Abstract
Completion of DNA replication needs to be ensured even when challenged with fork progression problems or DNA damage. PCNA and its modifications constitute a molecular switch to control distinct repair pathways. In yeast, SUMOylated PCNA (S-PCNA) recruits Srs2 to sites of replication where Srs2 can disrupt Rad51 filaments and prevent homologous recombination (HR). We report here an unexpected additional mechanism by which S-PCNA and Srs2 block the synthesis-dependent extension of a recombination intermediate, thus limiting its potentially hazardous resolution in association with a cross-over. This new Srs2 activity requires the SUMO interaction motif at its C-terminus, but neither its translocase activity nor its interaction with Rad51. Srs2 binding to S-PCNA dissociates Polδ and Polη from the repair synthesis machinery, thus revealing a novel regulatory mechanism controlling spontaneous genome rearrangements. Our results suggest that cycling cells use the Siz1-dependent SUMOylation of PCNA to limit the extension of repair synthesis during template switch or HR and attenuate reciprocal DNA strand exchanges to maintain genome stability.
Keywords: DNA repair synthesis, genome stability, PCNA SUMOylation, Srs2, SUMO interacting motif
Introduction
Replication of the genome is a fundamental and carefully controlled process shared by all organisms. Given the size of the genomes, the complexity of DNA replication and its interplay with other DNA transactions such as transcription, recombination and repair, the integrity of the genome is often undermined by replication stress (Jackson and Bartek, 2009). Both endogenous and exogenous damage during DNA replication can severely impede replication. Electron microscopy and measurements of the amount and size of newly synthesized DNA have revealed that upon DNA damage unreplicated single-stranded gaps are left behind after the completion of bulk chromosomal replication (Lopes et al, 2006). To cope with stalled replication forks and filling DNA gaps, cells have evolved a DNA damage tolerance (DDT) pathway that ensures replication completion. It is still unclear if DDT mechanisms operate predominantly at the stalled fork or single-stranded gaps. In addition, the pathway choice can be determined by the type of lesions and could be organism specific, since important differences have been found after treatments with various damaging agents and between vertebrates and yeasts (Edmunds et al, 2008; Jansen et al, 2009). Recent detailed studies in yeast have provided evidence that DDT can operate effectively even after chromosomal replication (Daigaku et al, 2010; Karras and Jentsch, 2010) underlining the importance of gap filling. Gap filling can either take place on stalled forks that have escaped S phase or after fork collapse via D-loop formation followed by repriming. At least three separable mechanisms have been proposed to rescue stalled forks or filling-in single-stranded gaps formed opposite DNA lesions. Two of them depend on the RAD6/RAD18-mediated ubiquitin conjugation system, whereas homologous recombination (HR) mediates the third mechanism. The first RAD6/RAD18 pathway directs damage bypass by translesion synthesis (TLS) polymerases to evade damaged DNA in an error-free or error-prone manner (Nelson et al, 1996; Johnson et al, 1999; Prakash et al, 2005, 2000). The second RAD6/RAD18-dependent pathway promotes a RAD5-mediated template switch (TS), in which the newly synthesized strand of the sister strand is used as a template (Torres-Ramos et al, 2002; Blastyák et al, 2007). The third pathway that rescues stalled replication forks includes the RAD52 epistasis group and utilizes the sister chromatid to achieve recombinational repair (Zhang and Lawrence, 2005; Gangavarapu et al, 2007; San Filippo et al, 2008; Krejci et al, 2012).
PCNA has been identified as a molecular switch that regulates DDT (Waga et al, 1994; Kelman, 1997). It is a homotrimeric ring-like protein that encircles DNA and functions as a sliding clamp on the DNA (Krishna et al, 1994; Tinker et al, 1994; Gulbis et al, 1996; Yao et al, 1996) and ensures the processivity of the replicative DNA polymerases (Prelich et al, 1987; Kelman, 1997). The regulatory role of PCNA depends on post-translational modifications reflecting the DNA damage status during replication (Hoege et al, 2002; Haracska et al, 2004; Pfander et al, 2005). Blocked DNA synthesis triggers PCNA ubiquitination. Rad18 monoubiquitinates PCNA (Ub-PCNA) on residue K164, thus activating TLS (Hoege et al, 2002). Furthermore, the RAD5-dependent post-replication repair (PRR) mechanism is responsible for the polyubiquitination of PCNA (polyUb-PCNA) on residue K164 leading to TS. The other major post-translational modification of PCNA is SUMOylation that occurs during S phase even in the absence of exogenous damage and results in modifications at residues K164 and K127 (Hoege et al, 2002). The residue K164 is the major SUMOylated site of PCNA in vivo and its modification depends mostly on the Siz1 E3 SUMO ligase (Pfander et al, 2005; Windecker and Ulrich, 2008). In yeast, it has been demonstrated that the foremost role of K164-SUMOylated PCNA (S-PCNA) is to recruit Srs2 to the replication fork (Papouli et al, 2005; Pfander et al, 2005). This is also the case in humans where PARI, a recently identified orthologue of Srs2, shares similar properties (Moldovan et al, 2012). This recruitment is believed to target the Srs2 antirecombinase activity to replication forks to prevent untimely HR that can trigger gross chromosomal rearrangements, cell-cycle arrest and cell death (Fabre et al, 1991; Smirnova and Klein, 2003; Marini and Krejci, 2010).
Srs2 is a DNA repair enzyme with ssDNA-dependent ATPase activity and a 3′ to 5′ helicase activity (Rong and Klein, 1993; Van Komen et al, 2003; Marini and Krejci, 2010, 2012) and the SRS2 gene was first identified as a suppressor of RAD6, the ubiquitin-conjugating enzyme (E2) involved in PRR. It was suggested to play role as an antirecombinase by preventing HR via channelling the lesion into the PRR pathway (Schiestl et al, 1990). This has been biochemically confirmed, as Srs2 was shown to efficiently dismantle the Rad51-presynaptic filament (Krejci et al, 2003; Veaute et al, 2003). In addition, Srs2 can translocate on ssDNA and physically interact with Rad51 protein (Krejci et al, 2004; Antony et al, 2009). These activities are essential for the Srs2 antirecombinase function (Krejci et al, 2004; Colavito et al, 2009; Seong et al, 2009). In fact, the interaction with Rad51 seems to not only target Srs2 to HR intermediates but also trigger ATP hydrolysis within the Rad51 filament to allow its coordinated disruption by Srs2 (Antony et al, 2009). In addition, Srs2 possesses a non-canonical PIP box requiring tandem receptor motifs for precise recognition of S-PCNA (Armstrong et al, 2012; Kim et al, 2012; Kolesar et al, 2012).
S-PCNA has the ability to recruit Srs2 to sites of replication where it can disrupt untimely formed Rad51 filaments and thus prevent HR. However, it is not known whether Srs2 or S-PCNA play further roles when replication stalls. In addition, the nature of the recombination events that are suppressed by S-PCNA and Srs2 remains elusive. Here, we report a novel mechanism by which S-PCNA together with Srs2 can block the extension of a recombination intermediate and limit its resolution associated with a crossing-over (CO). This activity requires the SUMO interaction motif (SIM) at the C-terminus of Srs2, but needs neither its translocase/helicase activity nor its interaction with Rad51. The molecular characterization of the underlying mechanism revealed that Srs2 interaction with S-PCNA dissociates Polδ or Polη from the repair synthesis ensemble. It does not affect Srs2 helicase or antirecombinase activities and represents a novel regulatory element controlling spontaneous genome rearrangements. Our results strongly support that cycling cells use the Siz1-dependent SUMOylation of PCNA to concentrate Srs2 at harmed replication forks, therefore limiting the extent of repair synthesis during TS or HR and downregulating reciprocal exchanges to maintain genome stability.
