Abstract
The application of mass spectrometry-based metabolomics in the field of drug metabolism has yielded important insights not only into the metabolic routes of drugs but has provided unbiased, global perspectives of the endogenous metabolome that can be useful for identifying biomarkers associated with mechanism of action, efficacy, and toxicity. In this report, a stable isotope- and mass spectrometry-based metabolomics approach that captures both drug metabolism and changes in the endogenous metabolome in a single experiment is described. Here the antioxidant drug tempol (4-hydroxy-2,2,6,6-tetramethylpiperidine-N-oxyl) was chosen because its mechanism of action is not completely understood and its metabolic fate has not been studied extensively. Furthermore, its small size (MW = 172.2) and chemical composition (C9H18NO2) makes it challenging to distinguish from endogenous metabolites. In this study, mice were dosed with tempol or deuterated tempol (C9D17HNO2) and their urine profiled using ultraperformance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Principal component analysis of the urinary metabolomics data generated a Y-shaped scatter plot containing drug metabolites (protonated and deuterated) that were clearly distinct from the endogenous metabolites. Ten tempol drug metabolites, including eight novel metabolites, were identified. Phase II metabolism was the major metabolic pathway of tempol in vivo, including glucuronidation and glucosidation. Urinary endogenous metabolites significantly elevated by tempol treatment included 2,8-dihydroxyquinoline (8.0-fold, P<0.05) and 2,8-dihydroxyquinoline-β-D-glucuronide (6.8-fold, P<0.05). Urinary endogenous metabolites significantly attenuated by tempol treatment including pantothenic acid (1.3-fold, P<0.05) and isobutrylcarnitine (5.3-fold, P<0.01). This study underscores the power of a stable isotope- and mass spectrometry-based metabolomics in expanding the view of drug pharmacology.
Keywords: Tempol, Stable Isotope, Metabolomics, Mass spectrometry, Drug Metabolism
INTRODUCTION
Metabolite identification in drug metabolism research typically includes in vitro incubation systems, in vivo animal studies, and human trials.1 In vitro assays using hepatocytes, liver microsomes, and recombinant human P450s have been widely used to investigate drug metabolism. Compared to an in vitro system, one major challenge of in vivo studies is the unequivocal identification of xenobiotic metabolites, especially for the identification of minor metabolites. More importantly, to elucidate the mechanism of drug action, in vivo investigations are essential to understand how endogenous metabolism is affected by xenobiotic metabolism. Mass spectrometry- and nuclear magnetic resonance (NMR)-based metabolomics are important tools to systematically identify xenobiotic and endogenous metabolites (i.e., biomarkers).2–4 Recent drug metabolism applications using mass spectrometry include studies of ifosfamide5, thioTEPA6, and procainamide metabolism.7 Studies of the endogenous metabolome with respect to understanding mechanisms of drug action include a urinary metabolomic study reporting a decrease in pantothenic acid and acylcarnitines as small molecule indicators of increased fatty acid β-oxidation due to the induction of genes involved in fatty acid transport and metabolism by the hypolipidemic drug fenofibrate.8 Additionally, serum metabolomics revealed that acetaminophen overdose is associated with an accumulation of long chain serum acylcarnitines, indicating that acetaminophen hepatotoxicity is associated with the inhibition of fatty acid β-oxidation.9 However, an ideal approach would be one that effectively captures both drug metabolism and changes in the endogenous metabolome in a single experiment.
The antioxidant drug tempol (4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl), a six-membered piperidine ring nitroxide, is a stable free radical stabilized by four methyl groups at the α-position.10 Tempol mainly exists in three forms; the nitroxide radical, the reduced hydroxylamine, and the oxidized oxoammonium cation.11 Different forms of tempol can be generated via one or two electron transfer reactions. During the transformation of the nitroxide radical to the oxoammonium cation, tempol is capable of dismuting two superoxide anion (O2•−) molecules by a direct reaction with O2•− or •OOH.12 Previous studies have demonstrated that tempol can ameliorate the effects of high-fat-diet induced-obesity in mice.13, 14 Tempol treatment can reduce plasma glucose levels, low-density lipoprotein cholesterol and triglyceride, and body weight.13–15 Additionally, some studies also reported that tempol can act as a chemopreventive agent in ataxia-telangiectasia mutated (ATM) deficient mice16 and reduce the age-related spontaneous tumor incidence in wild-type C3H mice.15 To date, only two metabolites, the hydroxylamine and the amine of tempol, were identified using gas chromatography coupled with mass spectrometry (GC-MS) and electron spin resonance.17, 18
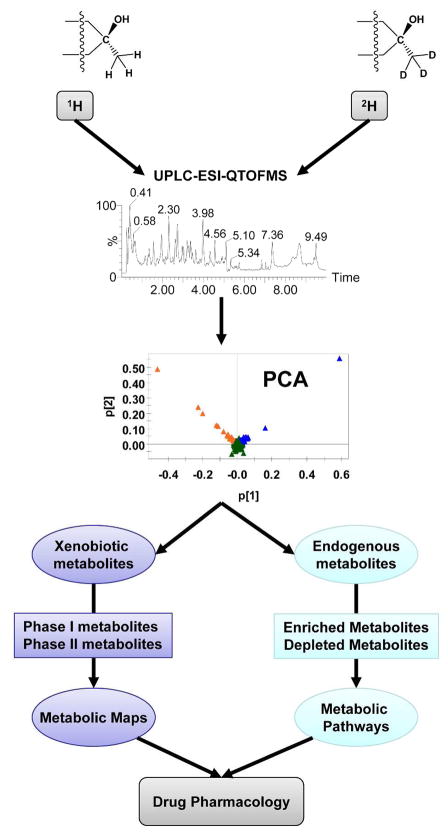
Mass spectrometry and stable isotope-labeled drugs has been applied to metabolite identification in in vivo and in vitro metabolism studies.19–21 However, not to be overlooked, the use of conventional approaches including radiolabeled tracers22 as well as making use of naturally occurring isotopes, such as 37Cl, have been instrumental in helping to detect and identify drug metabolites.5, 6 In this report, we hypothesized that stable isotope- and mass spectrometry-based metabolomics may simultaneously facilitate a better understanding of both the drug metabolism route as well as to identify endogenous biomarkers of drug action (Figure 1). To accomplish this, ultraperformance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOFMS)-based metabolomics combined with multivariate data analysis (MDA) was employed to analyze the metabolites generated from tempol and deuterated tempol and the impact of tempol on endogenous metabolism. Ten xenobiotic metabolites and four endogenous metabolites were determined in this study. Phase II metabolism was the major metabolic pathway of tempol in vivo, including glucuronidation and glucosidation. Fatty acid β-oxidation regulated by tempol may be involved in the anti-obesity and cancer chemoprevention effects of this compound. Overall, this study exemplifies the value of stable isotope-labeled drugs and mass spectrometry-based metabolomics for the simultaneous evaluation of drug metabolism and its effects on host metabolism.
