Abstract
A diverse group of animals, including members of most major phyla, have adapted to life in the perpetual darkness of caves. These animals are united by the convergence of two regressive phenotypes, loss of eyes and pigmentation. The mechanisms of regressive evolution are poorly understood. The teleost Astyanax mexicanus is of special significance in studies of regressive evolution in cave animals. This species includes an ancestral surface dwelling form and many con-specific cave-dwelling forms, some of which have evolved their recessive phenotypes independently. Recent advances in Astyanax development and genetics have provided new information about how eyes and pigment are lost during cavefish evolution; namely, they have revealed some of the molecular and cellular mechanisms involved in trait modification, the number and identity of the underlying genes and mutations, the molecular basis of parallel evolution, and the evolutionary forces driving adaptation to the cave environment.
Keywords: eye degeneration, loss of pigmentation, albinism, development, genetics
INTRODUCTION
A remarkably diverse group of animals has adapted to life in the perpetual darkness of caves (8, 38) (Figure 1). Just as remarkable are the convergent phenotypes that have evolved in these animals, namely the loss of eyes and pigmentation. The regressive phenotypes of cave animals have puzzled biologists since the time of Darwin (10), who famously remarked: “As it is difficult to imagine that eyes, though useless, could in any way be injurious to animals living in darkness, I attribute their loss solely to disuse.” After the advent of neo-Darwinism and the rediscovery of genetics, biologists attributed the evolution of cave-adapted traits either to natural selection or neutral evolution, although not providing much supportive evidence for either mechanism. In the past decade, new developmental and genetic approaches have positioned us to make progress in understanding the evolution of cave life. The emergence of the blind cavefish, Astyanax mexicanus, as a model system has been instrumental in this progress. In this review, we describe how Astyanax has been used to understand the evolution of cave animals in a developmental and genetic context.
Figure 1.

Examples of diverse cave animals lacking eyes and pigmentation. (a) Cave planarian. (b) Cave gastropod mollusk, Zospeum kusceri. (c) Cave isopod, Monolistra bolei. (d) Cave planthopper, Oliarus polyphemus. (e) Cave urodele amphibian, Proteus anguinus.
REGRESSIVE EVOLUTION IN CAVE ANIMALS
Regressive evolution refers to the loss of useless characters over time. It is a critical evolutionary process in all organisms. For example, humans would be as hairy and tailed as other primates if regressive evolution did not prune unused ancestral traits. The justification for considering regressive evolution as a separate form of evolution is that its mechanisms may be different from the evolution of novel traits by natural selection. This issue is still unclear, however, because the mechanisms of regressive evolution are poorly understood.
Cave animals represent one of the best examples of regressive evolution and offer some unique advantages for studying its mechanisms. First, the environmental cue leading to eye and pigment regression, the perpetual absence of light, has been constant throughout the evolutionary history of cave animals. Thus, in studying the evolution of cave animals, the mechanism of trait loss can be directly linked to the environmental conditions that have produced it. Second, the evolutionary timeframe in which regressive traits are lost is relatively short, allowing the earliest changes leading to trait reduction to be identified and studied. Third, each individual cave is a single experiment in evolution, and this experiment is replicated when additional caves are added to the study. Thus, cave animals offer an opportunity to study the mechanisms of convergent and parallel evolution of regressive traits that are driven by the same environmental cue.
The advantages of cave organisms would seem to make all of them excellent subjects for evolutionary studies. Unfortunately, this is not the case. The limiting factor is the inability of most cave animals to adapt to life in another constant habitat: the scientific laboratory. The subject of this review, the cavefish Astyanax mexicanus, is one of the few cave animals that can be maintained and propagated in the laboratory (65).
ASTYANAX IS AN EMERGING MODEL SYSTEM
The attribute of having cave and surface dwelling forms in the same species sets Astyanax apart from almost all other well studied cave animals (20, 61) (Figure 2). The feature that makes Astyanax the “white rat” of cave animals is its suitability as a laboratory animal. It can be raised on a simple diet, spawns frequently and abundantly, and has a generation time of approximately 4 to 6 months. Astyanax embryos are large, clear, and excellent subjects for experimental embryology. Astyanax surface fish and cavefish are fully interfertile (48), and cavefish can also be crossed for genetic complementation analysis (5, 60, 64). Cavefish can be compared with their surface fish con-specifics in the same way mutants are compared with wild-type phenotypes. Astyanax is therefore also the “fruit fly” of cave animals. The close phylogenetic relationship between Astyanax and zebrafish permits molecular techniques and resources to be shared between the two species. Gene knockdown and knockup procedures and transgenic methods modeled after their counterparts in zebrafish are also available in Astyanax. Thus, the evolution of regressive phenotypes can be studied by reverse as well as forward genetics. Astyanax and zebrafish have the same chromosome number (2N = 50) and share extensive blocks of syntenic DNA sequences throughout their genomes. There is currently no physical map of the Astyanax genome, as there is in zebrafish. However, Astyanax genetic maps have been produced (40–42) and anchored to the zebrafish physical map (16, 17), revealing candidates for genes underlying eye and pigment loss.
Figure 2.
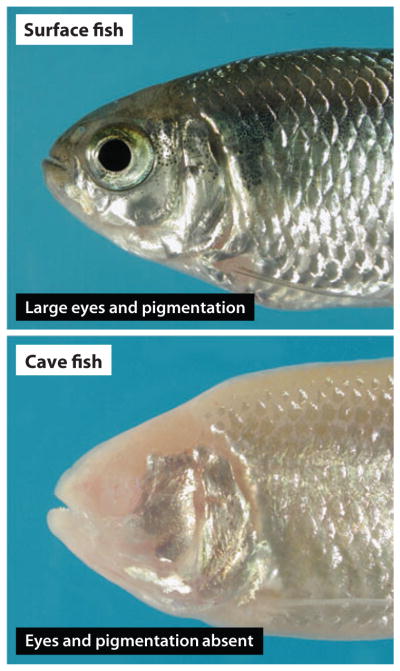
Astyanax mexicanus surface fish and cavefish.
EVOLUTIONARY HISTORY
There are 30 known caves harboring Astyanax cavefish populations in Mexico. We will focus on two major questions pertaining to the evolutionary history of regressive traits in Astyanax cavefish. First, did all cavefish populations originate from a single colonization (or entrapment) event, followed by dispersal to their present locations, or did they originate from multiple colonizations? Second, did regressive features evolve only once per colonization event, or did they evolve multiple times in parallel after dispersal and isolation of evolving cavefish?
Many additional Astyanax cavefish populations have been reported after the discovery of the first cavefish population in La Cueva Chica (2, 3, 14, 19, 33, 63). Almost all of these populations are located in northeastern Mexico. The exception is a single outlying cavefish population in the south central Mexican state of Guerrero. Located across the continental divide and separated from the northeastern caves by non-carbonate strata, the Guerrero population is almost certainly an independent line of cavefish evolution. The fact that Astyanax cavefish can arise independently is not surprising considering 86 cavefish species have been described in 19 teleost families (43).