Results
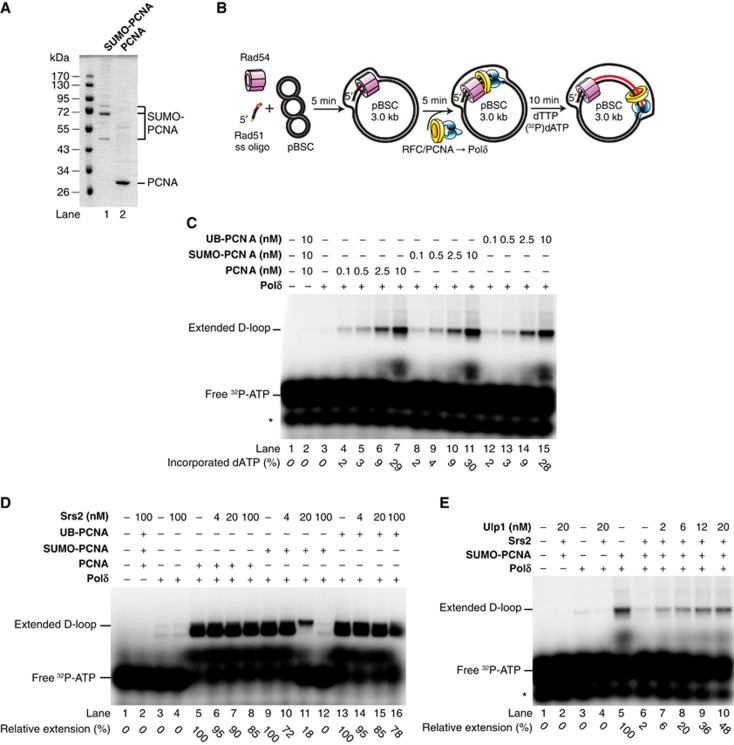
Srs2 inhibits the DNA polymerase activity of Polδ in a D-loop extension assay
To address the biological functions of S-PCNA, we carried out in vitro SUMOylation of yeast PCNA on lysine residue 164 (K164). The K164-S-PCNA was purified from an enzymatic reaction as described in the experimental procedures and a fraction containing only monomers of S-PCNA was used (Figure 1A). First, we tested the effect of S-PCNA on the DNA polymerase activity of Polη, a distributive TLS enzyme, as well as of the processive Polδ enzyme involved in repair synthesis (Maloisel et al, 2008; Li et al, 2009). Purified S-PCNA and unmodified PCNA were loaded on an oligonucleotide-based DNA substrate by the RFC complex and the effect on DNA primer extension was assessed. Under the same reaction conditions PCNA and S-PCNA stimulate the polymerase activity of both Polη and Polδ to the same extent (Supplementary Figure S1, compare lanes 4–10 and 14–20, respectively).
Figure 1.
Effect of S-PCNA on D-loop extension by Polδ. (A) Purified PCNA and S-PCNA. PCNA was SUMOylated and purified as described in the experimental procedures. (B) Schematic of the of the D-loop reaction linked with primer extension using a plasmid-based DNA substrate. (C) D-loop extension in the presence of PCNA, Ub-PCNA and S-PCNA. Rad51 (1 μM) and Rad54 (150 nM) mediated D-loops were mixed with Polδ (33 nM) and indicated amounts of PCNA, Ub-PCNA or S-PCNA in an equimolar complex with RFC, respectively. After 5 min incubation, the reaction was started with the addition of dTTP and [32-P]-dATP, followed by a 10-min extension. The reaction products were treated with proteinase K, resolved on a 0.8% agarose gel and analysed. The numbers at the bottom of the panel represent relative amounts of extension products. The percentage of incorporation is determined as an amount of incorporated dATP versus the total amount of [32P]-dATP. (D) Srs2-mediated inhibition of D-loop extension. Reactions were carried out as described in (C) except that 5 min after loading PCNA three concentrations of Srs2 (4, 20 and 100 nM) were added to the reactions. (E) Inhibition of D-loop extension by Srs2 requires SUMOylated PCNA. The D-loop extension reaction was performed in the presence of the D-loop substrate, RFC, Polδ, Srs2 and S-PCNA. After 5 min incubation with Srs2, increasing amounts of Ulp1 (2, 6, 12 and 20 nM) were added. Following 5 min incubation, the reactions were started with the addition of dTTP and radioactively labelled dATP. After a 10-min extension, the reactions were stopped and the products analysed. Relative extension represents an amount of incorporated [32P]-dATP in the extension products relative to the incorporation in the absence of Srs2, which was set as 100%.
Source data for this figure is available on the online supplementary information page.
The early steps of the Rad51-dependent rescue of a stalled replication fork include the formation of a D-loop structure and its extension. Therefore, we have tested the effects of PCNA, S-PCNA and Ub-PCNA on primer extension of a reconstituted plasmid-based D-loop DNA substrate (Figure 1B; Sebesta et al, 2011). In this assay, PCNA, S-PCNA and Ub-PCNA stimulate equally well the Polδ-mediated D-loop extension (Figure 1C, compare lanes 4–7, 8–11 and 12–15), indicating that PCNA stimulates the primer extension activity of Polδ regardless of the post-translational modification tested.
Since Srs2 was shown to interact with S-PCNA and to promote the synthesis-dependent strand annealing (SDSA) pathway (Ira et al, 2003; Robert et al, 2006), we wanted to test the effect of Srs2 interaction with S-PCNA on D-loop extension. The addition of increasing amounts of Srs2 results in at most a minor inhibition of the primer extension by Polδ in the presence of PCNA or Ub-PCNA (Figure 1D, compare lanes 5–8 and 13–16). However, addition of Srs2 in equimolar amounts to S-PCNA results in a five-fold reduction of D-loop extension (Figure 1D, lanes 7, 11 and 15), and further addition of Srs2 in the presence of S-PCNA leads to the complete abrogation of the primer extension by Polδ (Figure 1D, compare lanes 5–8, 9–12 and 13–16). The inhibition is efficient compared to that observed with unmodified PCNA and correlates with the fact that Srs2 binds S-PCNA with higher affinity than the other forms (Papouli et al, 2005; Pfander et al, 2005; Burgess et al, 2009; Armstrong et al, 2012).
To further underline the specificity towards S-PCNA in mediating the Srs2 inhibition, we tested whether the enzymatic removal of SUMO from PCNA will have any effect on D-loop extension. Hence, we have incubated the reaction mixture containing S-PCNA, Srs2 and Polδ with increasing concentrations of Ulp1, which mediates PCNA deSUMOylation (Parker et al, 2008). After 5 min incubation with Ulp1, the DNA polymerase reaction was started. As expected, the Ulp1-mediated removal of SUMO from PCNA restores the primer extension activity of Polδ (Figure 1E, compare lanes 6–10). This result indicates that the inhibition of D-loop extension by Srs2 depends on its increased local concentration by S-PCNA.
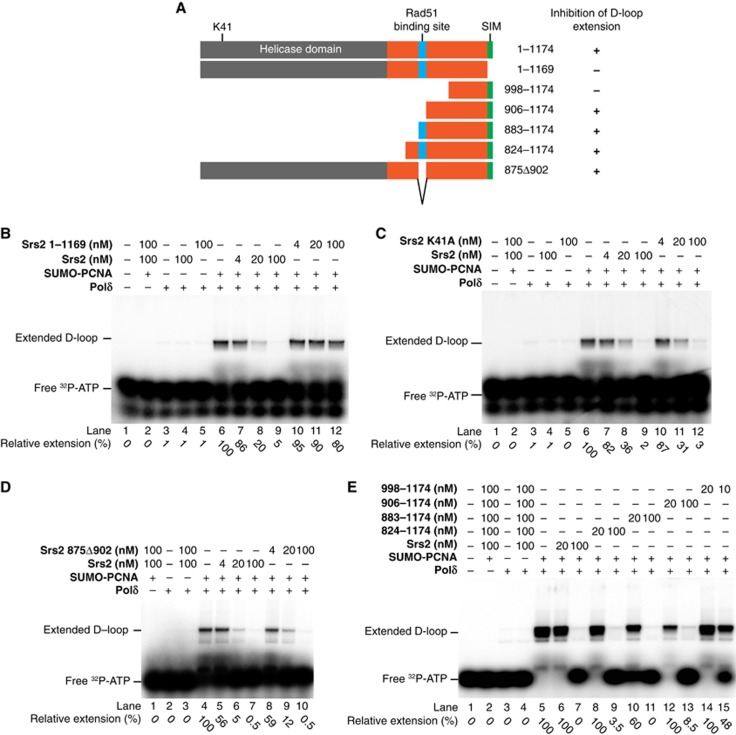
The Srs2 SUMO interacting motif is required for Polδ inhibition
The last five amino acids of Srs2 form a SUMO interacting motif (SIM) and are crucial for the interaction with S-PCNA (Pfander et al, 2005; Le Breton et al, 2008; Burgess et al, 2009). To determine whether this SIM is required for the inhibition of D-loop primer extension, we have tested the effect of a truncated Srs2 protein lacking the last five amino acids (Figure 2A). In contrast to full-length protein, this truncation does not show an inhibition of D-loop extension in the presence of S-PCNA (Figure 2B, compare lanes 7–9, and 10–12, respectively). To demonstrate that the Srs2 truncation is deficient only in binding S-PCNA, we have tested it for all known biochemical activities. This truncated Srs2 preserves DNA binding, ATPase and helicase activities as well as the ability to displace Rad51 protein form the presynaptic filament, indicating that its biochemical properties analysed are indistinguishable from those of full-length Srs2 protein (Supplementary Figures S2A and B). Similarly to full-length protein, the Srs2 truncation had a slight effect on the D-loop extension in the presence of unmodified PCNA (Supplementary Figure S2C), compare lanes 7–9 and 10–12, respectively). Finally, we tried to suppress the Srs2 inhibitory activity by competing it out with increasing amounts of free unconjugated SUMO. This addition has no effect on D-loop synthesis and cannot alleviate the inhibitory effect of Srs2 mediated by S-PCNA (Supplementary Figure S2D), therefore confirming the specific role of S-PCNA binding to Srs2.