Figure 1. Model of stable isotope and mass spectrometry-based metabolomics applied to drug metabolism.
The urinary metabolites from drug (1H) and deuterated drug (2H) treatment are analyzed using UPLC-ESI-QTOFMS. All xenobiotic and endogenous metabolites are distributed in the PCA model. The metabolic maps of the drug will be determined from its phase I and II metabolites, while the metabolic pathways regulated by the drug might be identified via the enriched and depleted endogenous metabolites. Drug pharmacology can be predicted based on these metabolic maps and pathways.
EXPERIMENTAL SECTION
Chemicals and reagents
Tempol, deuterated tempol, chlorpropamide, creatinine, pantothenic acid, and 2,8-dihydroxyquinoline were purchased from Sigma (St. Louis, MO). Isobutrylcarnitine was obtained from the Metabolic Laboratory, Vrije Universiteit Medical Center (Amsterdam, The Netherlands). All solvents and organic reagents were of the highest grade available.
Animal study
Male 6- to 8-week-old C57BL/6N mice were used for all the metabolomic studies described below. A urine sample was collected from all mice prior to saline (control), tempol, and D-tempol treatment to acclimate mice to the metabolic cages. Tempol (dissolved in 0.9% normal saline) was orally administered by gavage to mice at a dose of 50 or 250 mg/kg for 5 days. D-tempol, dissolved in 0.9% normal saline, was given by gavage at a dose of 50 mg/kg. Control mice were treated with normal saline alone. A 24 hr urine (room temperature) was collected individually in metabolic cages (Jencons, Leighton Buzzard, UK) at day 1, 3 and 5 following treatment. All the samples were stored at −80°C until analysis.
UPLC-ESI-QTOFMS analysis
Urine samples were prepared by adding 20 μL of urine to 180 μL 50% aqueous acetonitrile (50:50 water:acetonitrile). Samples were vortexed for 5 min and centrifuged at 18,000 x g for 20 min at 4 °C to remove particulates and precipitate protein. The supernatant was transferred to an autosampler vial for analysis. A 5 μL aliquot of the supernatant was chromatographed via ultra-performance liquid chromatography (Waters Corp, Milford, MA) using a 2.1×50mm Waters BEH C18 1.7 μm column and introduced via electrospray into a quadrupole time-of-flight mass spectrometer (UPLC-ESI-QTOFMS). The gradient mobile phase consisted of 0.1% formic acid solution (A) and acetonitrile containing 0.1% formic acid solution (B). The gradient was maintained at 100% A for 0.5 min, increased to 100% B over the next 7.5 min and returning to 100% A in the last 2 min. Data were collected in positive mode and negative mode, which was operated in full-scan mode from 100 to 1000 m/z. Nitrogen was used as both cone gas (50 liters/h) and desolvation gas (600 liters/h). Source temperature and desolvation temperature were set at 120 °C and 350 °C, respectively. The capillary voltage and cone voltage was 3000 and 20 V, respectively. Chlorpropamide (5 μM) was added in the sample as the internal standard.
Data processing and multivariate data analysis
Raw data were aligned using MarkerLynx software (Waters) to generate a data matrix consisting of peak areas corresponding to a unique m/z and retention time without normalization. The generated multivariate data matrix was exported into SIMCA-P+12.0 (Umetrics, Kinnelon, NJ) for principal component analysis (PCA). Tempol metabolites and endogenous metabolites in urine were initially determined by analyzing the ions in the loadings scatter plot.
Tempol metabolite identification and validation
After the putative tempol metabolites were determined by PCA analysis, tandem MS of selected metabolites was obtained by ramping collision energies from 15 to 40 V. The chemical structures of tempol metabolites were elucidated by interpretation of their fragmentation patterns but not by comparison with synthesized standards as indicated below with endogenous metabolites. It should be noted that unequivocal identification of drug metabolites would require comparison with synthesized drug metabolites and their conjugates and that was not done here. However, to provide further validation of tempol metabolites, corresponding metabolites containing D-tempol were searched for in the D-tempol treated mice. A drug metabolite was considered confirmed if the corresponding D-tempol metabolite (+17 m/z) was identified in the D-tempol portion of the loadings plot and had a similar retention time. The glucuronide conjugates were confirmed by hydrochloric acid hydrolysis as described.23 Briefly, a 50 μl urine sample from tempol treatment was incubated with 3M HCl in a final volume of 200 μl for 1 hr at 100°C and stopped by adding 1 M NaHCO3 pH 8. The reaction was centrifuged at 14,000 rpm for 15 min and the supernatant was dried down and reconstituted in 200 μl of water for tandem MS analysis. The tempol glucuronidation assay was performed as previously described.24
Endogenous metabolite identification and concentration measurements
To identify unknown metabolites, the METLIN database was searched. Seven Golden Rules25 were used to calculate the mass error based on the elemental compositions of each metabolite and to help reduce or eliminate unlikely candidates. To confirm the metabolite identification, the MS/MS fragmentation pattern and retention time of the putative endogenous metabolite was compared with that of an authentic compound.
Quantitation of the endogenous urinary metabolites was performed using the Xevo triple quadrupole tandem mass spectrometer (Waters Corp.) by multiple reaction monitoring (MRM) according to previous reports. 26, 27 Urine samples were diluted 1:20 in water for pantothenic acid measurements, or 1:2 for isobutrylcarnitine, 2,8-dihydroxyquinoline, and 2,8-dihydroxyquinoline-β-D-glucuronide. Five μl of diluted urine and standards ranging from 0 to 25 μM (dissolved in water) were chromatographed on an Acquity 1.7 μm C18 column. Standard curves with correlation coefficients greater than 0.99 were used to quantify the urine samples. The following MRM transitions were monitored: pantothenic acid (220 →116; ESI+), 2,8-dihydroxyquinoline (162 →116; ESI+), 2,8-dihydroxyquinoline-β-D-glucuronide (338 →162; ESI+), isobutrylcarnitine (232 →85; ESI+), creatinine (114 →86; ESI+), and chlorpropamide (277 →111; ESI+). Targetlynx software (Waters) was used to quantify the urine metabolites based on peak area. Chlorpropamide (0.5 uM) was used as the internal standard, and absolute peak areas (metabolite/chlropropamide) were used to quantitation. Results were corrected for the dilution, normalized by the concentration of urinary creatinine, and expressed as μmol/mmol creatinine.