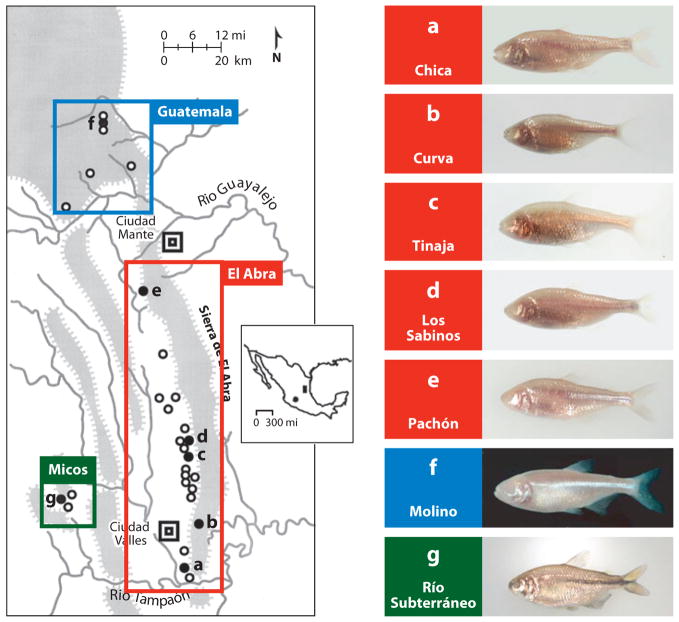
The best studied of the northeastern cavefish populations are Chica, Curva, Los Sabinos, Tinaja, Piedras, Japonés, Yerbaniz, Pachón, Molino, and Rio Subterráneo (Figure 3), which cluster into three geographical areas. The first eight populations, along with twelve others, are located in the Sierra de El Abra and referred to as the El Abra cluster. Molino and five other cavefish populations are located in the Sierra de Guatemala and the Sierra de Nicolás Pérez, which is called the Guatemala cluster. Río Subterráneo cavefish, along with two other populations, are referred to as the Micos cluster. The geographical distance between the clusters suggests at least three colonization events and independent lines of Astyanax cavefish evolution.
Figure 3.
Left: A sketch map showing the region containing 29 different Astyanax cavefish populations in northeastern Mexico. The spheres indicate the approximate position of caves with Astyanax cavefish. The shaded spheres (a–g) indicate specific cavefish populations shown on the right. The Guatemala, El Abra, and Micos clusters are indicated on the map. Inset: Mexico showing the northeastern region indicated in the sketch map (shaded rectangle) and the outlying Guerrero population (shaded sphere). Right: Examples of different cavefish shown in a–g on the left. Modified from Jeffery et al. (2003).
Phylogenetic analysis using DNA sequences has been the major approach used to determine the relationships between cavefish populations. As a rule, however, Astyanax cavefish populations show minimal genetic variation in the coding regions of nuclear genes relative to surface fish and each other. Therefore, phylogenetic inferences have been based on more divergent randomly amplified polymorphic DNAs (RAPDs), microsatellite sequences, and rapidly evolving mitochondrial DNA. The RAPD phylogeny supports at least two origins of Astyanax cavefish in northeastern Mexico: one in the El Abra cluster (Pachón, Tinaja, Curva, and Chica) and the other in the Micos cluster (Río Subterráneo) (13). Microsatellite analysis also suggests that three members of the El Abra cluster (Pachón, Los Sabinos, and Curva) had a single origin (53, 54). Studies using mitochondrial ND2 and cytochrome B gene sequences reach similar conclusions: three members of the El Abra cluster (Tinaja, Los Sabinos, and Curva) are closely related and different from a member of the Micos cluster (Río Subterráneo) (11, 53, 54). Pachón is a special case: Its nuclear microsatellite sequences are typical of other El Abra cavefish, but its mitochondrial ND2 and cytochrome B genes more closely resemble those of surface fish located outside the cave. Therefore, the Pachón mitochondrial genome may have been replaced by introgression, underscoring the importance of hybridization as well as parallel evolution in shaping some of the northeastern cavefish populations. It is unfortunate that Molino or other Guatemala populations were not included in these analyses. Nonetheless, the DNA sequence studies suggest at least two origins for Astyanax cavefish in northeastern Mexico, one in the Micos cluster and at least one other in the El Abra cluster, although this is likely a minimal estimate.
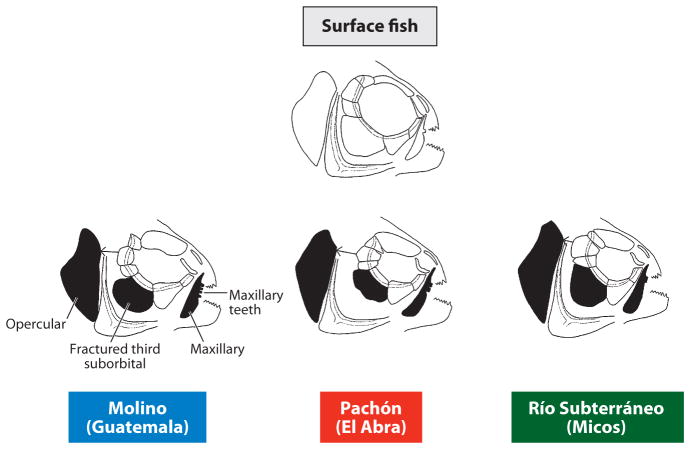
The number of times regressive traits arose within the northeastern cavefish populations is potentially greater than their number of origins. This is because once a cave has been colonized, fish can subsequently disperse horizontally and vertically through interconnected cave systems whilst evolving cave-adapted traits and becoming increasingly isolated from each other. Our understanding of the evolutionary history of regressive traits comes from two kinds of data: (a) morphological comparisons between the various cavefish populations and (b) genetics. Different cavefish populations have distinct morphologies (Figure 3a–f, Figure 4). Members of the Micos cluster have more intense pigmentation than members of the El Abra and Guatemala clusters, some of which have evolved albinism (49). Guatemala and Micos cavefish have streamlined body shapes, a trait resembling surface fish. In contrast, El Abra cavefish are more compressed along their anterior posterior axis, probably because of the loss of one or two vertebrae (11). As a result of orbital modifications related to eye reduction, changes in the size of the nasal aperture and operculum, and increases in the number of maxillary teeth, El Abra, Guatemala, and Micos cavefish have distinctive craniofacial morphologies (68) (Figure 4). Genetic complementation analysis also supports multiple independent origins of cavefish populations and their regressive phenotypes. For example, in a cross between two cavefish populations (see Figure 7), traits resembling those of ancestral surface fish are sometimes recovered, implying control by different genes and thus independent episodes of regressive evolution. The relevant genetic analysis will be considered later in this article.
Figure 4.