Figure 2.
The SIM but not the catalytic activity of the Srs2 mediates the inhibition of the D-loop extension by Polδ. (A) Schematic of the Srs2 domains and truncations used in this study. (B) D-loop extension by Polδ in the presence of S-PCNA and Srs2 or its SIM deleted truncation (Srs2 (1–1169)). Reactions were assembled as described earlier and incubated with increasing amounts of Srs2 or Srs2 (1–1169). (C) Inhibition of D-loop extension by Polδ is independent of the Srs2 ATPase activity. Reactions were carried out as described in the experimental procedures using wild-type Srs2 and Srs2-K41A mutant. (D) The inhibition is independent of the Rad51 binding site of Srs2. The Rad51 interaction-deficient mutant of the Srs2 875Δ902 inhibits the D-loop extension. Reactions were carried out as described using increasing amounts (4, 20 and 100 nM) of Srs2 and Srs2 875Δ902 proteins. (E) Minimal Srs2 region required for inhibition of extension. Reactions were carried in the presence of full-length Srs2 as well as Srs2 C-terminal fragments ranging from 824 to 998 until the end of the protein. The numbers at the bottom of the panels are the relative amounts of extension product representing an amount of incorporated [32P]-dATP in the extension products relative to the incorporation in the absence of Srs2, which was set as 100%.
Source data for this figure is available on the online supplementary information page.
Srs2 possesses DNA helicase activity and is capable of dislodging Rad51 from the presynaptic filament; thus, we have tested the effect of the Srs2-K41A mutant deficient in ATPase and translocase activities (Krejci et al, 2004). Surprisingly, the K41A mutant is as competent as wild-type Srs2 in inhibiting D-loop extension (Figure 2C, compare lanes 7–9 and 10–12, respectively), indicating that the helicase and/or translocase activities of Srs2 are not required for this inhibition. We next analysed whether Srs2 directly inhibits D-loop extension using a labelled oligonucleotide. The analysis of the products by denaturing gels confirms that the extent of DNA synthesis decreases with increasing concentrations of Srs2 (Supplementary Figure S3).
To rule out the possibility that Srs2 inhibits DNA synthesis by unwinding the D-loops, we have performed the experiment using a radioactively labelled oligonucleotide. As indicated in Supplementary Figure S4A and our previous publication (Sebesta et al, 2011), Srs2 does not unwind the D-loop, indicating that the inhibition is mediated by a protein–protein interaction with S-PCNA.
We have shown previously that the region 875–902 of Srs2 is responsible for the interaction with Rad51 (Colavito et al, 2009). In our experimental system, the Srs2 875Δ902 mutant is fully capable to inhibit the extension of the D-loop by Polδ in the presence of S-PCNA (Figure 2D, lanes 8–10). The extent of inhibition is almost identical to that of a wild-type protein (Figure 2D, lanes 5–7), suggesting that the Rad51 interaction domain is not required for this inhibition. This is further supported by replacement of Rad51 and Rad54 by RecA protein in the D-loop formation. In the RecA-mediated D-loop DNA substrate, we observe the same extent of inhibition as detected using a Rad51-based D-loop, indicating that the strand extension inhibition is independent of Rad51 and Rad54 proteins (Supplementary Figure S4B, compare lanes 4–7 to lanes 11–14). To further narrow down the region required for blocking the extension reaction, we tested a set of N-terminally truncated versions of Srs2 (Figures 2A and E). Similarly to the full-length protein, most of the Srs2 fragments inhibit D-loop extension. Specifically, even the Srs2 fragment spanning residues 906–1174 and lacking the Rad51 interacting domain is fully capable of mediating the S-PCNA-dependent inhibition (Figure 2E, lanes 12 and 13). On the other hand, the shorter truncation fragment of Srs2 (998–1174) no longer sustains the inhibitory effect (Figure 2E, lanes 14 and 15), a result that could reflect the loss of an additional interaction site or of a proper conformation.
S-PCNA does not affect the Srs2 activities and forms a complex with Srs2
To address whether S-PCNA influences known Srs2 biochemical activities, we tested its helicase activity in the presence of PCNA or S-PCNA. As shown in Supplementary Figure S4C, neither PCNA nor S-PCNA has any effect on the ability of Srs2 to unwind 3′ overhangs. Similarly, PCNA or S-PCNA did not alter the ability of Srs2 to displace Rad51 from a nucleoprotein filament (Supplementary Figure S4D). However, this displacement of Rad51 is titrated out by increasing amounts of DNA molecules loaded with S-PCNA before the addition of Srs2. This result suggests that the interaction of Srs2 with S-PCNA loaded onto a separate DNA molecule can compete out the capacity of Srs2 to dislodge Rad51 (Supplementary Figure S4E). Furthermore, this suppression is stoichiometric since equimolar amounts of S-PCNA were sufficient to reduce by half the inhibitory effect of Srs2 on D-loop formation (Supplementary Figure S4E, compare lanes 1 and 5). This is in agreement with a pull-down experiment showing that Srs2 can bind simultaneously Rad51 and either PCNA or S-PCNA (Supplementary Figure S4F).
To gain further insight into the mechanism of the Srs2 and S-PCNA mediated inhibition, we tested the effect of Srs2 addition at different stages of the D-loop extension assay (Supplementary Figure S5). First, Srs2 was added together with Polδ and S-PCNA, (Stage I). Alternatively, Srs2 was added 5 min after Polδ or at the same time when dNTPs start the extension reaction (Stages II and III, respectively). Srs2 exhibits a similar inhibitory effect on the extension reaction regardless of the order of addition (Supplementary Figure S5, compare lanes 5–8, 9–12 and 13–16), indicating that S-PCNA and Srs2 readily form an elongation inhibitory complex.
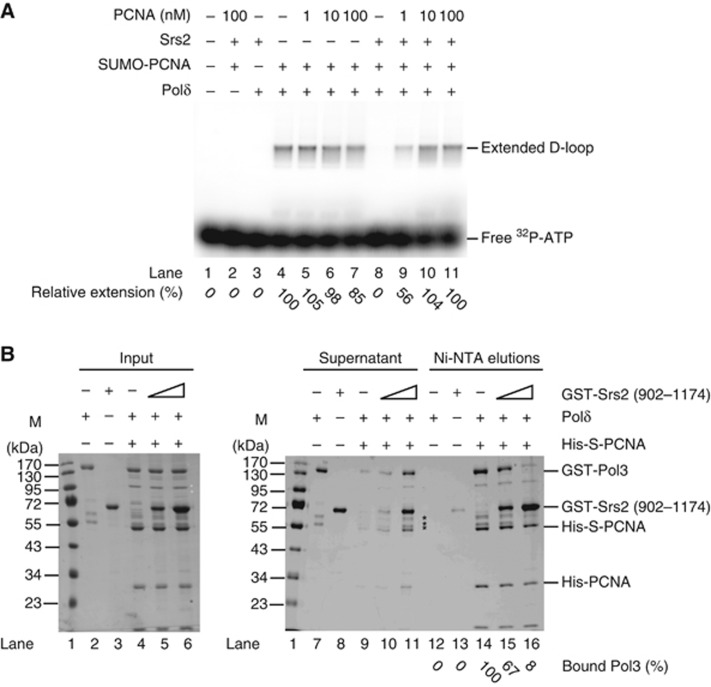
PCNA SUMOylation triggers the disassembly of Polδ/PCNA complex by Srs2
To test the possible mechanisms by which Srs2 inhibits D-loop-mediated strand extension, we monitored the amount of the free Polδ in the reaction. A three-fold molar excess of S-PCNA over Polδ was used in order to ensure that all Polδ molecules are in a complex with S-PCNA. This is indeed the case, since the addition of increasing amounts of unmodified PCNA to the reaction does not further increase D-loop extension in the absence of Srs2 (Figure 3A, lanes 4–7). On the other hand, in the presence of Srs2, the addition of unmodified PCNA reverses the blockage of D-loop extension (Figure 3A, lanes 8–11), indicating that Srs2 triggers the release of Polδ from the complex with S-PCNA.
Figure 3.
Srs2 inhibits D-loop extension by disruption of the Polδ/S-PCNA interaction. (A) Addition of free PCNA can alleviate the inhibition of D-loop extension. Reactions were carried out as described earlier except that after 5 min incubation, indicated amounts of unmodified PCNA were added into the reaction containing S-PCNA/RFC (20 nM), Polδ (33 nM) and, where indicated, 100 nM of Srs2. Following 10 min incubation at 30°C, the reactions were treated with proteinase K and resolved on a 0.8% agarose gel. Extension products were quantified, and the percentage of relative extension is given at the bottom of the figure. (B) Srs2 disrupts the interaction between S-PCNA and Polδ. S-PCNA from the in vitro SUMOylation reaction containing both PCNA and S-PCNA (with the hexahistidine tag, 5 μg) was mixed with 5 μg of purified polymerase δ and increasing amounts of Srs2 (5 or 12.5 μg). After incubation, the beads were washed and treated with SDS to elute bound proteins. The supernatants with unbound proteins and the SDS elution were analysed by SDS–PAGE. Lanes 2, 3, 7 and 8 are controls where only Srs2 or Polδ was incubated with Ni-NTA beads. * points at Polδ degradation products. Relative quantification of GST-Pol3 on Ni-NTA beads is indicated.