Data analysis
Experimental values are presented as mean ± SD. Statistical analysis was processed using two-tailed Student’s t-test. P-values of less than 0.05 were considered significant.
RESULTS AND DISCUSSION
PCA modeling of tempol treatment, D-tempol treatment, and control groups
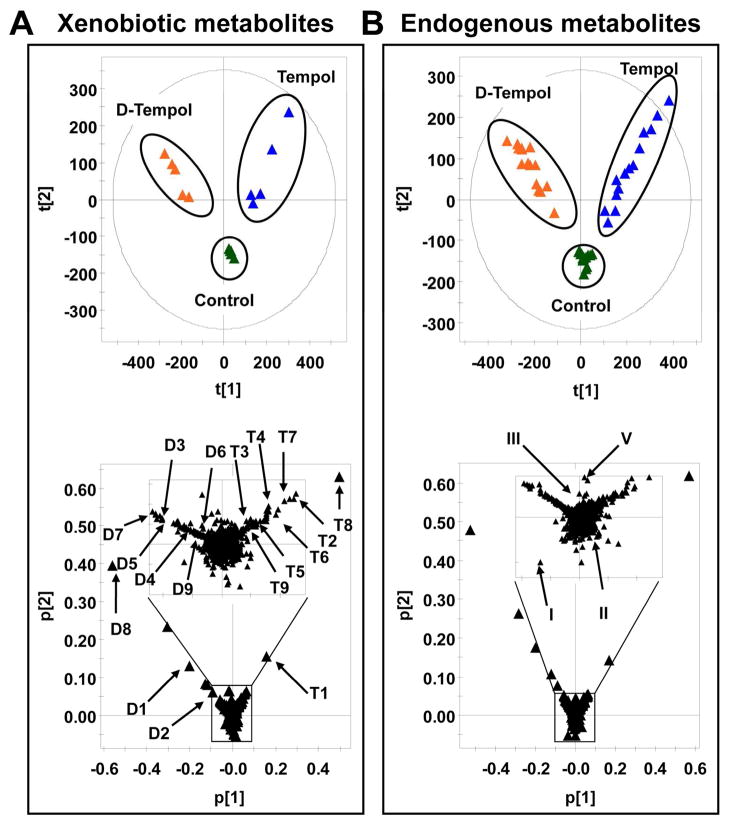
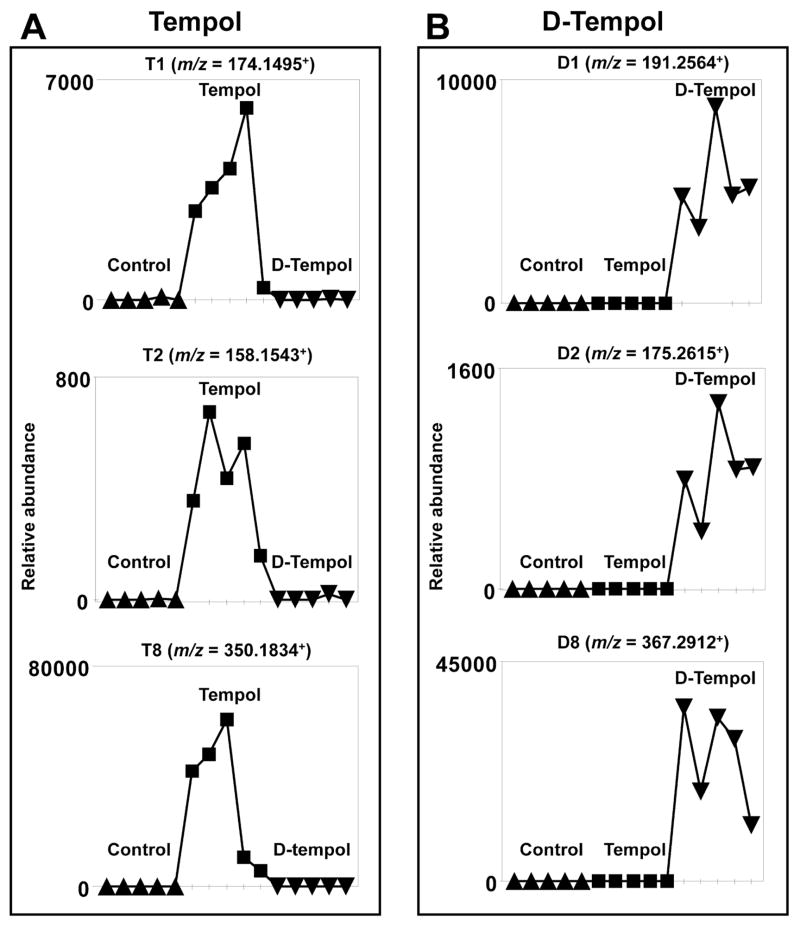
UPLC-ESI-QTOFMS analysis of urine coupled with PCA modeling was used to profile the urinary metabolome from control, tempol treated, and D-tempol treated mice. All the samples were injected into the UPLC-ESI-QTOFMS at random, and quality control samples including blanks, pooled samples, and a cocktail of standards, showed good separation and reproducibility in the PCA model (Figure S1) suggesting that UPLC-ESI-QTOFMS was accurate, precise, and reproducible during the analysis. Tempol treatment (50 mg/kg), D-tempol treatment (50 mg/kg), and control mouse urines from day 1 were separated in component 1, showing a “Y” shape in the scores scatter plot (Figure 2A, top). Tempol metabolites (T1 to T9) were distributed in the first quadrant (upper right), whereas D-tempol metabolites (D1 to D9) were distributed in the second quadrant (upper left) (Figure 2A, bottom). As shown in the Figure 3A and 3B, the trend plots of these ions are useful for determining which ions are tempol metabolites (T1, T2, and T8) and D-tempol metabolites (D1, D2, and D8) as they are expected to be absent in the control urines. Despite the isotope effect of deuterium labeling, no significant difference in the metabolism of tempol and D-tempol was observed (Figure S7B–C).28 The endogenous metabolites were clustered in the center of the scores scatter plot and distinct from the cloud of ions associated with the drug metabolites. This effect is essential to clearly identify altered endogenous metabolites following tempol treatment.
Figure 2. Metabolomic analysis of xenobiotic and endogenous metabolites in mouse urine.