Surface fish and cavefish exhibit differences in craniofacial morphology. Shaded bones show the most change from surface fish to cavefish and amongst the Guatemala, El Abra, and Micos cavefish clusters. Modified from Yamamoto et al. (2003).
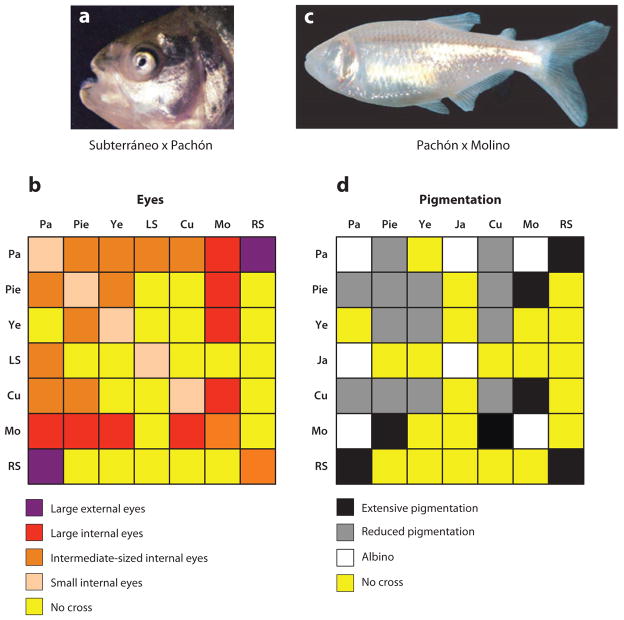
Figure 7.
Genetic complementation tests for eye and pigment regression. (a,b) Extent of eye formation in F1 hybrids produced by crosses between different cavefish populations. (a) A Río Subterráneo x Pachón F1 hybrid with large external eyes. (b) Punnett square illustrating eye size in F1 hybrids of crosses between various cavefish populations. (c,d) Extent of pigmentation in F1 hybrids produced by crosses between different cavefish populations. (c) Albinisim is present in a Pachón x Molino F1 hybrid. (d) Punnett square illustrating the status of pigmentation in F1 hybrids of crosses between different cavefish populations. LS: Los Sabinos. Pa: Pachón (albino). Pie: Piedras. Ye: Yerbaniz. Ja: Japonés (albino). Cu: Curva. Mo: Molino (albino). RS: Río Subterráneo. (a) Y. Yamamoto and W.R. Jeffery (unpublished). (c) From Protas et al. (2006).
In summary, the questions about evolutionary history introduced above can be answered as follows: (a) multiple colonizations have generated at least three (Guerrero, Micos, and El Abra) and possibly four (Guatemala) different Astyanax cavefish founder populations, and (b) multiple episodes of regressive evolution have occurred in some of these founder populations, resulting in parallel evolution of cave-related phenotypes.
DEVELOPMENTAL BASIS OF EYE REGRESSION
Although appearing eyeless, adult cavefish actually have small nonfunctional eyes buried deep within their orbits. Different cavefish populations show various degrees of eye degeneration: Curva, Los Sabinos, and Pachón have smaller vestigial eyes than Río Subterráneo or Molino cavefish (61, 62, 64). The molecular and cellular mechanisms of eye degeneration have been studied most extensively in Pachón cavefish.
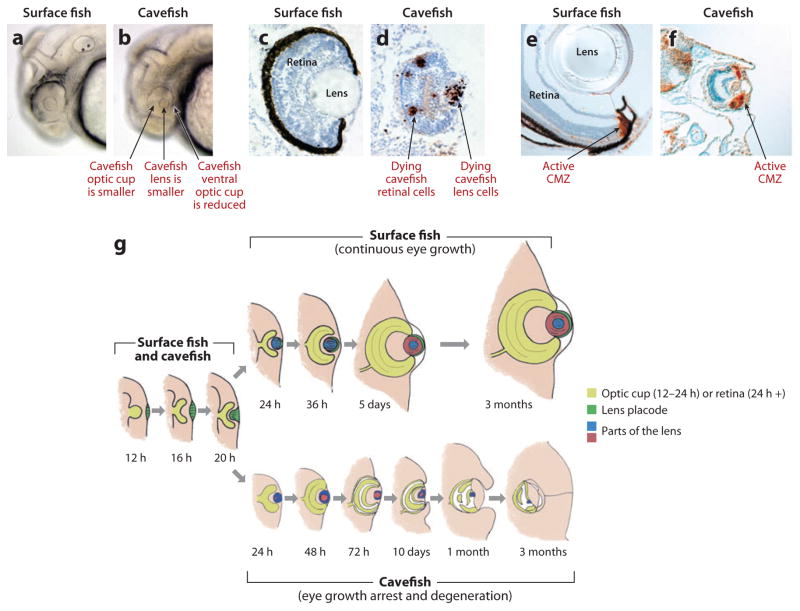
Small eye primordia are formed in Pachón embryos, but they subsequently arrest, degenerate, and sink into the orbits (7, 67) (Figure 5). Compared with surface fish, the Pachón embryonic lens is relatively small, and the optic cup is missing its ventral sector (Figure 5a,b). The same defects are seen in Chica, Tinaja, Curva, and Los Sabinos cavefish (23). Later in development, fiber cells do not differentiate in the cavefish lens, which becomes progressively smaller and vestigial during later development (29, 52, 58, 61). In contrast, a normally differentiated retina initially develops from the cavefish optic cup, including the typical layers containing neuronal, glial, and photoreceptor cells (1, 22, 29, 66). Subsequently, photoreceptor cells disappear, however, and are not replaced (29, 55, 66). The retina continues to grow but becomes highly disorganized, and further growth eventually ceases.
Figure 5.
Eye development and degeneration in surface fish and cavefish respectively. (a,b) The lens and optic cup are smaller, and the ventral side of the optic cup is missing in Pachón cavefish embryos (b) relative to surface fish embryos (a). (c,d) TUNEL assay shows apoptosis at 2 days postfertilization (dpf) in the lens and retina of Pachón cavefish embryos (d) but not in surface fish embryos (c). (e, f) PCNA labelling shows active cell proliferation in the CMZ of surface fish (e) and Pachón cavefish (f) retinas at 10 dpf. (g) A diagram showing the events of Astyanax eye development and degeneration. Left. Early events of eye primordium formation are the same in surface fish and cavefish until approximately 1 dpf. Top: In surface fish, the eye differentiates and the eye parts grow in concert with increased body growth. Bottom. In cavefish, the eye primordium grows for a while, then arrests, degenerates, and is internalized by overgrowth of the body. (a,b) from Yamamoto & Jeffery (2000). (c,d) from Strickler et al. (2007). (e, f) from Strickler et al. (2002).