Source data for this figure is available on the online supplementary information page.
To confirm that Srs2 interaction with S-PCNA is capable of releasing Polδ from the DNA polymerizing complex, we used a pull-down assay. We immobilized S-PCNA on Ni-NTA beads via a His tag on PCNA and tested the effect of Srs2 (902–1174) on the ability of Polδ to interact with S-PCNA. The addition of Srs2 to the reaction mixture containing Polδ resulted in the disruption of the interaction between Polδ and S-PCNA (Figure 3B). Alternatively, we carried out the experiment with the full-length Srs2. We immobilized the S-PCNA/Polδ complex on GTH-beads via a GST tag present on the Pol3 subunit of Polδ (Supplementary Figure S6, lane 7) and challenged the complex by addition of increasing amounts of Srs2 protein. As expected, the addition of Srs2 to the reaction mixture results in the disruption of the interaction between Polδ and S-PCNA and the release of S-PCNA from beads (Supplementary Figure S6, compare lanes 7, 9 and 11), further supporting the Srs2 inhibition of D-loop extension by disrupting the Polδ/S-PCNA complex.
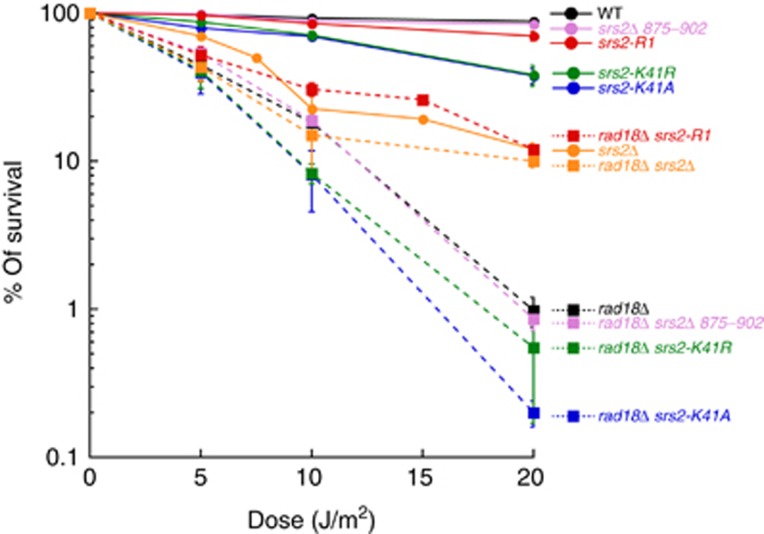
The interaction between Srs2 and S-PCNA is responsible for the sensitivity of rad18Δ cells
To determine the relative importance of Srs2 binding to Rad51, its translocase activity and its capacity to disrupt the interaction between the polymerase and S-PCNA during the rescue of stalled replication forks, we have tested the ability of different srs2 mutants to suppress the UV sensitivity of the PRR-deficient rad18Δ mutants. We have used the srs2-K41A and srs2-K41R mutants deficient for their helicase activity, the srs2 875Δ902 mutant that no longer interacts with Rad51 as well as the srs2-R1 allele that abolishes the interaction with S-PCNA. Unexpectedly, the srs2-K41A, srs2-K41R and srs2 875Δ902 mutants fail to suppress the rad18 sensitivity to UV irradiation, indicating that the sensitivity of rad18Δ mutants is not a consequence of preventing recombinational repair (Figure 4). Interestingly, srs2-R1 and srs2Δ are the only tested alleles of SRS2 that partially suppress the sensitivity of rad18Δ mutants to UV (Figure 4). These data are in agreement with previously published observations (Colavito et al, 2009) in which srs2Δ, srs2 (1–860), srs2ΔPIP or srs2ΔSIM but not srs2 875Δ902 was able to suppress the rad18Δ sensitivity (Colavito et al, 2009; Armstrong et al, 2012; Kolesar et al, 2012).
Figure 4.
Suppression of the UV sensitivity of rad18Δ mutants by different alleles of SRS2. Haploid strains in a RAD18 (filled circles, solid lines) and rad18Δ (filled squares, dotted lines) background were grown to stationary phase in liquid YEPD medium and exposed to UV irradiation at 0.5 J/m2/s as described in Supplementary data. Each point on the curves is the average of three experiments.
It is therefore the ability of Srs2 to disrupt the interaction between the polymerase and S-PCNA in a context where PCNA cannot be ubiquitinated that is responsible for the sensitivity of rad18Δ cells. This result also suggests that in the absence of the Rad18-dependent ubiquitination, maintenance of S-PCNA may prevent the TLS rescue of the stalled replication fork by recruiting Srs2.
PCNA SUMOylation is required for genome stability
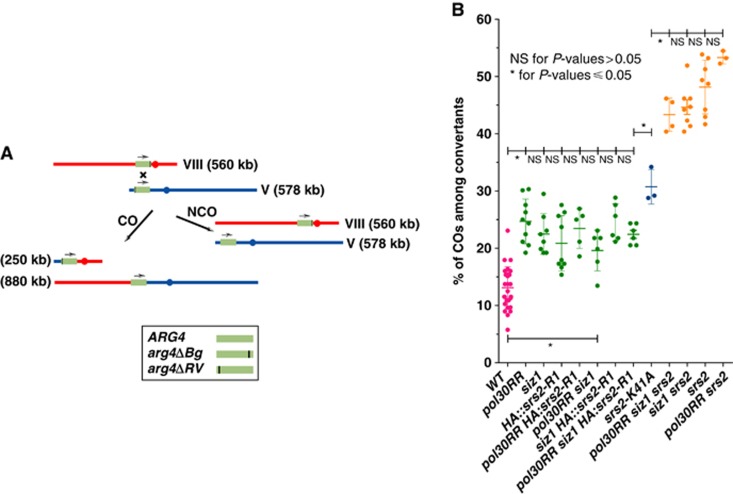
To address the biological significance of the disruption of the S-PCNA/Polδ complex in the context of a D-loop extension, we reasoned that it should reduce the length of the repair tracts during either TS or recombinational repair (HR). A known outcome of longer repair synthesis tracts is their more often association with reciprocal exchanges and genetic instability (Inbar and Kupiec, 1999). Because reciprocal exchanges involving sister chromatids cannot be measured genetically, we tested this idea using an assay measuring reciprocal exchanges between two mutant alleles of a gene, one located at its native locus while the other is inserted at an ectopic locus in an orientation that allows the recovery of reciprocal translocations (Figure 5A; Robert et al, 2006). Using this system, we have shown previously that the absence of the SIM within Srs2 (Srs2-R1 mutant) indeed leads to an elevated percentage of reciprocal exchanges between non-allelic homologous sequences in unchallenged cycling cells (Le Breton et al, 2008). However, the percentage of COs was lower than that monitored in the absence of Srs2. Therefore, if the CO levels observed in Srs2-R1 mutants correspond to the fraction triggered by the interaction with S-PCNA, we should mimic this result in strains in which PCNA is not SUMOylated, that is, cells lacking the E3 ligase Siz1 responsible for most of the SUMOylation of PCNA (Hoege et al, 2002) or alternatively in cells containing mutations of PCNA that prevent the modification of the major SUMOylated lysine residues (K127R and K164R, pol30RR). To test this prediction, we constructed isogenic strains carrying either the siz1Δ::HIS3MX6 or the pol30RR mutations and measured the spontaneous CO levels. We found that srs2-R1 yields CO levels that are undistinguishable from those measured in either siz1Δ or pol30RR strains. Furthermore, the combination of srs2-R1 with siz1Δ, pol30RR or both does not lead to any further increase in the level of COs (Figure 5B). Our observation strongly suggests that these mutations inactivate the same pathway and that unchallenged cells use the Siz1-dependent SUMOylation of PCNA to regulate genome stability by recruiting and concentrating Srs2 to D-loop structures. Importantly, the absence of Srs2 alone leads to elevated levels of genome instability that are not reduced by siz1Δ, pol30RR or both confirming that Srs2 prevents genome rearrangements in both a S-PCNA-dependent and independent fashion (Figure 5B; Le Breton et al, 2008). The S-PCNA-independent pathway could involve the Srs2 helicase activity, since the elevated levels observed in the null mutant are the sum of the combined defects of both the helicase activity and the ability to interact with S-PCNA (Figure 5B).
Figure 5.