(A) Scores plot of a PCA model and PCA loadings scatter plot of urinary ions from tempol treatment (50 mg/kg), D-tempol treatment (50 mg/kg), and control group at day 1. Each point represents an individual mouse urine sample (top) and a urinary ion (bottom). Tempol and D-tempol metabolites are labeled in the loadings scatter plot (T1 to T10 and D1 to D10). (B) Scores plot of a PCA model and PCA loadings scatter plot of urinary ions in tempol treatment (50 mg/kg), D-tempol treatment (50 mg/kg), and control group from day 1 to 5. Each point represents an individual mouse urine sample (top) and a urinary ion (bottom). The regulated-endogenous metabolites are labeled in the loadings scatter plot (I to IV). The t[1] and t[2] correspond to principal components 1 and 2, respectively. The p[2] values represent the interclass difference and p[1] values represent the relative abundance of the ions. All the data were obtained in positive mode (ESI+).
Figure 3. Trend plots of ions in control, tempol and D-tempol groups.
(A) Tempol metabolites with m/z values of 174.1495+ (T1), 158.1543+ (T2), and 350.1834+ (T8). (B) D-tempol metabolites with m/z values of 194.2564+ (D1), 175.2615+ (D2), and 367.2912+ (D8). Note the presence of suspected tempol and D-tempol metabolites in the tempol treatment and D-tempol group (n = 5), respectively. Metabolite codes correspond to those in Table 1.
To determine the potential endogenous metabolites associated with tempol treatment, all the mouse urines from the three groups at days 1, 3, and 5 were analyzed together (Figure 2B, top). In this “Y” shape loading scatter plot (Figure 2B, bottom), the ions that deviated from the cloud of ions in the center are those considered putative endogenous biomarkers of tempol treatment. Two ions (I and II) in the top of ion cloud were depleted following tempol treatment, whereas two ions (III and IV) in the bottom of the ion cloud were enriched (Figure 2B, bottom).
Structural identification of tempol metabolites in urine
D-tempol, obtained from Sigma, contains 12 2H in the methyl groups and 5 2H in the six-membered piperidine ring. The mass differences between the metabolites from tempol and D-tempol would therefore be 17 mass units. The representative interpretation of the MS/MS spectrum of the metabolites of tempol and D-tempol are presented in Figure S2. Thus, according to this property, tempol metabolites and their counterpart metabolites of D-tempol could be found in the “Y” shape loadings scatter plot providing unequivocal evidence that these metabolites indeed were generated from tempol. Along with the mass differences between the drug metabolites, these metabolites also shared similar retention times. The chemical compositions and retention times of tempol and D-tempol metabolites are listed in the Table 1. Except for the hydroxylamine metabolite (T1) and dehydroxylation metabolites (T2), other metabolites, T3 to T10, are considered new metabolites. MS/MS spectra and the structural interpretation of those metabolites are presented in Figures S3 and S4.
Table 1.
Identified tempol and endogenous metabolites in mouse urine.
| Symbol | RT (min) | [M+H]+ | Formula | Mass Error (ppm) | Identity |
|---|---|---|---|---|---|
| Tempol Metabolites | |||||
| T1 | 1.58 | 174.1495 | C9H19NO2 [H+] | 0.6 | Tempol hydroxylamine |
| T2 | 0.45 | 158.1543 | C9H19NO [H+] | −1.3 | 1-Dehydroxytempol (Amine) |
| T3 | 2.21 | 156.1329 | C9H17NO[H+] | −5.8 | 4-Dehydroxytempol |
| T4 | 1.21 | 190.1442 | C9H19NO3 [H+] | −0.5 | 3-Hydroxytempol |
| T5 | 1.84 | 332.1704 | C15H25NO7 [H+] | −1.5 | 4-Dehydroxytempol 1-O-glucuronide |
| T6 | 0.35 | 334.1865 | C15H27NO7 [H+] | −0.2 | 1-Dehydroxytempol 4-O-glucuronide |
| T7 | 1.48 | 336.2018 | C15H29NO7 [H+] | −1.2 | Tempol 4-O-glucoside |
| T8 | 1.31 | 350.1834 | C15H27NO8 [H+] | 5.4 | Tempol 4-O-glucuronide |
| T9 | 1.69 | 512.2334 | C21H37NO13 [H+] | −1.8 | Tempol 1-O-glucoside and 4-O-glucuronide |
| T10 | 1.41 | 512.2349 | C21H37NO13 [H+] | 1.2 | Tempol 1-O-glucuronide and 4-O-glucoside |
| D-Tempol Metabolites | |||||
| D1 | 1.54 | 191.2564 | C9H2D17NO2 [H+] | 1.5 | D-tempol hydroxylamine |
| D2 | 0.44 | 175.2615 | C9H2D17NO [H+] | 1.7 | 1-Dehydroxy-D-tempol |
| D3 | 2.80 | 172.2374 | C9H1D16NO [H+] | −11.0 | 4-Dehydroxy-D-tempol |
| D4 | 1.04 | 206.2447 | C9H3D16NO3 [H+] | 0.0 | 3-Hydroxy-D-tempol |
| D5 | 1.81 | 348.2699 | C15H9D16NO7 [H+] | −4.3 | 4-Dehydroxy-D-tempol 1-O-glucuronide |
| D6 | 0.35 | 351.2920 | C15H10D17NO7 [H+] | −3.7 | 1-Dehydroxy-D-tempol 4-O-glucuronide |
| D7 | 1.45 | 353.3111 | C15H12D17NO7 [H+] | 6.2 | D-tempol 4-O-glucoside |
| D8 | 1.21 | 367.2912 | C15H10D17NO8 [H+] | 8.1 | D-tempol 4-O-glucuronide |
| D9 | 1.64 | 529.3385 | C21H20D17NO13 [H+] | −4.7 | D-tempol 1-O-glucoside and 4-O-glucuronide |
| D10 | 1.36 | 529.3394 | C21H20D17NO13 [H+] | −3.0 | D-tempol 1-O-glucuronide and 4-O-glucoside |
| Endogenous Metabolites | |||||
| I | 1.40 | 220.1183 | C9H17NO5 [H+] | −0.9 | Pantothenic acid |
| II | 1.58 | 232.1547 | C11H21NO4 [H+] | −0.8 | Isobutyrylcarnitine |
| III | 2.73 | 162.0539 | C9H7NO2 [H+] | −9.8 | 2,8-Dihydroxyquinoline |
| IV | 2.49 | 338.0874 | C15H15NO8 [H+] | 0.6 | 2,8-Dihydroxyquinoline-β-D-glucuronide |
Identification and quantitation of endogenous metabolites in mouse urine
The chemical compositions and identities of the altered endogenous ions were determined by Seven Golden Rules25 and by searching the METLIN database.29 Two top increased ions, 162.0539+ and 338.0874+, were found in the top of the central portion of the loadings plot. Chemical formula calculations on those ions showed that the ions corresponded to 2,8-dihydroxyquinoline (C9H7NO2, III) and its glucuronide (C15H15NO8, IV). Further analysis indicated that the ions 162.0539+ (RT = 2.73) and 338.0874+ (RT = 2.49) were indeed 2,8-dihydroxyquinoline and 2,8-dihydroxyquinoline-β-D-glucuronide in comparison with authentic standards. Analysis of the top-decreased ions identified 220.1183+ as pantothenic acid (C9H17NO5, I) and 232.1547+ as isobutyrlcarnitine (C11H21NO4, II) by comparison with authentic standards. The endogenous metabolites identified are listed Table 1. The MS/MS fragments of endogenous metabolites pantothenic acid (I), isobutyrlcarnitine (II), 2,8-dihydroxyquinoline (III), and 2,8-dihydroxyquinoline-β-D-glucuronide (VI) are listed in Figure S5.
