generated no amplification. Specificity was confirmed by analysing the melting curves of PCR products. RT–qPCR results were repeated at least three times in independent experiments and expressed as mean values: P-value was determined by Student’s t-test. Sequences of the oligonucleotides used are listed in Supplementary data.
GST-pull down
GST fusions of hCAF1 and hCCR4 were expressed in Escherichia coli and purified over glutathione sepharose beads. Binding assays were carried out as previously described (Prevot et al, 2001).
Luciferase reporter assay
Cells, plated in 24-well plates 24 h before transfection, were transfected with 50 ng of reporter plasmid and 25 ng of pRL-TK Renilla luciferase vector (Promega, Madison, WI) used as an internal control using ExGen 500 (Euromedex) as described (Morel et al, 2003). The luciferase activity was determined by a dual luciferase assay kit (Promega) with a Luminoskan Ascent luminometer (Thermo Scientific, Waltham, MA).
Assessment of apoptosis
For annexin V binding, cells (200 000) were harvested by trypsin treatment. After inactivation of trypsin by addition of the supernatant, the cells were washed and resuspended in annexin V binding buffer, incubated with propidium iodide and FITC-labelled annexin V (AbCys; according to manufacturer’s instructions), and then analysed by flow cytometry using a FACS Canto II flow cytometer (Becton Dickinson).
Proliferation assay
Cells (1.5 × 103) were seeded in triplicate in 96-well plates. One plate was prepared each day during the time course. At each time point, cells were treated with Uptiblue (Interchim) and incubated for 3 h at 37°C. Fluorescence intensity was monitored at 530–560 nm excitation wave length and 590 nm emission wavelength (CytoFluor, PerSeptive Biosystem).
Immunofluorescence
Cells (10 × 104) were grown on coverslips in 12-well plates. For IFN treatment, cells were incubated for 1 h in fresh medium containing 5 ng/ml of IFNγ, and then treated as previously described (Robin-Lespinasse et al, 2007) and analysed with a Confocal SP5 Leica microscope.
Proximity ligation assay
PLA was performed using the Duolink kit (Eurogentech) according to manufacturer’s instructions. Cells (2 × 105) were fixed in ice-cold methanol for 5 min, washed twice in PBS and incubated with primary antibodies for 1 h at 37°C in a pre-heated humidity chamber. After two washes in PBS-Tween 0.1%, cells were incubated with the appropriate PLA probes for 1 h at 37°C then washed in PBS-T, and the ligation solution was added on the coverslips and incubated for 30 min at 37°C. Finally, the amplification solution containing a DNA polymerase was added and incubated with the cells for 100 min at 37°C. After a final wash, the cells were mounted on glass slides in a mounting solution of Dapi, and imaging was performed on a fluorescence microscope.
Viral infection
In all, 2 × 105 cells were infected by SeV at a final concentration of 20 HAU/ml in complete DMEM. After 2 h, the inoculum was removed and replaced with fresh medium. At different times post infection, cells were lysed in TRI-Reagent (Sigma) for total RNA extraction. SeV genome has been detected and quantified by RT-qPCR using the following primers: 5′GCCGGGTCCCACGAATCTAGG-3′ and 5′-CCCAATCACGGCGCCGAAGAA-3′ Mean values of duplicate measurements were calculated according to the –ΔΔCt quantification method using 36B4 gene expression as a reference for normalization. Cell viability was measured at 0, 48 and 96 h post infection by Uptiblue (Interchim). Fluorescence intensity was monitored at 530–560 nm excitation wave length and 590 nm emission wavelength (CytoFluor, PerSeptive Biosystem).
Chromatin immunoprecipitation
Cells treated or not with 5 ng/ml of IFNγ for 1 h were cross-linked with 1% formaldehyde at room temperature for 10 min, then treated with 0.125 M glycine for 5 min, and washed with ice-cold PBS. ChIP assays were carried out using the kit from Upstate Biotechnology. Briefly, chromatin was shared by sonication in a Diagenode Bioruptor (10 pulses x 30′′). ChIP assays were performed with anti-STAT1α p91 (sc-345), with the anti-acetyl-Histone H4 (06-598) or rabbit IgG as a negative control. The antibody–chromatin reactions were precipitated with salmon sperm DNA/protein A agarose beads for 3 h by rotation. Unbound chromatin was removed according to manufacturer’s instructions. Samples were extracted twice with 250 μl of elution buffer and heated at 65°C overnight to reverse cross-links. After DNA purification, qPCRs were performed in triplicate in a 96-well optical reaction plate using SYBR Green PCR Master Mix (Life Technology). The –ΔΔCt values for each locus were calculated with respect to the ChIP input DNA, normalized to a reference locus (3′ downstream region of the GAPDH gene). The sequences of primers used to amplify ChIP-enriched DNA are listed in Supplementary data.
Restriction enzyme hypersensibility assay
Restriction enzyme hypersensibility experiments were adapted from Ni et al, (2005). Briefly, cells were washed with ice-cold PBS and nuclei were isolated with lysis buffer (10 mM Tris pH 7.4, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 0.1% NP-40, 0.3 M sucrose). 2 × 106 nuclei were suspended in 200 μl of digestion buffer (10 mM Tris pH 7.9, 50 mM NaCl, 10 mM MgCl2, 0.3M sucrose, 1 mM DTT), and incubated with 2 U or 20 U of Pst1 (Fermentas) at 37°C for 30 min. The reaction was stopped by adding two volumes of nuclear lysis buffer (0.3 M CH3COONa, 5 mM EDTA, 0.5% SDS, and 0.1 mg of proteinase K/ml) and overnight incubation at 55°C. DNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in water. The undigested DNA was measured by quantitative real-time PCR with primers flanking the Pst1 site. PCR in the last exon of IFI27 gene with primers that did not flank any Pst1 restriction site was carried out to correct for equal loading of the samples.