Reciprocal exchange is enhanced by the loss of interaction between S-PCNA and Srs2. (A) Genetic system is used to determine the proportion of COs among spontaneous recombinants. Bg and RV indicate the mutation of the BglII and EcoRV sites that invalidate the ARG4 genes. Note that the converted allele leading to arginine prototrophy has been arbitrarily attributed to one of the arg4 mutant copies. (B) Plot of the percentage of CO-associated recombination events in various mutant backgrounds. Each dot represents an experiment carried out on 156 spontaneous events. NS, not significant for P-values >0.05; *, significant at P-values ≤0.05.
Close together markers co-convert more frequently in srs2-R1 or pol30RR mutants
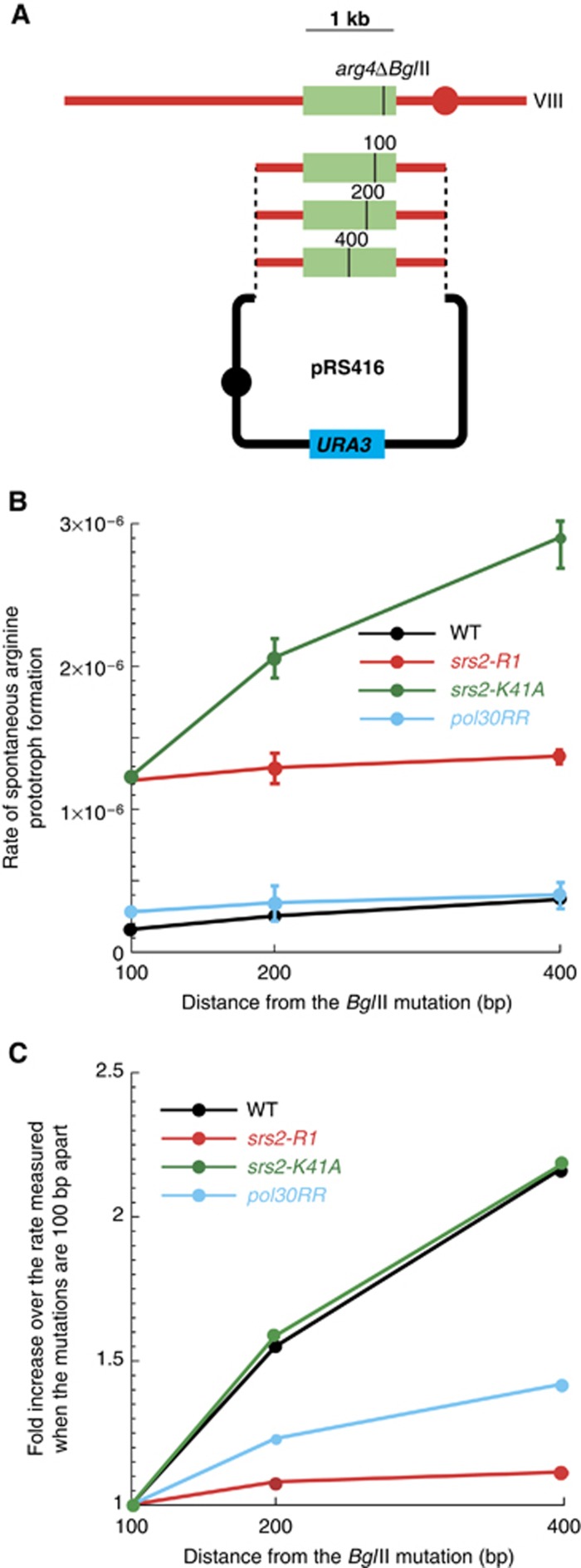
Conversion tracts tend to co-convert closely spaced mutations. Thus, a prediction for longer conversion tracts is that fewer recombinants should be formed. We tested this idea by measuring the spontaneous rates of arginine prototroph formation between the chromosomal copy containing the BglII mutation and three centromeric plasmids in which mutations were introduced 100, 200 and 400 bp upstream of the BglII site, respectively (Figure 6A). The constructs were introduced into wild type, srs2-R1, pol30RR and srs2-K41A backgrounds and subjected to fluctuation analyses. We found that srs2-K41A and srs2-R1 exhibit elevated recombination levels compared to pol30RR and the wild-type control (Figure 6B). However, when we monitored the recombination increase as a function of the distance from the BglII site, we found that, as predicted, srs2-R1 and pol30RR exhibit a very modest increase in spontaneous recombinant formation whereas the srs2-K41A and the wild-type control show a similar robust increase under the same conditions (Figure 6C). This result supports the idea that failure to interact with S-PCNA yields longer conversion tracts, a feature not observed in the helicase-dead mutant.
Figure 6.
srs2-R1 and pol30RR reduce the spontaneous conversion rate for closely spaced mutations. (A) A two-kb chromosomal fragment of the ARG4 locus was inserted into a centromeric plasmid containing the URA3 marker. Three plasmids containing a frameshift mutation located at 100, 200 and 400 bp upstream of the BglII site were generated and transformed into wild-type, pol30RR, srs2-K41A and srs2-R1 strains containing the arg4ΔBglII mutation at the ARG4 locus. (B) Rates of arginine prototroph formation. (C) Relative effect to that observed when the mutations are 100 bp apart.
Srs2/S-PCNA interaction as a general model for extension regulation
As is suggested by the suppression experiments, the mechanism that we have uncovered may not be restricted to DNA synthesis during D-loop extension. To test this idea, we used a standard primer extension assay and a singly primed ΦX-174 circular ssDNA. Since wild-type Srs2 can partially dismantle the primer extension assay substrate (Sebesta et al, 2011), we used the Srs2-K41R mutant in this experiment (Supplementary Figure S7A). We found that Srs2-K41R inhibits preferentially DNA synthesis in the presence of S-PCNA (Supplementary Figure S7A, compare lanes 7–10 with lanes 11–14). Similarly, Srs2 also inhibits DNA synthesis preferentially when S-PCNA is loaded on the ΦX-174 substrate (Supplementary Figure S7B, compare lanes 5–8 and 9–12). Since Srs2-K41R and Srs2 (902–1174) are as proficient as wild-type Srs2 in this reaction (data not shown), we rule out the possibility that Srs2 interferes with the stability of the ΦX-174 substrate.
Next, we investigated whether the S-PCNA/Srs2 interaction is limited only to the extension by Polδ and carried out the D-loop extension assay using Polη. Since Polη does not discriminate well the incorporation of ribonucleotides and because ATP is present in the reaction, we used [32-P]-dCTP to follow the D-loop extension reaction in the presence of PCNA and S-PCNA (Supplementary Figure S7C). In lower salt conditions where the extension of a D-loop by Polη is PCNA independent (Sebesta et al, 2011), Srs2 inhibits the Polη-dependent D-loop extension only to the level corresponding to a PCNA-independent extension (Supplementary Figure S7D, lanes 4–8). However, at the physiologically higher salt conditions, Srs2 inhibits strongly the extension of the D-loop by Polη (Supplementary Figure S7D, lanes 9–13). The effect is again specific for S-PCNA, since the inhibition is much weaker in the presence of unconjugated PCNA (a five-fold versus a two-fold inhibition). These experiments strengthen our view that Srs2 inhibits DNA extension by interfering with the interaction between S-PCNA and a polymerase.
Discussion
Collision of the replication fork with other machineries, exogenously induced damage or proteins tightly associated with DNA is thought to occur frequently in cycling cells. Such events can cause serious threats to the cells and need to be quickly detected and efficiently repaired to preserve genome integrity (Friedel et al, 2009). PCNA acts as a molecular switch that controls how DNA lesions are processed during replication. In the presence of DNA damage, PCNA is modified at the conserved lysine residue 164 by either mono-ubiquitin or a lysine-63-linked poly-ubiquitin chain, which induce error-prone or error-free replication bypass of the lesions, respectively (Hoege et al, 2002). PCNA can also undergo SUMOylation during replication in the absence of DNA damage (Hoege et al, 2002), a modification that contributes to the recruitment of the Srs2 helicase to the replication fork to prevent untimely recombination during S phase (Papouli et al, 2005; Pfander et al, 2005). However, our current understanding of how S-PCNA-dependent recruitment of Srs2 results in decreased levels of recombination at replication forks is not clear. In this study, we have investigated the biochemical and biological consequences of PCNA SUMOylation and have uncovered a novel molecular mechanism that can modulate DNA repair and recombination at stressed replication forks and thus limits genome instability.