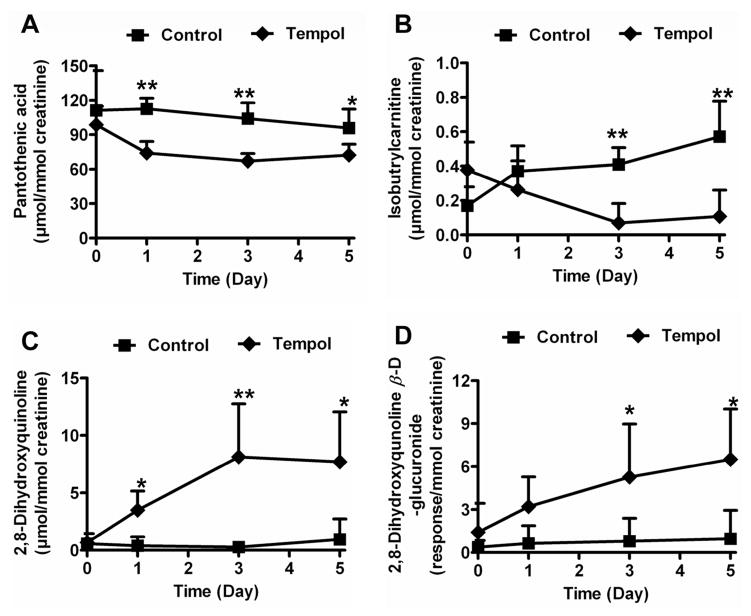
For the endogenous metabolites identified in Table 1, concentration measurements were estimated against calibration curves. A previous study demonstrated large variation in metabolite concentration in both human and animal urine indicating that creatinine might offer the best internal standard for correcting urine volume effects.30 Here, however, the concentration of creatinine was not significantly different between tempol treatment and control group (data no shown). However, all the endogenous metabolites were normalized by creatinine to account for any subtle differences in kidney function. Pantothenic acid, isobutrylcarnitine, 2,8-dihydroxyquinoline and its glucuronide concentrations were measured by triple quadrupole operating in MRM mode. Although the trend plots showed a decrease in pantothenic acid following tempol treatment from day 1 to day 5 (Figure S6A), there were no significant differences for the concentration of pantothenic acid in control and low dose tempol group (50 mg/kg) from day 1 to 5 (Figure S6B). This result exemplifies the importance of accurate quantitation following global profiling experiments.31 However, LC-MS/MS quantitation demonstrated that the excretion of pantothenic acid was significantly decreased from day 1 to day 5 after high dose tempol treatment (250 mg/kg) (Figure 4A). The excretion of isobutrylcarnitine was decreased at day 3 (5.9-fold, P<0.01) and 5 (5.3-fold, P<0.01) following the low dose tempol treatment (Figure 4B). In contrast, 2,8-dihydroxyquinoline and its glucuronide were significantly increased at day 3 and 5 following low dose tempol treatment (Figures 4C–D). The concentration of 2,8-dihydroxyquinoline was increased 8.0-fold (P<0.05) in urine at day 5 of tempol treatment, while 2,8-dihydroxyquinoline-β-D-glucuronide was increased 6.8-fold (P<0.05).
Figure 4. Quantitation of urinary pantothenic acid, isobutyrylcarntine, 2,8-dihydroxyquinoline and 2,8-dihydroxyquinoline-β-D-glucuronide.
Results were normalized to creatinine concentration. (A) Pantothenic acid in high dose tempol (250 mg/kg) and control mice from day 0 to 5. (B) Isobutyrylcarnitine in low dose tempol treatment (50 mg/kg) and control mice from day 0 to 5. (C) 2,8-Dihydroxyquinoline in low dose tempol treatment and control mice from day 0 to 5. (D) 2,8-Dihydroxyquinoline-β-D-glucuronide in low dose tempol treatment and control mice from day 0 to 5. P-values were calculated using an unpaired t-test. *P < 0.05, **P < 0.01.