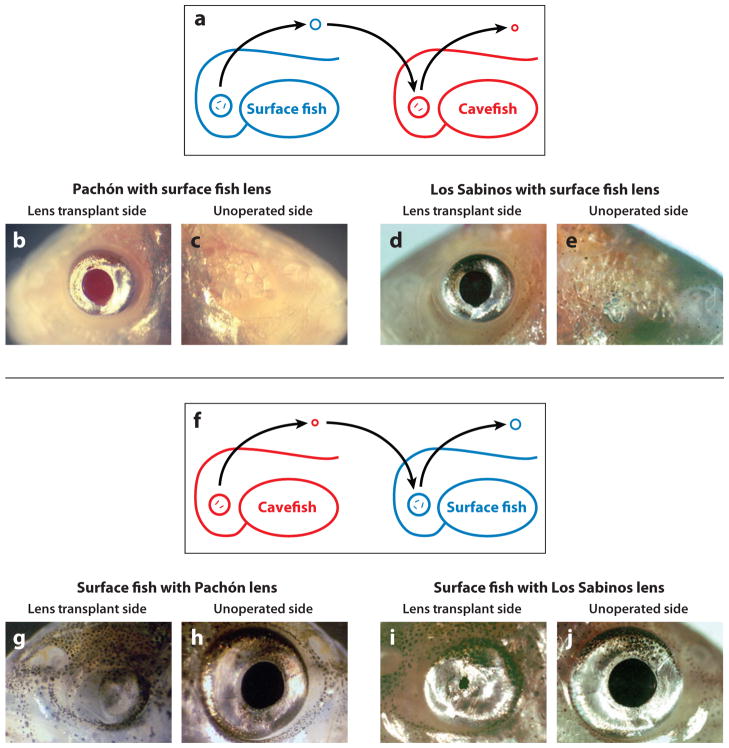
The arrest in cavefish eye growth is probably a consequence of the dysfunction of a crucial regulatory component(s) in the developing eye. What is this component(s)? To determine its identity, eye development was compared in surface fish and Pachón cavefish using TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) and other methods to detect apoptosis (1, 21, 58). Apoptosis was detected in several different eye tissues (Figure 5c,d). However, the embryonic lens starts to die several days before any other eye tissue, earmarking it for a possible regulatory role (21, 58). A similar apoptotic event occurs in the developing lens of Los Sabinos cavefish (23). Later, retinal and RPE (retinal pigment epithelium) cells also undergo apoptosis (58). Lens transplantation procedures were developed to determine the role of the lens in sustaining Astyanax eye development (23, 66) (Figure 6). In these experiments, a surface fish lens was transplanted into the Pachón or Los Sabinos optic cup, and reciprocally, a cavefish lens was transplanted into the surface fish optic cup. The transplants were done on only one side of the embryo, allowing the other side to serve as a control. Transplanted lenses behave autonomously: The cavefish lens died on schedule in the surface fish host, and the surface fish lens survived and grew in the cavefish host. These experiments showed that a transplanted surface fish lens can restore the formation of a normal (although smaller) eye in Pachón or Los Sabinos cavefish, including a growing retina with photoreceptor cells (Figure 6), suggesting that the lens has a fundamental role in regulating cavefish eye degeneration.
Figure 6.
Lens transplantation. (a–e) Transplantation of a surface fish embryonic lens into the optic cup of a Pachón (b,c) or Los Sabinos (d,e) cavefish embryo after their own lens is removed rescues the eye in adults. (b,d) Lens transplant side. (c,e) Unoperated side. (f–j) Transplantation of a Pachón (g,h) or Los Sabinos (i, j) embryonic lens into the optic cup of a surface fish embryo after its own lens is removed causes retardation of eye development in adults. (g,i) Lens transplant side. (i, j) Unoperated side. From Yamamoto and Jeffery (2000) and Jeffery et al. (2003).
Retinal development is known to be dependent on a functional lens in teleosts (27), but the underlying mechanisms are poorly understood. Does the lens normally influence the retina by providing an instructional signal, a permissive signal, or both? The teleost retina continues to grow throughout life by adding new cells from two stem cell niches: the ciliary marginal zone (CMZ), which produces new cells for most of the retinal layers and RPE, and the inner nuclear layer, which replenishes the photoreceptor cells (24, 37). Thus, inhibition of retinal stem cell proliferation, possibly owing to lack of lens function, would seem to be a prime target for arrest of retinal development in cavefish. Surprisingly, however, several different markers showed that cell proliferation was not affected throughout the period in which retinal structure is being dismantled, and its growth is arrested in the adult (1, 57, 58) (Figure 5e, f). If new retinal cells are continually produced in cavefish, why is retinal growth arrested? Given that apoptosis was also seen in the cavefish retina, an alternative possibility is that the new cells die before they can differentiate and contribute to the retina. This possibility was tested in a BrdU pulse-chase labeling experiment (58). In this experiment, labeled cells appeared in the retinal growth zones in both surface fish and cavefish during the BrdU pulse but could only be detected during the chase in surface fish. The addition of new cells to the cavefish retina is cancelled by apoptosis. Next, a surface fish lens was transplanted into the cavefish optic cup, and the BrdU experiment was repeated (58). In contrast to the previous results, retinal cells labelled during the BrdU pulse were retained during the chase, indicating that the transplanted lens protected the cavefish retina from apoptosis. Thus, a normal lens appears to protect the retina from apoptosis and sustain its growth. When the cavefish lens fails to develop normally its protective function is compromised and newly born retinal cells die rather than differentiate.
The destruction of the lens is not the only defect in the cavefish eye. As soon as it appears in the early embryo, the cavefish optic vesicle is smaller than its surface fish counterpart, and the optic cup is missing its ventral sector (7, 67) (Figure 5a,b). These changes in the optic cup have impacts later in development, as evidenced by a degree of independence between lens and retinal degeneration in some cavefish populations (61, 62). Lens manipulation experiments also address this point. When a surface fish lens is deleted, there is no induction of retinal apoptosis (58), as would be expected if the lens is the sole protector of the retina. In surface fish, there may be a second or redundant influence emanating from another part of the eye that sustains retinal growth. Therefore, it is likely that defects other than lens apoptosis also operate to curtail eye development in cavefish.
GENETIC BASIS OF EYE REGRESSION
Astyanax cavefish populations are interfertile with surface fish and amongst themselves (48). The eyes of Pachón or Molino cavefish x surface fish F1 hybrids are smaller than normal but completely functional (61, 64). In contrast, the F2 generation shows a variety of eye sizes, ranging from nearly normal to almost as completely reduced as those of cavefish. The results of these crosses show that eye degeneration is a multi-factorial trait. From the distribution of eye phenotypes in the F2 generation, it was estimated that approximately 6 different genes are directly responsible for eye regression (61). The multigenic control of eye regression was tested by linkage mapping with RAPD (6) or microsatellite (41) markers. Both mapping procedures identified multiple quantitative trait loci (QTL) for eye loss: three QTL from the RAPD analysis and 12 QTL from the more sensitive microsatellite analysis. Thus, many different genes control eye degeneration.