Statistical analysis
Data are presented as mean±s.d. or mean±s.e.m. Statistical analyses were carried out with Student’s t-tests. Differences were considered statistically significant at P<0.05.
Accession codes
Human Exon 1.0 ST Array data are available in the Gene Expression Omnibus repository under accession number GSE43334.
Supplementary Material
Acknowledgments
We are grateful to Alain Puisieux for his precious support. We thank Cécile Languilaire, Sarah Benoit and Amandine Mosnier for helpful technical assistance, J Lykke-Andersen for giving the anti-DCP1a antibody and ‘Angloscribe’ for reading the manuscript. This study was partially supported by the Ligue Nationale Contre le Cancer (Equipe labellisée 2009), the ‘Association pour la Recherche sur le Cancer’ (ARC) and Région Rhône-Alpes ARC1-Santé 2012. CC is supported by a fellowship from the French Ministry of Research and ARC and CK by the Ligue Nationale Contre le Cancer and by the Comité de la Loire.
Author contributions: LC, SS, CC and CK conceived the study, designed the experiments and analysed data. CC and CK carried out most of the experiments, except that DA performed the microarray experiments and with PDLG conducted bioinformatics’ analysis of microarray data. MLR and KC contributed in result discussions and provided material. LC coordinated the project. The manuscript was written by LC, with contributions from CC, CK and SS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS (2009) The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell 20: 3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M, Yamamoto T (2010) The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell 1: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C, Morera AM, Asensio MJ, Chauvin MA, Morel AP, Dijoud F, Magaud JP, Durand P, Rouault JP (2004) CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol Cell Biol 24: 5808–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L (2005) Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 11: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan JA, Adams-Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, Stoltenborg JK, Dicker IB (1998) Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J 336: Pt 2471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M (2011) Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Bromberg J, Darnell JE Jr. (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19: 2468–2473 [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19: 4111–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H, Stark GR (2009) Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci USA 106: 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D (2007) P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J Cell Sci 120: 3262–3270 [DOI] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO (2012) The Ccr4–NOT complex. Gene 492: 42–53 [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HT (2004) The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Denis CL, Chen J (2003) The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD (2006) Interferons, immunity and cancer immunoediting. Nat Rev Immunol 6: 836–848 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009) Deadenylation is a widespread effect of miRNA regulation. RNA 15: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK (2007) PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89: 819–830 [DOI] [PubMed] [Google Scholar]
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB (2007) Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 27: 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N (2009) Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty S, Mahajan MA, Samuels HH (2008) Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J Biol Chem 283: 6806–6816 [DOI] [PubMed] [Google Scholar]
- Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J (2010) Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol 30: 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T (1999) Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 18: 7432–7441 [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK (2005) Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Roizman B, Weichselbaum RR (2012) Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin Cancer Res 18: 3015–3021 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE Jr (2002) Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3: 651–662 [DOI] [PubMed] [Google Scholar]
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C (2009) The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 19: 307–316 [DOI] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B (2008) The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 27: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Reese JC (2012) Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47: 315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin L, Puisieux A (2005) C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene 358: 73–81 [DOI] [PubMed] [Google Scholar]
- Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L (2003) BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci 116: 2929–2936 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, Inoue K, Ajima R, Miyasaka T, Yoshida Y, Ogura A, Toshimori K, Noce T, Yamamoto T, Noda T (2004) Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet 36: 528–533 [DOI] [PubMed] [Google Scholar]
- Ni Z, Karaskov E, Yu T, Callaghan SM, Der S, Park DS, Xu Z, Pattenden SG, Bremner R (2005) Apical role for BRG1 in cytokine-induced promoter assembly. Proc Natl Acad Sci USA 102: 14611–14616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko OO, Collart MA (2011) Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol 31: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Pillai RS (2005) MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11: 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L (2001) Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem 276: 9640–9648 [DOI] [PubMed] [Google Scholar]
- Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR (2000) Complex roles of Stat1 in regulating gene expression. Oncogene 19: 2619–2627 [DOI] [PubMed] [Google Scholar]
- Ramana CV, Gil MP, Schreiber RD, Stark GR (2002) Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol 23: 96–101 [DOI] [PubMed] [Google Scholar]
- Robin-Lespinasse Y, Sentis S, Kolytcheff C, Rostan MC, Corbo L, Le Romancer M (2007) hCAF1, a new regulator of PRMT1-dependent arginine methylation. J Cell Sci 120: 638–647 [DOI] [PubMed] [Google Scholar]
- Rouault JP, Prevot D, Berthet C, Birot AM, Billaud M, Magaud JP, Corbo L (1998) Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem 273: 22563–22569 [DOI] [PubMed] [Google Scholar]
- Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C (2009) The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res 37: 5511–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y (2008) Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y (2011) Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet 7: e1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX (2008) Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6: 775–781 [DOI] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, Roizman B, Bergh J, Pawitan Y, van de Vijver MJ, Minn AJ (2008) An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA 105: 18490–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SJ, Lim SJ, Shi S, Dutta P, Li WX (2011) Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J 25: 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Stark GR (2008) Roles of unphosphorylated STATs in signaling. Cell Res 18: 443–451 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB (2008) Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.