The effect of tempol has been widely evaluated in various in vivo and in vitro models, 32 however, its metabolic pathway remains unclear. Previous studies reported that two metabolites, the hydroxylamine and the amine of tempol, were detected in rat liver microsomes 17 or human keratinocyte cells.18 In the present study, tempol metabolism was investigated using UPLC-ESI-QTOFMS-based metabolomics. Different from previous metabolomic studies on drug metabolism,5, 6 deuterated tempol was used to distinguish tempol metabolites from endogenous metabolites. Especially for mass spectrometry-based applications, it should be pointed out that this approach is expected to work for other isotopes including 15N. A “Y” shape of the PCA model was generated from those groups, and tempol metabolites, D-tempol metabolites, and endogenous metabolites were specifically distributed in different areas of “Y” shape of loading scatter plot (Figure 2B, bottom). A total of 10 tempol metabolites, including 8 novel metabolites, were identified by this metabolomic approach (Table 1). As shown in Figure S7B and 7C, more than 95% of total tempol and D-tempol is metabolized to its glucuronide and glucoside metabolites, and there were no significant differences for the generation of phase I and II metabolites between tempol and D-tempol groups. Glucuronide metabolites were the major metabolite of tempol, which is common in animal drug metabolism.33 Interestingly, several glucoside conjugates were found in vivo including a mixed monoglucoside monoglucuronide conjugate. Similar mixed monoglucosides monoglucuronides have been reported for bilirubin and epigallocatechin gallate.34, 35
In addition to xenobiotic metabolites, four endogenous metabolites affected by tempol treatment were identified in the “Y” loading scatter plot (Figure 2B). The excretion of pantothenic acid and isobutrylcarnitine in urine was significantly decreased by tempol treatment. In the cytosol, pantothenic acid can be metabolized to generate coenzyme A (CoA) that is an essential cofactor for numerous enzymatic reactions, especially for fatty acid β-oxidation in the mitochondria and peroxisomes. Acylcarnitines are generated to transport fatty acid across the mitochondrial membranes followed by generation of the CoA intermediate. A previous study reported that the depletion of pantothenic acid and isobutrylcarnitine reflected an increase in β-oxidation during fenofibrate treatment, which can reduce lipid levels via activation of peroxisome proliferator-activated receptor α (PPARα).8 Additionally, the excretion of 2,8-dihydroxyqunoline and its glucuronide was significantly increased in urine by tempol treatment. 2,8-Dihydroxyqunoline glucuronide was reported to show higher levels in mouse urine during Wy-14,643 treatment which also can increase peroxisomal fatty acid β-oxidation through activation of PPARα.36 A recent study indicated that the excretion of dihydroxyquinoline was increased in rat urine after treated with medium-fat and high-fat diet.37 These results provide compelling evidence that fatty acid β-oxidation might be induced following tempol treatment. The formation of dihydroxyquinoline might be related to gut microbiota metabolism. Indeed, it was reported that dihydroxyquinoline can be produced from quinoline under the effect of gut bacteria of the Pseudomonas species.38, 39 Another study showed that dihydroxyquinoline glucuronide can be detected only in conventional mouse serum, not in germ-free mice.40 The mechanism regarding how tempol treatment can cause an increase in 2,8-dihydroxyqunoline and its glucuronide warrants further investigation.
The change of four endogenous metabolites associated with fatty acid oxidation suggests that tempol might affect fatty acid β-oxidation. Generally, fatty acid β-oxidation occurs in mitochondria and peroxisomes while ω-oxidation is prevalent in microsomes. Both peroxisomal β-oxidation and microsomal ω-oxidation lead to a reduction of molecular oxygen (O2) to hydrogen peroxide (H2O2).41 H2O2 is one of the reactive oxygen species (ROS) produced during cellular metabolism. Under oxidative stress, H2O2 is a contributor to oxidative damage. Tempol, a representative nitroxide, possesses superoxide dismutase- and catalase-mimetic activity that protects cells and animals against a variety of oxidative insults.32 Previous studies have demonstrated that tempol is effective for metabolizing cellular O2 and H2O2, and protects cells from damaging effects of the hydroxyl radical (•OH).11, 42 Therefore, it is reasonable to find changes in endogenous metabolites associated with fatty acid oxidation during tempol treatment. Various studies have demonstrated that the disruption of fatty acid oxidation was detected in obesity and cancer models.43–46 These findings may point to a potential mechanism for the effect of tempol on obesity and cancer.
Several studies have validated that the stable isotope-based metabolomics approach can facilitate the identification of xenobiotic metabolites during drug metabolism. One example is that deuterated acetaminophen (APAP), [acetyl-2H3] APAP, was used to determine APAP toxicity metabolites.19 Three novel APAP metabolites, S-(5-acetylamino-2-hydroxyphenyl) mercaptopyruvic acid, 3,3-biacetaminophen, and benzothiazine compound, were revealed to be related to APAP-induced toxicity based on mass isotopomer analysis. In another study, orthogonal partial least squares (OPLS) analysis of urinary ions from ethanol (C2H6O) and deuterated ethanol (C2D6O) treatment revealed that N-acetyltaurine was a novel metabolite of ethanol that can function as a biomarker of hyperacetatemia.47 Xenobiotic metabolism also can dramatically influence the level of endogenous metabolites that might be candidates for a biomarker that reflects the pharmacological effects or toxicity of xenobiotics. Since the endogenous metabolites affected by unlabeled or labeled xenobiotics are very similar, the ions contributing to the separation of unlabeled group, labeled group, and untreated group in the scores plot (“Y” shape) are due to xenobiotic and endogenous metabolites. More importantly, xenobiotic metabolites and endogenous metabolites are distributed in different quadrants of the scores plot where endogenous metabolites are clearly distinct from xenobiotic metabolites (labeled and unlabeled). It is particularly important to determine the enriched endogenous metabolites affected by the xenobiotic, which usually are overlapped by xenobiotic metabolites. Taken together, stable isotope and mass spectrometry-based metabolomics is a useful tool to identify xenobiotic and endogenous metabolites. The metabolic maps of drug and metabolic pathways regulated by drug treatment can be used to predict drug pharmacology in vivo, including pharmaceutical effects and toxicity.
In summary, stable isotope- and mass spectrometry-based metabolomics was found to be effective for the identification of xenobiotic and endogenous metabolites during drug treatment or xenobiotic exposure. Drug metabolite profiling of tempol-treated mice in this study indicated that glucuronidation and glucosidation were the major metabolic pathways of tempol in vivo. A comprehensive in vivo metabolic map of tempol was generated through use of metabolomics (Figure 5). The change in endogenous metabolites indicated that tempol has pharmacological activities in vivo via its effect on fatty acid oxidation that may be involved in the anti-obesity and cancer chemoprevention effects of this compound.
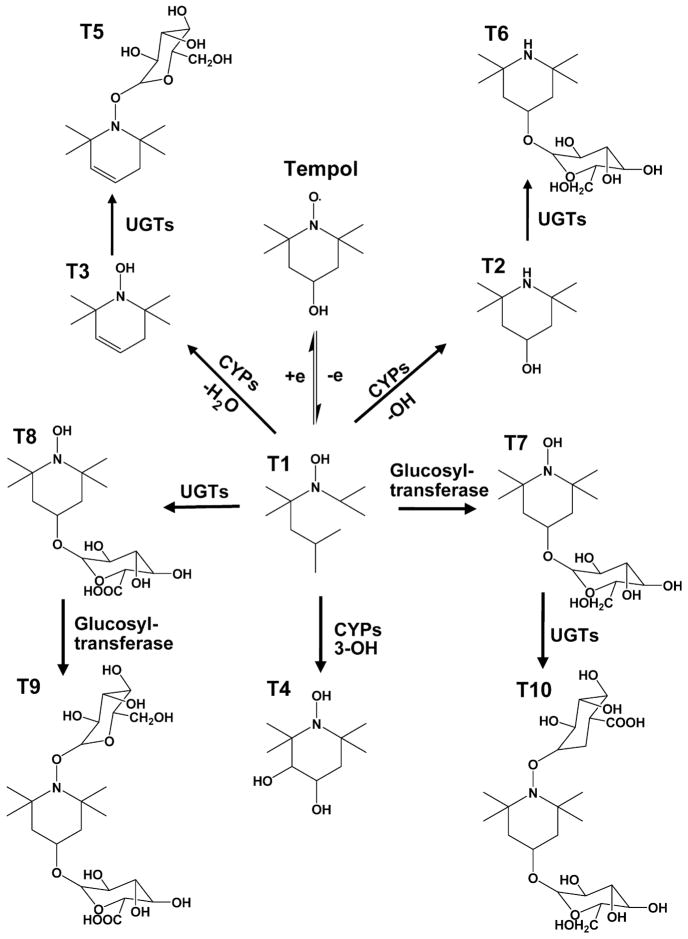
Figure 5. Major metabolic pathways of tempol in vivo.