S-PCNA does not prevent polymerase activity
Since PCNA monoubiquitination promotes the recruitment of TLS polymerases (Hoege et al, 2002), we analysed the influence of post-translational modifications of PCNA on the DNA synthesis activity of replicative and TLS polymerases. S-PCNA indeed stimulates the polymerase activity of both Polδ and Polη, but only to an extent similar to that reported for unmodified or ubiquitinated PCNA (Haracska et al, 2006), indicating that the presence of ubiquitin or SUMO residues on PCNA does not affect significantly the competence of DNA polymerases to elongate a linear template. Since S-PCNA recruits the Srs2 antirecombinase to the replication fork where it could regulate HR (Fabre et al, 2002; Krejci et al, 2003; Veaute et al, 2003), we tested the direct effect of S-PCNA on a recombination reaction. However, since previous findings have indicated that the canonical antirecombination activity of Srs2 is independent of its interaction with S-PCNA (Le Breton et al, 2008), we did not expect to find a strong effect of S-PCNA on the ability of Srs2 to dismantle Rad51 filaments. Indeed, we detected normal removal of Rad51 by Srs2 in the presence of PCNA or S-PCNA. Concomitantly, when we loaded S-PCNA on streptavidin-biotin blocked DNA substrates we observed a decrease in the ability of Srs2 to dismantle Rad51 protein from ssDNA, most probably due to a titration of Srs2 by the S-PCNA-loaded DNA substrate. This is supported by the pull-down experiments showing that the interaction with Rad51 and PCNA is independent of each other. Therefore, S-PCNA sequesters Srs2 at a position or in a state that outcompetes the interaction with Rad51. Altogether this indicates that recruiting Srs2 to replication forks could serve yet another purpose in the HR process.
Specific S-PCNA interaction with Srs2 leads to the inhibition of repair synthesis
Since D-loop formation and its extension is a prerequisite for the Rad51-dependent fork rescue and because the TS mechanism shares similarities with HR (Higgins et al, 1976; Goldfless et al, 2006), we expected the Srs2/S-PCNA complex to inhibit repair synthesis. The D-loop reaction linked with primer extension (Sebesta et al, 2011) is insensitive to the presence of unmodified or ubiquitinated PCNA. On the contrary, under the same conditions, the Srs2 protein fully inhibits D-loop extension in the presence of S-PCNA. This observation indicates that, rather than displacing Rad51 filaments, targeting Srs2 to the fork through PCNA SUMOylation confers an inhibitory effect on D-loop extension. Several independent experiments further support this conclusion. First, introduction of the deSUMOylating enzyme Ulp1 into the reaction mixture restores the extension by Polδ. Second, an Srs2 protein missing its SIM loses the ability to inhibit the D-loop extension reaction, indicating a requirement for direct protein–protein interaction between Srs2 and S-PCNA. Third, the ability of Srs2 to dismantle Rad51 filaments might not be essential during the rescue of stalled replication forks since the interaction of Srs2 with S-PCNA is dispensable for this activity (Le Breton et al, 2008). Fourth, our data show that the helicase activity and ability to interact with Rad51, the hallmark of antirecombinase activity (Krejci et al, 2004; Antony et al, 2009; Colavito et al, 2009), are neither required for the inhibitory effect observed in the D-loop extension assay nor involved in the suppression of the sensitivity to UV of a RAD18 deletion. This also explains the findings of a previous genetic screen for suppressors of rad18Δ UV sensitivity (Palladino and Klein, 1992). Two classes of srs2 alleles have been described, one of which affects recombination. Interestingly, out of the five alleles suppressing the UV sensitivity, four are located outside the helicase domain, farthermost at the C-terminus of the protein that we show to be responsible for the inhibitory effect on DNA extension (Palladino and Klein, 1992). Fifth, since Srs2 is capable of inhibiting RecA-mediated D-loop extension, the Srs2 inhibition is independent of the interaction with Rad51 and Rad54.
Srs2 disrupts S-PCNA/Polδ elongation complex
A possible mechanism by which Srs2 mediates the inhibition of DNA synthesis could be by disrupting the interaction between Polδ and S-PCNA. In agreement with this hypothesis, addition of saturating concentrations of unmodified PCNA to the reaction relieves the Srs2 inhibition, allowing free Polδ to engage in de novo DNA synthesis, a result confirmed by pull-down experiments with purified proteins. This observation suggests that the interaction of Srs2 with S-PCNA could affect the accessibility of the interdomain connector loop (IDCL) of PCNA (Eissenberg et al, 1997). The IDCL serves as an interaction surface of the PIP box containing proteins and hence could also affect the binding of PCNA to other proteins through this motif. Indeed, Srs2 does not only inhibit Polδ, the DNA polymerase involved during repair synthesis (Maloisel et al, 2008; Li et al, 2009; Brocas et al, 2010), but also specifically inhibits extension of S-PCNA/Polη complex, indicating that this regulatory mechanism could be more general.
Model of stalled fork repair
Based on genetic evidence, several models for repair of stalled replication forks have been proposed (Unk et al, 2010). The data presented here extend these models in several ways and put forward a possible molecular mechanism for the role of S-PCNA and Srs2 during replication. It is widely accepted that PCNA and its post-translational modifications serve as a molecular switch that can regulate the processing of stalled or collapsed replication forks. Under undisturbed conditions, both PCNA and S-PCNA stimulate the processivity of the replicative DNA polymerases. We do not know the nature of the signal that triggers PCNA SUMOylation during S phase, however in the presence of DNA damage, PCNA undergoes mono- or poly-ubiquitination (Hoege et al, 2002; Torres-Ramos et al, 2002; Haracska et al, 2004). Mono-ubiquitination of PCNA on residue K164 directly promotes the Rad18-dependent damage tolerance repair pathway (Haracska et al, 2004). In addition, polyubiquitination of PCNA results in error-free damage tolerance mediated by Rad5-dependent TS/gap repair mechanism (Blastyák et al, 2007; Branzei et al, 2008; Unk et al, 2010). On the other hand, SUMOylation of PCNA on residues K164 and K127 also promotes replication through damaged DNA, and targets Srs2 to the replication fork to downregulate the HR pathway (Papouli et al, 2005; Pfander et al, 2005). However, in light of several recent observations, we believe that the ability of Srs2 to dismantle the Rad51 filament might be required preferentially at recombination foci rather than at replication forks since this activity is not dependent on the interaction with S-PCNA (Le Breton et al, 2008; Burgess et al, 2009). In addition, it has been shown that both Rad18/Rad5 and Rad51-dependent pathways promote the formation of X-shaped structures that resemble pseudo double Holliday junctions at damaged replication forks (Liberi et al, 2005; Branzei et al, 2006; Minca and Kowalski, 2011). Furthermore, Rad18 and Rad51 cooperate to promote TS and the Ubc9-dependent SUMOylation pathway regulates this cooperation (Branzei et al, 2008). Interestingly, the Rad51 recombinase is important for proper replication in Xenopus extracts and is required for the formation of intermediates that accumulate in rad5 and rad18 mutants during the repair of stalled or collapsed replication forks (Postow et al, 2009; Hashimoto et al, 2010). Our data also show that interaction of Srs2 with S-PCNA can outcompete the interaction with Rad51 and thus limit the ability of Srs2 to dismantle the filament. In such a scenario, S-PCNA targets Srs2 to the stalled fork and limits the extent of Rad51-dependent repair synthesis by releasing Polδ from its complex with S-PCNA, thus blocking the further extension of the 3′ end of the invading primer strand and limiting the extent of conversion (Figures 6 and 7). Since CO formation is more dependent on homology length than gene conversion (Inbar and Kupiec, 1999; Prado and Aguilera, 2003) shorter DNA repair stretches keep reciprocal exchanges at a low level. Indeed, as PCNA becomes increasingly SUMOylated, Srs2 turns out to be gradually concentrated at D-loops where it can block the extension of repair synthesis, providing a mechanism limiting the extent of conversion tracts. As proposed earlier (Ira et al, 2003; Robert et al, 2006), Srs2 promotes SDSA and limits the occurrence of potentially harmful COs, but the direct molecular mode of action is not clear. First, Srs2 can promote SDSA by blocking second end capture. Alternatively, Srs2 could directly unwind D-loops (Dupaigne et al, 2008) or promote an even more efficient D-loop unwinding by targeting the efficient Mph1 helicase (Prakash et al, 2009; Sebesta et al, 2011). Finally, through its direct interaction Srs2 could also mark/target the D-loop for an effective dissolution by Sgs1 or further processing by the Mus81/Mms4 proteins (Fabre et al, 2002; Chiolo et al, 2005; Robert et al, 2006; LK, unpublished data).
Figure 7.