Crosses between different cavefish populations were done to discover whether these genes are the same in various cavefish populations (Figure 7). The results were dramatic: Eye size in some of the resulting hybrids was significantly larger than in the parents (60, 64). For example, crosses between Molino and several members of the El Abra cluster yielded F1 progeny with large eyes, although they were still internalized during development. But crosses between Río Subterráneo and Pachón gave large eyes that remained on the exterior of adult fish and appeared to be functional (Figure 7a) (Y. Yamamoto and W.R. Jeffery, unpublished). Recent studies have shown that crosses between different cavefish populations can rescue vision, restoring sight to cavefish lineages that have not been able to see for a million years or more (5). Therefore, complementation indicates that different sets of genes are responsible for eye degeneration in Molino, Río Subterráneo, Pachón, and to a lesser extentin other El Abra cavefish (Figure 7b). Assuming that approximately 12 gene loci are responsible for eye degeneration in most cavefish populations, it has been estimated that loss of function at three or four of these loci is enough to abolish vision (5).
In addition to establishing the multiplicity of genes involved in eye regression, the cavefish crosses also provide insights about the regressive evolution of eyes. It was suggested above that episodes of regressive evolution are potentially more numerous than the number of original cavefish founder populations. Genetic complementation of eye phenotypes in the Guatemala, Micos, and El Abra clusters supports this idea, revealing at least three or four and possibly even more independent episodes of eye regression.
GENES CONTROLLING EYE REGRESSION
To determine the molecular basis for eye regression, the genes involved in this process need to be identified. These genes could be regulatory genes functioning at or near the beginning of eye developmental pathways, subsidiary genes of various sorts within the framework of the pathways, or genes encoding enzymes or structural proteins that function at or near the end of the pathways. In addition, some genes could be mutated and directly affect eye development, whereas other genes could act downstream of the mutated genes and changes in their expression could indirectly mediate eye degeneration.
Candidate Gene Analysis
Many genes with important roles in vertebrate eye development are already known, therefore a candidate gene approach can be used to identify eye genes whose expression is changed in Pachón cavefish relative to surface fish. Some eye genes surveyed by candidate gene analysis show little or no expression changes, and thus may not be involved in eye degeneration (1, 22, 57, 59). However, only a few of these genes have been completely sequenced in both forms of Astyanax. Therefore, it is still possible that changes in their protein sequence might lead to altered functions and contribute to eye degeneration. In contrast, the expression of some genes are changed in cavefish relative to surface fish. Gamma-M crystallin and rhodospin gene expression is reduced in the cavefish lens and retina, respectively (29, 55). These changes are consistent with respective lack of lens fiber cell differentiation and degeneration of the retinal photoreceptor layer (29). The αA-crystallin gene, which encodes an potent antiapoptotic factor, is strongly down-regulated in the embryonic lens of Piedras (4) and Pachón cavefish (59). In contrast, expression of the hsp90α gene is specifically activated in the lens of Pachón, Chica, and Tinaja cavefish embryos just prior to apoptosis (18). The expression of hsp90α is not affected outside the lens, and the expression of its close relative hsp90β is similar between cavefish and surface fish (18). Pharmacological inhibition of Hsp90 function suppresses lens apoptosis and to some extent rescues lens differentiation, suggesting that Hsp90α in the lens may be a proapoptotic factor. Thus, cavefish lens apoptosis may be controlled by the activation of proapoptotic factors and suppression of antiapoptotic factors.
Some of the changes in gene expression are subtler than those described above but nonetheless important in cavefish eye degeneration. For example, expression of the major eye regulatory gene pax6 is changed in Pachón, Los Sabinos, and Curva cavefish (56). In surface fish embryos, pax6 expression domains in the bilateral optic fields connect across the midline. In cavefish embryos, however, the corresponding pax6 domains are diminished in size and show a gap across the midline (56). In mice, the division of the optic vesicle into the optic cup and stalk is controlled by reciprocal antagonistic interactions between Pax6, Pax2, and Vax1 transcription factors (51). Pax6 directs optic cup development, whereas Pax2 and Vax1 control optic stalk development. Accordingly, reduction of pax6 levels (or elevation in pax2 and vax1 levels) increases the optic stalk at the expense of the optic cup. Thus, reduction of pax6 expression coupled with overexpression of the pax2a and vax1 genes accounts for the ventrally reduced optic cup in cavefish embryos (67). Later in development, the vax1 gene is expressed on the ventral side of the surface fish retina, but it is absent in the ventral portion of the cavefish retina (1), showing that upregulation or downregulation of Vax1 has contrasting effects on cavefish optic cup and retina development respectively.
The wider gap between pax6-expressing optic fields in the cavefish neural plate provides a clue as to how eye degeneration might be controlled. During vertebrate development, Hedgehog (Hh) signals emanating from the underlying prechordal plate inhibit pax6 expression along the midline to divide the original eye domain into bilateral eyes (12, 30) (Figure 8a). Teleosts have two-hh midline signaling genes, sonic hedgehogA (shhA) and shhB, which show overlapping expression patterns and possibly redundant functions (12). The shhA and shhB midline expression domains are expanded in Pachón, Chica, and Los Sabinos cavefish relative to surface fish (see Figure 8a,b for shhA in Pachón). Later in cavefish development, shh expression is also expanded in the developing forebrain (32, 46) and the presumptive oral pharyngeal area (69). The expression patterns of genes acting downstream of shhA and shhB in the Hh midline signaling pathway, such as patched1 and patched2, encoding Shh receptors; and nkx2.1a and nkx2.1b, encoding Shh dependent transcription factors, are also expanded (69), suggesting that a general increase in midline Hh signaling has evolved in cavefish.
Figure 8.
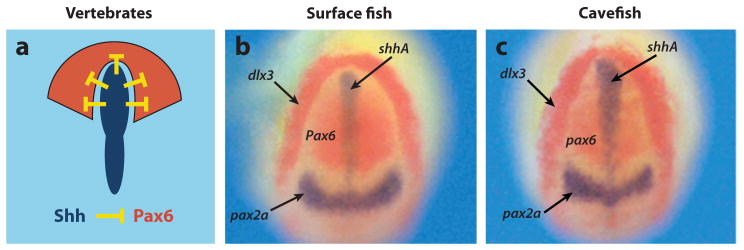
Negative relationship between midline Shh signaling and Pax6 controls eye development. (a) Diagram of the neural plate and underlying anterior midline of a typical vertebrate embryo showing the negative relationship between Shh (blue) and Pax6 (red). (b,c) Expanded shhA expression along the anterior midline of (c) Pachón cavefish embryo with respect to (b) surface fish embryo hybridized in situ at the neural plate stage. Dorsal views of the anterior neural plate showing shhA expression (blue) compared with dlx3, pax6, and pax2a markers. From Yamamoto et al. (2004).