Tempol and hydroxylamine (T1) can be transformed from each other. Under the effect of cytochrome P450 enzymes, hydroxylamine (T1) first is converted to T2 and T3 through the elimination of hydroxyl group and water, respectively. T1 also can be metabolized to T4 via the hydroxylation. Under the effect of UDP-glucuronosyltransferases (UGTs), T2 and T3 can be conjugated with glucuronic acid to generate their glucuronides, T5 and T6, respectively. In addition, T1 can be directly transformed into its glucoside (T7) and glucuronide (T8). Also, T7 and T8 can further be converted to T10 and T9 by conjugation with glucuronic acid and glucose, respectively.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (F.J.G. and J.B.M.) and ES022186 (A.D.P.).
Footnotes
Supplemental metabolites identification and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ekins S, Andreyev S, Ryabov A, Kirillov E, Rakhmatulin EA, Bugrim A, Nikolskaya T. Computational prediction of human drug metabolism. Expert Opin Drug Metab Toxicol. 2005;1(2):303–24. doi: 10.1517/17425255.1.2.303. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39(2–3):581–97. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patterson AD, Lanz C, Gonzalez FJ, Idle JR. The role of mass spectrometry-based metabolomics in medical countermeasures against radiation. Mass Spectrom Rev. 2010;29(3):503–21. doi: 10.1002/mas.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crockford DJ, Maher AD, Ahmadi KR, Barrett A, Plumb RS, Wilson ID, Nicholson JK. 1H NMR and UPLC-MS(E) statistical heterospectroscopy: characterization of drug metabolites (xenometabolome) in epidemiological studies. Anal Chem. 2008;80(18):6835–44. doi: 10.1021/ac801075m. [DOI] [PubMed] [Google Scholar]
- 5.Li F, Patterson AD, Hofer CC, Krausz KW, Gonzalez FJ, Idle JR. Comparative metabolism of cyclophosphamide and ifosfamide in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem Pharmacol. 2010;80(7):1063–74. doi: 10.1016/j.bcp.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Patterson AD, Hofer CC, Krausz KW, Gonzalez FJ, Idle JR. A comprehensive understanding of thioTEPA metabolism in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem Pharmacol. 2011;81(8):1043–53. doi: 10.1016/j.bcp.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Patterson AD, Krausz KW, Dick B, Frey FJ, Gonzalez FJ, Idle JR. Metabolomics reveals the metabolic map of procainamide in humans and mice. Biochem Pharmacol. 2012;83(10):1435–44. doi: 10.1016/j.bcp.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson AD, Slanar O, Krausz KW, Li F, Hofer CC, Perlik F, Gonzalez FJ, Idle JR. Human urinary metabolomic profile of PPARalpha induced fatty acid beta-oxidation. J Proteome Res. 2009;8(9):4293–300. doi: 10.1021/pr9004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Krausz KW, Shah YM, Idle JR, Gonzalez FJ. Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chem Res Toxicol. 2009;22(4):699–707. doi: 10.1021/tx800464q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishna MC, DeGraff W, Hankovszky OH, Sar CP, Kalai T, Jeko J, Russo A, Mitchell JB, Hideg K. Studies of structure-activity relationship of nitroxide free radicals and their precursors as modifiers against oxidative damage. J Med Chem. 1998;41(18):3477–92. doi: 10.1021/jm9802160. [DOI] [PubMed] [Google Scholar]
- 11.Soule BP, Hyodo F, Matsumoto K, Simone NL, Cook JA, Krishna MC, Mitchell JB. The chemistry and biology of nitroxide compounds. Free Radic Biol Med. 2007;42(11):1632–50. doi: 10.1016/j.freeradbiomed.2007.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishna MC, Grahame DA, Samuni A, Mitchell JB, Russo A. Oxoammonium cation intermediate in the nitroxide-catalyzed dismutation of superoxide. Proc Natl Acad Sci U S A. 1992;89(12):5537–41. doi: 10.1073/pnas.89.12.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebenezer PJ, Mariappan N, Elks CM, Haque M, Francis J. Diet-induced renal changes in Zucker rats are ameliorated by the superoxide dismutase mimetic TEMPOL. Obesity (Silver Spring) 2009;17(11):1994–2002. doi: 10.1038/oby.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.San Martin A, Du P, Dikalova A, Lassegue B, Aleman M, Gongora MC, Brown K, Joseph G, Harrison DG, Taylor WR, Jo H, Griendling KK. Reactive oxygen species-selective regulation of aortic inflammatory gene expression in Type 2 diabetes. Am J Physiol Heart Circ Physiol. 2007;292(5):H2073–82. doi: 10.1152/ajpheart.00943.2006. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell JB, Xavier S, DeLuca AM, Sowers AL, Cook JA, Krishna MC, Hahn SM, Russo A. A low molecular weight antioxidant decreases weight and lowers tumor incidence. Free Radic Biol Med. 2003;34(1):93–102. doi: 10.1016/s0891-5849(02)01193-0. [DOI] [PubMed] [Google Scholar]
- 16.Schubert R, Erker L, Barlow C, Yakushiji H, Larson D, Russo A, Mitchell JB, Wynshaw-Boris A. Cancer chemoprevention by the antioxidant tempol in Atm-deficient mice. Hum Mol Genet. 2004;13(16):1793–802. doi: 10.1093/hmg/ddh189. [DOI] [PubMed] [Google Scholar]
- 17.Iannone A, Bini A, Swartz HM, Tomasi A, Vannini V. Metabolism in rat liver microsomes of the nitroxide spin probe tempol. Biochem Pharmacol. 1989;38(16):2581–6. doi: 10.1016/0006-2952(89)90541-8. [DOI] [PubMed] [Google Scholar]
- 18.Kroll C, Langner A, Borchert HH. Nitroxide metabolism in the human keratinocyte cell line HaCaT. Free Radic Biol Med. 1999;26(7–8):850–7. doi: 10.1016/s0891-5849(98)00268-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem. 2008;283(8):4543–59. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wind M, Gebhardt K, Grunwald H, Spickermann J, Donzelli M, Kellenberger L, Muller M, Fullhardt P, Schmitt-Hoffmann A, Schleimer M. Elucidation of the in vitro metabolic profile of stable isotope labeled BAL19403 by accurate mass capillary liquid chromatography/quadrupole time-of-flight mass spectrometry and isotope exchange. Rapid Commun Mass Spectrom. 2007;21(7):1093–9. doi: 10.1002/rcm.2926. [DOI] [PubMed] [Google Scholar]