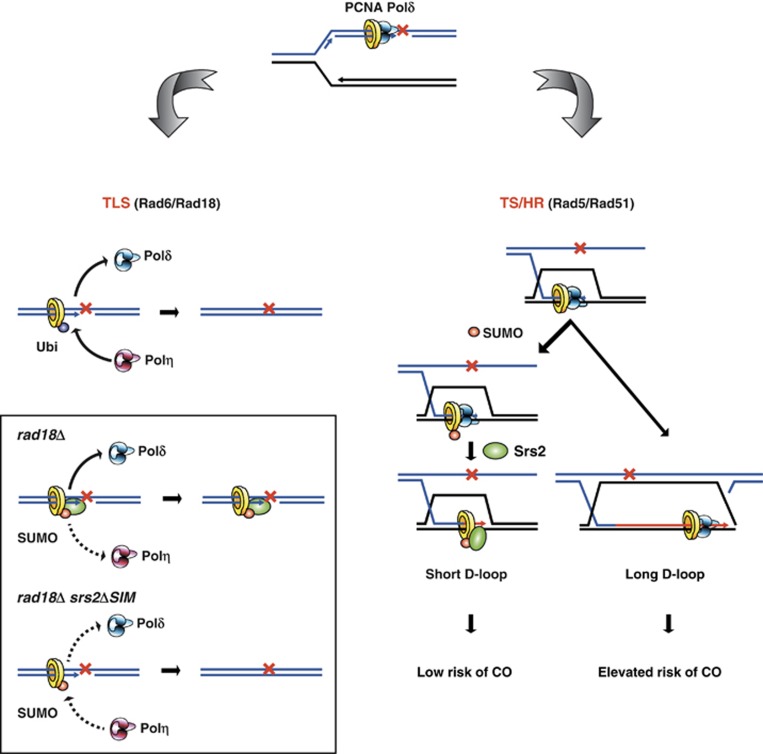
Schematic view of the roles of S-PCNA and Srs2 in the rescue of stalled replication forks. Left panel, monoubiquitination of PCNA on residue K164 promotes the Rad6/Rad18-dependent TLS pathway. Therefore, in the absence of Rad18 (boxed area) the cells become extremely sensitive to UV damage. On one hand, absence of ubiquitination no longer triggers the replacement of Polδ by Polη while on the other hand the unconjugated lysine K164 allows SUMOylation to recruit Srs2 that in turn dislodges either Polδ or Polη. This sensitivity is partially alleviated when Srs2 can no longer interact with S-PCNA (srs2ΔSIM). The dotted arrows indicate suboptimal processes. Right panel, polyubiquitination of PCNA results in error-free Rad5-dependent TS. Furthermore, Rad51 cooperates to promote TS regulated by the Ubc9-dependent SUMOylation pathway (TS/HR). SUMOylation of PCNA on residues K164 and K127 targets Srs2 to the replication fork to downregulate the HR pathway. As PCNA becomes increasingly SUMOylated, Srs2 is gradually concentrated at D-loops where it can block the further extension of repair synthesis and limit the extent of conversion tracts by releasing Polδ from its complex with S-PCNA. As a consequence, shorter stretches of DNA synthesis are easier to become destabilized through various activities therefore promoting NCO outcomes. Finally, Srs2 could also stimulate SDSA directly, thus lowering the risk of CO occurrence.
Siz1 is required for genomic stability during S phase
The association of Srs2 with recombination foci seems to be S-PCNA independent (Burgess et al, 2009). This is also supported by the observation that deletion of SIZ1 does not affect the elimination of toxic intermediates (Stelter and Ulrich, 2003; Pfander et al, 2005). However, we confirm that Siz1 is required for genome stability (Xhemalce et al, 2004; Paek et al, 2010). In its absence, ectopic recombination intermediates formed spontaneously in the cell are no longer resolved with a strong bias towards non-COs (NCOs), a result similar to that observed when both lysine residues of PCNA (pol30RR) that can become SUMOylated are replaced by arginine residues. This result strongly argues that the effect on genome stability in unchallenged cells is mediated principally through PCNA SUMOylation, since the absence of the ubiquitination pathway has no effect on the resolution of recombination intermediates either by itself or in a siz1 and pol30RR background (SG, unpublished results). Because the formation of a CO following a DSB induced out of S phase is not affected by the absence of components associated with replication forks (Supplementary Figure S8), we believe that during S-phase S-PCNA actively prevents the formation of double Holliday junctions and their resolution by inhibiting or terminating the extension of the D-loop. Because they can lead to loss of heterozygosity and chromosomal aberrations, COs are potentially dangerous intermediates that occur much less frequently in mitotic than in meiotic cells (Esposito, 1978; Beumer et al, 1998; Stark and Jasin, 2003). Indeed, COs have been associated with many human disorders (Shaw and Lupski, 2004) supporting the need for tight regulation and control. In yeast, at least three mechanisms characterized by three different helicases, Srs2, Sgs1 and Mph1, can operate to suppress CO formation (Prakash et al, 2009; Marini and Krejci, 2010).
In summary, our findings provide a novel insight into the mechanism by which S-PCNA/Srs2 complex may operate in the rescue of a stalled replication fork. While PCNA acts as a molecular switch responsible for engaging the cell into a given repair pathway through various post-translational modifications, Srs2 acts as a molecular caretaker that efficiently transduces PCNA signalling. Together with S-PCNA, Srs2 is an additional control point in the fork rescue mechanism providing flexibility by limiting the amount of toxic DNA structures and promoting the gap-filling/TS function of Rad18-Rad5 through reversible recombination intermediates (Branzei et al, 2008).
Yet, such a mechanism involving S-PCNA and Srs2 can become toxic for the cells under certain circumstances. Indeed, the overproduction of either Srs2 or its helicase-dead variant results in a synthetic lethal phenotype in combination with many replication-associated genes, an activity that depends on the S-PCNA interaction motif (León Ortiz et al, 2011), and emphasizes the need for the S-PCNA-mediated activity to be under a very tight control. Accordingly, it has recently been reported that in the absence of Elg1, an alternative subunit of the RFC clamp loader that interacts preferentially with S-PCNA, both Srs2 and S-PCNA accumulate on chromatin (Parnas et al, 2010). This raises the possibility for a role of Elg1 in unloading S-PCNA from DNA after the completion of a repair event in unchallenged cells, an intriguing mechanism to address in future studies. Finally, it will be extremely interesting to determine whether PARI or other human translocases behave like Srs2 in a reconstituted DNA synthesis assay with human S-PCNA.
Materials and methods
PCNA SUMOylation and purification of S-PCNA
In vitro SUMOylation reaction of PCNA was carried out in 2 ml of P0 buffer (40 mM Tris–HCl pH 7.5; 8 mM MgCl2; 100 μg/ml BSA; 10% glycerol; 100 μM ATP) in the presence of PCNA (40 μg), GST-Aos1 (30 μg), GST-Uba2 (30 μg), GST-Ubc9 (30 μg), GST-Siz1 (1–465) (90 μg), Smt3-GG (600 μg), RFC (4 μg), DNA (50 μg) for 2 h at 30°C.
The crude PCNA and S-PCNA containing reaction mixture has been applied to a 1-ml MonoQ 5/50 column (GE Healthcare) and extensively washed with buffer A (70 mM NaCl, 40 mM Tris–HCl pH 7.5, 0.01% Nonidet P-40, 10% glycerol). Reaction enzymes have been washed out with a 3-ml gradient to 33% of buffer B (1 M NaCl, 40 mM Tris–HCl pH 7.5, 0.01% Nonidet P-40, 10% glycerol). S-PCNA has been eluted with a 5-ml gradient of buffer B from 33 to 50%. The S-PCNA containing fractions have been pooled, diluted and concentrated using a 200-μl MiniQ PC3.2 column by eluting the S-PCNA with a 3-ml gradient to 100% of buffer B. Peak fractions have been collected and used directly in subsequent assays. Mass spectrometry analysis revealed that 95% of PCNA SUMOylation is found on K164 and 5% on K127. Ub-PCNA was purified as described previously (Haracska et al, 2006).
D-loop extension assay
The D-loop assay was performed essentially as described previously (Krejci et al, 2004), and the details are included in Supplementary data. The primer extension reaction was assembled as described in Sebesta et al (2011). Briefly, standard 30 μl reaction mixture containing 12 μl from the D-loop reaction was supplemented with 660 nM RPA, 10 nM PCNA or SUMO- or Ubiquitin-PCNA, and 33 nM Polδ in buffer O (20 mM Tris–HCl pH 7.5, 5 mM DTT, 0.1 mM EDTA, 150 mM KCl, 40 μg/ml BSA, 8 mM MgCl2, 5% glycerol, and 75 μM each of dGTP and dCTP). PCNA loading reaction was incubated at 30°C for 5 min. Reaction was stopped by cooling on ice followed by addition of Srs2 and other indicated proteins (Ulp1). Reaction was continued at 30°C for an additional 5 min. DNA synthesis was initiated by addition of buffer O containing 75 μM dTTP and 0375 μCi [α-32P] dATP or 75 μM. After 10 min extension at 30°C, the reactions were stopped, deproteinized and loaded onto a 0.8% (w/v) agarose gel. The gel was either directly analysed for fluorescent DNA species or dried on DE81 paper and exposed to a Phosphorimager screen and imaged in Fuji FLA 9000 imager with the Multi Gauge software (Fuji).
Recombination assays
The determination of spontaneous CO and DSB-induced CO frequencies have been previously reported (Bartsch et al, 2000; Robert et al, 2006) and are described in Supplementary data.
UV survival curves
Haploid strains were grown for 3 days in YEPD to saturation. Cells were serially diluted and plated on YEPD. Plates with the lids removed were exposed to the UV light source for increasing times at 0.5 J/m2/s. To prevent photoreactivation, exposure to the UV light source was conducted in the dark and the plates were subsequently kept in the dark following irradiation. Plates were incubated at 30°C for 2 days prior to determining cell survival. The experiments were repeated three times and the results shown for each point are the average with the standard deviation.