The role of enhanced Hh signaling in cavefish eye development was investigated by increasing shh expression in surface fish embryos (67). When shhA mRNA was injected into one side of a cleaving embryo, pax6 expression was downregulated in the corresponding optic field. Surface fish larvae that developed from embryos with elevated Shh levels were missing an eye on one side of the head. Importantly, lens apoptosis was also induced by shh overexpression in surface fish embryos (67). Thus, a phenocopy of blind cavefish can be obtained in surface fish by increasing the levels of shh gene expression, demonstrating the key role for Shh signaling in eye degeneration.
QTL Analysis
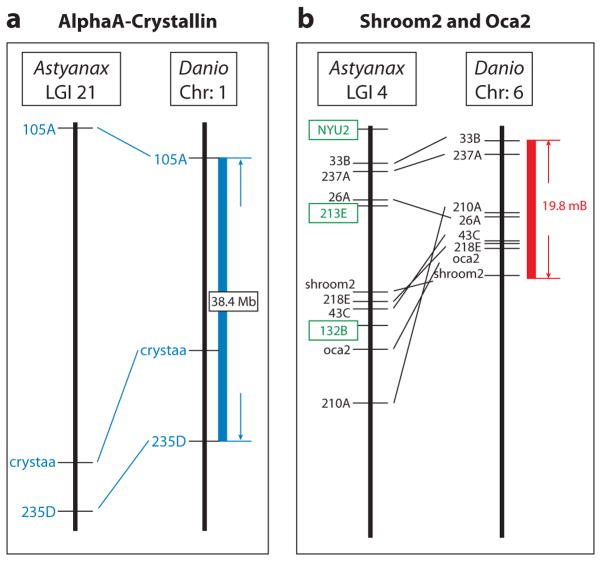
Linkage analysis is a more direct approach than candidate gene analysis to identify genes that are mutated to cause eye degeneration. None of the approximately 12 known eye size QTL are located near the pax6, shhA, shhB, or other known hedgehog gene loci on the Astyanax genetic map (41). Although what shows up in QTL analysis may not reflect the entire spectrum of altered genes, these results suggest that these genes (or their cis-regulatory regions) are not mutated in cavefish. Instead, they are probably subsidiary genes that act downstream of genes that were mutated to initiate eye reduction. As in all QTL studies, the challenge is to find the relevant gene(s) within the identified QTL. To do this, the Astyanax microsatellite map was anchored to the zebrafish physical map (16). The assemblage of multiple linkage maps with zebrafish revealed numerous large blocks of synteny between the two species, many of which are shared across critical regions of identified eye QTL. The anchored map revealed three candidate genes for eye QTL (16) (Figure 9). The first candidate is the αA-crystallin gene, which is tightly linked to the eye QTL on linkage group 21 (Figure 9a). As mentioned above, the anti-apoptotic factor αA-crystallin was previously predicted to be involved in eye degeneration by its substantial downregulation in Piedras and Pachón cavefish (4, 59). There are no significant differences in the coding sequences of cavefish and surface fish αA-crystallin (4), implying that the relevant mutation may be in a regulatory region. A second candidate gene is shroom2, which maps very close to an eye QTL located near the albinism trait and oca2 gene (see below) on linkage group 5 (Figure 9b). The shroom2 gene, which is also closely linked to oca2 in zebrafish and other teleosts, has a role in the synthesis and localization of melanin granules in the RPE (15). The third candidate gene is rom1, which is tightly linked to an eye QTL on linkage group 1. The rom1 gene encodes a structural protein required for maintaining the flattened shape of the outer discs of rod photoreceptor cells (35). It will be important to confirm the role of these genes in eye regression by identifying mutations and performing functional analysis.
Figure 9.
An anchored linkage map reveals eye and pigment candidate genes in Astyanax QTL and corresponding syntenic regions in Danio (zebrafish). (a) The position of the αA-crystallin gene on Astyanax linkage group 21 and syntenic region on Danio chromosome 1. (b) The positions of the shroom2 and oca2 genes on Astyanax linkage group 4 and syntenic region on Danio chromosome 6. From Gross et al. (2008).
The three candidate genes revealed by QTL map anchoring are probably expressed near the end of developmental pathways leading to eye lens and retina differentiation. This raises the possibility that some genes at the termini of development pathways may be affected during eye regression in Astyanax. If so, this resembles tail loss in anural ascidian larvae, another well-known case of regressive evolution, in which muscle actin structural genes become nonfunctional via mutations in their coding regions, but earlier-expressed regulatory genes are still functional (28). Thus far, only a few of the many eye QTL have been anchored to candidate genes. It seems likely that some of the other eye QTL may contain genes that act upstream in eye development, perhaps even early enough to regulate the Shh signaling pathway.
DEVELOPMENTAL BASIS OF ALBINISM AND REDUCED PIGMENTATION
The reduction or loss of skin pigmentation is another major regressive trait in Astyanax cavefish (44). In surface dwelling animals, pigmentation is used for protection from sunlight, camouflage, mimicry, and species and sex recognition (39), all of which are irrelevant in the dark cave environment. Astyanax has three types of pigment cells: black melanophores, silver iridophores, and orange xanthophores. The light skin of cavefish is due to fewer numbers of melanophores and a reduction or loss of melanin pigment. Most Pachón and Molino cavefish are albinos and lack melanin pigmentation in their bodies and eyes. As discussed below, however, colorless melanophores or their precursors are present in albino cavefish. In predominantly nonalbino cavefish populations, there has been a reduction in the number of melanophores and in their melanin content, making these pigment cells appear brown rather than black. Brown melanophores are found in Curva, Los Sabinos, Piedras, Yebanez, Japonés, and in nonalbino Pachón cavefish, but not in Molino cavefish (17, 50). Reduced numbers of colorless, brown, and black melanophores are found in all cavefish populations with respect to surface fish. Río Subterráneo cavefish have the highest numbers of melanophores; Tinaja, Curva, and Los Sabinos cavefish have intermediate numbers; and Pachón cavefish have the lowest numbers of (colorless) melanophores (31). Thus, as in the case of eyes, different cavefish populations have distinctive melanophore and melanin phenotypes, supporting multiple independent origins of regressed pigmentation.
All types of body pigment cells are derived from the neural crest. Neural crest cells have many critical derivatives in addition to pigment cells; for example, the peripheral nervous system, facial cartilage and bone, and parts of the heart and endocrine organs. The neural crest cells that generate pigment cells migrate from the dorsal neural tube along a specific pathway to the skin. Several lines of evidence indicate that this subset of neural crest cells and their migration pathway is not altered in Pachón cavefish (31). First, these cells appear to migrate normally when traced with a vital dye. Second, the number of xanthophores, which are derived from the same migratory neural crest derivatives as melanophores, is not markedly changed in cavefish. Third, when the cavefish neural tube is cultured in vitro, pigment-like cells migrate from the explant. Thus, melanophore reduction is not due to the absence of pigment cell precursors or effects on their ability to select the proper migration pathway. Instead, specific changes are likely to occur in the sequence of differentiation events after the divergence of pigment cell types from their common neural crest precursors.