- 21.Athersuch TJ, Nicholson JK, Wilson ID. Isotopic enrichment enhancement in metabonomic analysis of UPLC-MS data sets. Journal of Labelled Compounds & Radiopharmaceuticals. 2007;50(5–6):303–307. [Google Scholar]
- 22.Haubner R, Wester HJ. Radiolabeled tracers for imaging of tumor angiogenesis and evaluation of anti-angiogenic therapies. Curr Pharm Des. 2004;10(13):1439–55. doi: 10.2174/1381612043384745. [DOI] [PubMed] [Google Scholar]
- 23.Mueller M, Kolbrich-Spargo EA, Peters FT, Huestis MA, Ricaurte GA, Maurer HH. Hydrolysis of 3,4-methylenedioxymethamphetamine (MDMA) metabolite conjugates in human, squirrel monkey, and rat plasma. Anal Bioanal Chem. 2009;393(6–7):1607–17. doi: 10.1007/s00216-009-2607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyland R, Osborne T, Payne A, Kempshall S, Logan YR, Ezzeddine K, Jones B. In vitro and in vivo glucuronidation of midazolam in humans. Br J Clin Pharmacol. 2009;67(4):445–54. doi: 10.1111/j.1365-2125.2009.03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Patterson AD, Krausz KW, Tanaka N, Gonzalez FJ. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor alpha in bile acid homeostasis. J Lipid Res. 2012;53(8):1625–35. doi: 10.1194/jlr.M027433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Li F, Patterson AD, Wang Y, Krausz KW, Neale G, Thomas S, Nachagari D, Vogel P, Vore M, Gonzalez FJ, Schuetz JD. Abcb11 deficiency induces cholestasis coupled to impaired beta-fatty acid oxidation in mice. J Biol Chem. 2012;287(29):24784–94. doi: 10.1074/jbc.M111.329318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R, Strelevitz TJ, Gao H, Clark AJ, Schildknegt K, Obach RS, Ripp SL, Spracklin DK, Tremaine LM, Vaz AD. Deuterium isotope effects on drug pharmacokinetics. I. System-dependent effects of specific deuteration with aldehyde oxidase cleared drugs. Drug Metab Dispos. 2012;40(3):625–34. doi: 10.1124/dmd.111.042770. [DOI] [PubMed] [Google Scholar]
- 29.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 30.Saude EJ, Adamko D, Rowe BH, Marrie T, Sykes BD. Variation of metabolites in normal human urine. Metabolomics. 2007;3(4):439–451. [Google Scholar]
- 31.Patterson AD, Gonzalez FJ, Idle JR. Xenobiotic metabolism: a view through the metabolometer. Chem Res Toxicol. 2010;23(5):851–60. doi: 10.1021/tx100020p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilcox CS. Effects of tempol and redox-cycling nitroxides in models of oxidative stress. Pharmacol Ther. 2010;126(2):119–45. doi: 10.1016/j.pharmthera.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson GG, Skett P. Introduction to drug metabolism. 3. viii. Nelson Thornes Publishers; Cheltenham, UK: 2001. p. 256. [Google Scholar]
- 34.Sang S, Yang CS. Structural identification of novel glucoside and glucuronide metabolites of (-)-epigallocatechin-3-gallate in mouse urine using liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(22):3693–9. doi: 10.1002/rcm.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommerer U, Gordon ER, Goresky CA. Microsomal specificity underlying the differing hepatic formation of bilirubin glucuronide and glucose conjugates by rat and dog. Hepatology. 1988;8(1):116–24. doi: 10.1002/hep.1840080123. [DOI] [PubMed] [Google Scholar]
- 36.Zhen Y, Krausz KW, Chen C, Idle JR, Gonzalez FJ. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol Endocrinol. 2007;21(9):2136–51. doi: 10.1210/me.2007-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fardet A, Llorach R, Martin JF, Besson C, Lyan B, Pujos-Guillot E, Scalbert A. A liquid chromatography-quadrupole time-of-flight (LC-QTOF)-based metabolomic approach reveals new metabolic effects of catechin in rats fed high-fat diets. J Proteome Res. 2008;7(6):2388–98. doi: 10.1021/pr800034h. [DOI] [PubMed] [Google Scholar]
- 38.Shukla OP. Microbial transformation of quinoline by a Pseudomonas sp. Appl Environ Microbiol. 1986;51(6):1332–42. doi: 10.1128/aem.51.6.1332-1342.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla OP. Microbiological degradation of quinoline by Pseudomonas stutzeri: the coumarin pathway of quinoline catabolism. Microbios. 1989;59(238):47–63. [PubMed] [Google Scholar]
- 40.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu S, Rao S, Reddy JK. Peroxisome proliferator-activated receptors, fatty acid oxidation, steatohepatitis and hepatocarcinogenesis. Curr Mol Med. 2003;3(6):561–72. doi: 10.2174/1566524033479537. [DOI] [PubMed] [Google Scholar]
- 42.Wilcox CS, Pearlman A. Chemistry and antihypertensive effects of tempol and other nitroxides. Pharmacol Rev. 2008;60(4):418–69. doi: 10.1124/pr.108.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaak EE. Basic disturbances in skeletal muscle fatty acid metabolism in obesity and type 2 diabetes mellitus. Proc Nutr Soc. 2004;63(2):323–30. doi: 10.1079/PNS2004361. [DOI] [PubMed] [Google Scholar]
- 44.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–56. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9(3):230–4. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 46.Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe de Angelis M, Klingenspor M, Buck A, Schwaiger M, Saur D, Schmid RM, Kloppel G, Sipos B, Greten FR, Arkan MC. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A. 2009;106(9):3354–9. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi X, Yao D, Chen C. Identification of N-acetyltaurine as a novel metabolite of ethanol through metabolomics-guided biochemical analysis. J Biol Chem. 2012;287(9):6336–49. doi: 10.1074/jbc.M111.312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.