Determination of spontaneous recombination rates in strains harbouring a plasmid
A colony from the strains to be tested was inoculated in synthetic medium lacking uracil and grown to saturation for 3 days at 30°C at 220 r.p.m. Next, the cultures were diluted to a concentration of 500 cells per ml and dispatched into twelve 2.5 ml cultures and grown to saturation for 5 days at 30°C at 220 r.p.m. The concentration of each individual culture was measured by plating out appropriate amounts of a dilution of the cultures onto synthetic medium lacking uracil. Appropriate volumes of each culture were plated onto synthetic medium lacking uracil and arginine. After 4 days at 30°C, the number of colonies were counted and subjected to a rate analysis (Spell and Jinks-Robertson, 2004).
Supplementary Material
Acknowledgments
This study was supported by Wellcome International Senior Research Fellowship WT076476, the Ministry of Education Youth and Sport of the Czech Republic grants (ME 10048), grants from Czech Science Foundation (GACR 13-26629S, GACR301/09/317, GACR 203/09/H046 and GACR P207/12/2323), European Regional Development Fund—Project FNUSA-ICRC (No CZ.1.05/1.1.00/02.0123), and FEBS Short Term Fellowship (PB), The Hungarian Science Foundation [OTKA 101225, GOP-1.1.1-11-2011-0026, GOP-1.1.1-11-2012-0030 (LH) and GOP-1.1.1.-09/1-2009-0021] (LH); IPA Cross-border Co-operation Programme [HUSRB/1002/214/126] (LH), ANR-07-BLAN-0350-01 (SG) and la Ligue Nationale Contre le Cancer (SG). We thank Stéphane Marcand, Jiri Bartek, Karine Dubrana and Julianne Smith for the critical reading of the manuscript.
Author contributions: LK, PB and SG designed the research; PB performed biochemical experiments shown in Figures 1, 2 and 3A and Supplementary Figures S1, S2C, D, S4C, S5 and S7A. MS purified proteins and performed the experiments shown in Supplementary Figures S2A, B, S3, S4A, B, D, E and S7B–D. AS performed experiments shown in Figure 3B and Supplementary Figures S4F and S6. SG performed experiments shown in Figures 4, 5 and 6. PB, VS, LP and PK provided SUMO-PCNA used in the study. NP, TR and VM contributed new reagents/analytical tool; LK, SG, LH, MS and PB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AA, Mohideen F, Lima CD (2012) Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch S, Kang LE, Symington LS (2000) RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol 20: 1194–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Pimpinelli S, Golic KG (1998) Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics 150: 173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L (2007) Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M (2006) Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Brocas C, Charbonnier J-B, Dhérin C, Gangloff S, Maloisel L (2010) Stable interactions between DNA polymerase δ catalytic and structural subunits are essential for efficient DNA repair. DNA Repair (Amst) 9: 1098–1111 [DOI] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R (2009) Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol 185: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JHJ, Foiani M, Liberi G (2005) Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol 25: 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, Krejci L, Sung P (2009) Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res 37: 6754–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD (2010) Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X (2008) The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol Cell 29: 243–254 [DOI] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE (2008) PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ayyagari R, Gomes X, Burgers PM (1997) Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol 17: 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M (1978) Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci USA 75: 4436–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Boulet A, Faye G (1991) Possible involvement of the yeast POLIII DNA polymerase in induced gene conversion. Mol Gen Genet 229: 353–356 [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer W-D, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Gangavarapu V, Prakash S, Prakash L (2007) Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol 27: 7758–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Lovett ST (2006) DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell 21: 595–604 [DOI] [PubMed] [Google Scholar]
- Gulbis J, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87: 297–306 [DOI] [PubMed] [Google Scholar]
- Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L (2004) Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol 24: 4267–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Prakash L, Prakash S (2006) Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA 103: 6477–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B (1976) A model for replication repair in mammalian cells. J Mol Biol 101: 417–425 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Inbar O, Kupiec M (1999) Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol 19: 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N (2009) Mammalian polymerase ζ is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst) 8: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, poleta. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Karras GI, Jentsch S (2010) The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Kelman Z (1997) PCNA: structure, functions and interactions. Oncogene 14: 629–640 [DOI] [PubMed] [Google Scholar]
- Kim SO, Yoon H, Park SO, Lee M, Shin J-S, Ryu K-S, Lee J-O, Seo Y-S, Jung HS, Choi B-S (2012) Srs2 possesses a non-canonical PIP box in front of its SBM for precise recognition of SUMOylated PCNA. J Mol Cell Biol 4: 258–261 [DOI] [PubMed] [Google Scholar]
- Kolesar P, Sarangi P, Altmannova V, Zhao X, Krejci L (2012) Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res 40: 7831–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X (2012) Homologous recombination and its regulation. Nucleic Acids Res 40: 5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Macris MA, Li Y, Komen SV, Villemain J, Ellenberger T, Klein HL, Sung P (2004) Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem 279: 23193–23199 [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein HL, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Krishna T, Kong X, Gary S, Burgers PM, Kuriyan J (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Le Breton C, Dupaigne P, Robert T, Le Cam E, Gangloff S, Fabre F, Veaute X (2008) Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res 36: 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León Ortiz AM, Reid RJD, Dittmar JC, Rothstein R, Nicolas A (2011) Srs2 overexpression reveals a helicase-independent role at replication forks that requires diverse cell functions. DNA Repair (Amst) 10: 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer W-D (2009) PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol Cell 36: 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Maloisel L, Fabre F, Gangloff S (2008) DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol Cell Biol 28: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V, Krejci L (2010) Srs2: the ‘Odd-Job Man’ in DNA repair. DNA Repair (Amst) 9: 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V, Krejci L (2012) Unwinding of synthetic replication and recombination substrates by Srs2. DNA Repair (Amst) 11: 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D (2011) Replication fork stalling by bulky DNA damage: localization at active origins and checkpoint modulation. Nucleic Acids Res 39: 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G-L, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD (2012) Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 45: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J, Lawrence CW, Hinkle DC (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Paek AL, Jones H, Kaochar S, Weinert T (2010) The role of replication bypass pathways in dicentric chromosome formation in budding yeast. Genetics 186: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Klein HL (1992) Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132: 23–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD (2008) SUMO modification of PCNA is controlled by DNA. EMBO J 27: 2422–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O, Zipin-Roitman A, Pfander B, Liefshitz B, Mazor Y, Ben-Aroya S, Jentsch S, Kupiec M (2010) Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J 29: 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Postow L, Woo E, Chait B, Funabiki H (2009) Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 284: 35951–35961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (2003) Control of cross-over by single-strand DNA resection. Trends Genet 19: 428–431 [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein HL, Haber JE, Sung P, Ira G (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Washington MT, Haracska L, Kondratick C, Prakash L (2000) Role of yeast and human DNA polymerase eta in error-free replication of damaged DNA. Cold Spring Harb Symp Quant Biol 65: 51–59 [DOI] [PubMed] [Google Scholar]
- Prelich G, Tan C, Kostura M, Mathews M, So A, Downey K, Stillman B (1987) Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature 326: 517–520 [DOI] [PubMed] [Google Scholar]
- Robert T, Dervins D, Fabre F, Gangloff S (2006) Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J 25: 2837–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Klein HL (1993) Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem 268: 1252–1259 [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein HL (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Prakash S, Prakash L (1990) The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124: 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Haracska L, Krejci L (2011) Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst) 10: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong C, Colavito S, Kwon Y, Sung P, Krejci L (2009) Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J Biol Chem 284: 24363–24371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13 Spec No 1: R57–R64 [DOI] [PubMed] [Google Scholar]
- Smirnova M, Klein HL (2003) Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat Res 532: 117–135 [DOI] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S (2004) Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol 262: 3–12 [DOI] [PubMed] [Google Scholar]
- Stark JM, Jasin M (2003) Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol Cell Biol 23: 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Tinker R, Williams K, Kassavetis G, Geiduschek E (1994) Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell 77: 225–237 [DOI] [PubMed] [Google Scholar]
- Torres-Ramos CA, Prakash S, Prakash L (2002) Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol 22: 2419–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Blastyák A, Haracska L (2010) Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 9: 257–267 [DOI] [PubMed] [Google Scholar]
- Van Komen S, Reddy MS, Krejci L, Klein HL, Sung P (2003) ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J Biol Chem 278: 44331–44337 [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Waga S, Bauer G, Stillman B (1994) Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem 269: 10923–10934 [PubMed] [Google Scholar]
- Windecker H, Ulrich HD (2008) Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J Mol Biol 376: 221–231 [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Seeler J-S, Thon G, Dejean A, Arcangioli B (2004) Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J 23: 3844–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Turner J, Kelman Z, Stukenberg P, Dean F, Shechter D, Pan Z, Hurwitz J, O’Donnell M (1996) Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1: 101–113 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lawrence CW (2005) The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA 102: 15954–15959 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.