GENETIC BASIS OF PIGMENTATION REGRESSION
Genetic crossing experiments indicate that pigment regression in Astyanax is a polygenic trait (50, 61). Similar to eyes, in some cases complementation tests result in F1 hybrids with greater pigmentation (61, 64), which is suggestive of different genes, whereas in others there is noncomplementation, indicating that the same genes are involved (40, 64) (Figure 7c,d). Recently, 18 different QTL have been identified that affect the amount and quality of melanophore-based pigmentation in Pachón cavefish (41, 42). Among the many QTL and predicted genes that define aspects of melanophore and pigment reduction, there are two relatively simple traits whose underlying genes and mutations have been identified and characterized: the albinism gene (49) and the so-called brown gene (50).
Albinism and the OCA2 Gene
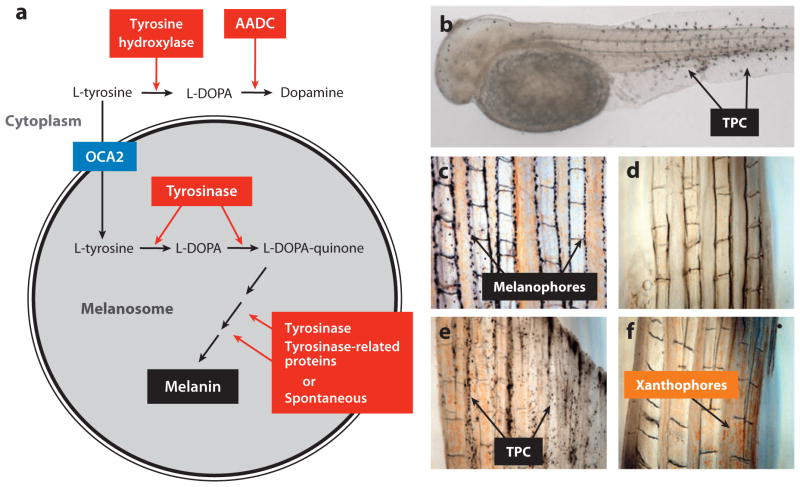
Albinism, the absence of melanin pigment, is predominant in Pachón and Molino cavefish. The melanin synthesis pathway is well known (Figure 10a). It involves the transport of L-tyrosine substrate into the melanosome, the conversion of L-tyrosine to L-DOPA and L-DOPA to L-DOPAquinone by the rate limiting enzyme tyrosinase, and a series of downstream reactions, some catalyzed by tyrosinase or tyrosinase-related proteins, and others that occur spontaneously, to produce black melanin pigment. The steps in this pathway were examined in Pachón cavefish to determine the lesion in melanin biosynthesis (31). The provision of exogenous L-DOPA to fixed specimens resulted in the deposition of melanin in otherwise colorless melanophores, indicating that tyrosinase is present and functional (Figure 10d–e). The same approach resulted in the detection of tyrosinase in colorless melanophores in Chica, Los Sabinos, Tinaja, and Curva cavefish, explaining why they appear to have fewer normally differentiated melanophores (31). Because of the presence of active tyrosinase in Pachón cavefish, L-tyrosine would also be expected to be converted to L-DOPA, but this is not the case: Provision of excess L-tyrosine to fixed specimens instead of L-DOPA did not result in melanin production. Thus, melanogenesis appears to be blocked at the step in which L-tyrosine is transferred into the melanosome.
Figure 10.
Cavefish tyrosinase-positive albinism. (a) A summary of steps in which L-tyrosine is converted to melanin in the melanosome and dopamine in the cytoplasm showing the key roles of tyrosinase, other enzymes, and the presumed role of OCA2 as an L-tyrosine transporter. (b) Melanophores revealed in an albino Pachón cavefish embryo after conversion of exogenous L-DOPA to melanin by active tyrosinase. TPC: tyrosinase positive cells (melanophores). (c,d) Explants of surface fish (c) albino Pachón adult tail fin (d) showing melanophores in the former but not the latter. (e) Albino Pachón adult tail fin explant in which tyrosinase positive cells are revealed by L-DOPA treatment and melanin deposition. (f) Albino Pachón adult tail fin explant showing inability to convert exogenous L-tyrosine to melanin. The presence of xanthophores is also indicated in albino Pachón adult tail fin explants. (a–f) from McCauley et al. (2004).
Early genetic studies showed that albinism is controlled by a single recessive gene in Pachón cavefish (49). Accordingly, linkage analysis in surface fish x Pachón or Molino pedigrees located a single albinism QTL(oca2) on the Astyanax microsatellite map (40) (Figure 9b). The albinism gene mapped to the same position in both cavefish populations. This result could be explained by the same mutation in the same gene, different mutations in the same gene, or mutations in different but very closely linked genes. A complementation test was performed between Pachón and Molino cavefish to address this issue (40). Albino progeny were obtained in the F1 generation (Figure 7c), indicating that the same gene is responsible for albinism in both cavefish populations. A candidate gene approach, in which several genes regulating the melanogenesis cascade were placed on the microsatellite map, showed that the gene responsible for albinism is oculocutaneous albinism2 (oca2), which encodes a putative 12-pass melanosome membrane protein. Mutations in oca2 are also responsible for the most frequently observed human albinisms (25) and cause albinism in mice, in which the mutant gene was originally named pink-eyed dilution (47). In humans, oca2 is a very large gene, consisting of 24 exons spanning 345 kb (36).
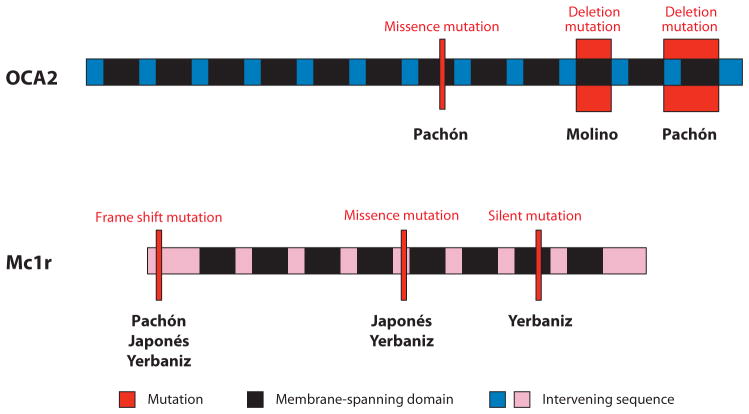
The molecular basis of the hypomorphic albinism phenotype was determined by identifying cavefish oca2 mutations (40). Three differences from surface fish were discovered in Pachón oca2: two point mutations resulting in conserved amino acid substitutions and a large deletion extending from within intron 23 through most of exon 24. Because of this deletion, the Pachón OCA2 protein would contain only a part of intron 23 and lack most of exon 24 (Figure 11). In Molino cavefish, there was a single change, another large deletion encompassing exon 21 that would significantly shorten the OCA2 protein (Figure 11). Both deletions are in regions predicted to be parts of membrane spanning domains and expected to cause loss of function. To identify the mutations responsible for albinism, DNA constructs containing wild-type surface fish oca2 and copies of cavefish oca2 individually containing each of the point and deletion mutations were prepared and introduced into a mouse OCA2-deficient melanocyte cell line (40). The surface fish oca2 DNA construct and two Pachón cavefish oca2 DNA constructs with amino acid polymorphisms were able to rescue melanogenesis, indicating that the corresponding point mutations are neutral. In contrast, DNA constructs containing the large deletions found in Pachón and Molino oca2 could not rescue melanin synthesis, implying that they are the mutations responsible for oca2 loss of function.
Figure 11.
Diagrams indicating the approximate positions (red bars) and types of mutations in genes encoding the OCA2 and Mc1r proteins in different cavefish populations. In OCA2, black regions indicate the 12 membrane spanning domains indicate and blue regions indicate the intervening protein sequences. In Mc1r, black regions indicate the seven membrane spanning domains indicate and pink regions indicate the intervening protein sequences.
The oca2 gene appears to be responsible for loss of melanin pigment in two cavefish populations, but the mutations are distinct, implying that Pachón and Molino evolved the albinism phenotype independently. There is also evidence that albino Japonés cavefish have evolved albinism via changes in the oca2 gene (40). However, no differences could be detected in the oca2 coding region sequence, suggesting that a regulatory region may be mutated in Japonés cavefish.
The Brown Phenotype and the MC1R Gene
The brown phenotype is another simple trait affecting body and eye pigmentation in Chica, Los Sabinos, Pachón, Piedras, Japonés, and Yerbaniz cavefish (17, 50, 64). The mutant allele responsible for this trait appears to be absent in Molino, a further indication of separate origins for pigment loss. The brown phenotype decreases the number of body melanophores and changes eye pigmentation from black to brown by reducing melanin synthesis. Complementation tests indicate that the same gene is affected to produce the brown phenotype in different cavefish populations (64). Segregation analysis in crosses of surface fish and Pachón cavefish predicted that a single recessive gene is responsible for this trait. Consistent with this idea, linkage analysis of a Pachón x surface fish cross has revealed a single QTL, and no QTLs were detected in a Molino x surface fish pedigree (17). To determine the identity of the brown locus within the QTL, the relevant Astyanax linkage group was anchored to the zebrafish physical genome, and a candidate gene approach based on genes with known melanogenic activity was applied. This analysis recognized only two of several candidate genes, transient receptor potential melastatin (trpm7) and melanocortin 1 receptor (Mc1r), within the QTL. When these genes were placed on the map, trpm7 mapped outside the QTL, whereas Mc1r was positioned in the middle of the QTL, making it the strongest candidate for the brown gene.
The Mc1r gene encodes a receptor for the melanogenesis regulating ligand MSHα (45). Mc1r activation after ligand binding leads to stimulation of downstream effectors, including adenyl cyclase and the target gene mitf, which controls pigmentation. The Mc1r protein has an N-terminal domain, seven transmembrane domains, and a carboxyl terminal domain. The potential role of Mc1r in the brown phenotype was determined by sequencing the gene in several different cavefish populations followed by functional analysis (17). In Pachón, a 2-bp deletion was detected at the extreme 5′ end of the transcript, which would introduce a frame shift and premature stop codon in the translated protein prior to the transmembrane domain (Figure 11). The same frame-shift mutation was revealed in Yerbaniz and Japonés cavefish but in heterozygous form. In addition, a missense mutation changing an arginine to a cysteine residue was discovered in these two cavefish populations, and a silent mutation was uncovered in Yerbaniz. No sequence differences in the Mc1r coding region have been detected in Piedras or Curva cavefish, implying that the relevant mutations may be in gene regulatory regions (17). Functional analysis was conducted in zebrafish. Astyanax Mc1r DNA constructs containing the wild-type surface fish allele, the Pachón deletion mutant allele, or the Yerbaniz missence mutant allele were introduced into zebrafish embryos whose endogeneous functional Mc1r was knocked down with a specific morpholino (17). The Astyanax surface fish Mc1r allele rescued pigmentation, but neither of the cavefish mutant alleles did so, suggesting that these changes account for loss of function in the two cavefish populations. Mutations in the Mc1r gene, including arginine to cysteine transitions at the same position, are also known as the source of pigment modification in a wide range of vertebrates, including humans (45). Therefore, Mc1r is another example of a single gene that is repeatedly targeted by different mutations to change a complex phenotype during parallel or convergent evolution.
REGRESSIVE EVOLUTION OF PIGMENTATION
Pigment regression has at least three causes in Astyanax cavefish. First, the predominantly albino Pachón and Molino cavefish have accumulated different mutations in the oca2 gene, causing defective melanin synthesis. Second, some cavefish populations, including some of those with albinism, have also accumulated different mutations in the Mc1r gene causing reduced numbers of melanophores and melanin pigmentation due to defective MSHα signaling. The oca2 and Mc1r genes are probably epistatic in Astyanax, as they are in humans (26), but it is impossible to tell which of them mutated first in the cave populations where they coexist. Third, mutations in additional genes that remain to be identified also contribute to the reduction of melanophores. Thus, certain genes are repeatedly targeted for mutations leading to evolutionary changes in cavefish pigmentation. Why is this? For oca2, the answer may be related to properties of the gene itself or its potentially restricted function in melanogenesis. One is its very large size (36), which might be expected to render it a more frequent target for mutation. Another may be its chromosomal position, which is in a region associated with rearrangements and deletions in humans (70). Finally, oca2 could be a nonpleiotropic gene with no function other than pigmentation, allowing it to be targeted for mutation without affecting other traits. The reasons for repetitive mutation in Mc1r are less clear. In mice and humans, the MSHα-Mc1r signaling system may have critical physiological functions other than control of pigmentation (34), indicating that it may not be a simple nonpleiotropic gene. Further information about the genetic basis of pigmentation regression in cavefish may be revealed by clarifying the identity and function of the other genes involved in loss of pigmentation.
EVOLUTIONARY FORCES
As described above, Darwin saw difficulties in explaining the regressive features of cave animals by natural selection. Thus, one of the most interesting questions in the evolutionary biology of cave animals is whether natural selection has a role in the evolution of regressive traits, and if not, what are the alternatives? There are two major competing hypotheses to explain regressive evolution in Astyanax cavefish and other cave animals (9). The first hypothesis is that natural selection can account for these losses because they are beneficial for life in the cave environment. The competing hypothesis is neutral mutation and genetic drift: Genes controlling the development of regressive features accumulate mutations, which eventually result in loss of function and disappearance of the trait. Genetic analysis has shown that the regression in eyes and pigmentation occur independently (61), and thus these two recessive traits could be controlled by different evolutionary mechanisms. It should be emphasized that the current data does not allow us to conclude unequivocally that either selection or neutral mutation is responsible for regressive evolution. Although neither hypothesis has been proved in the case of Astyanax cavefish, the developmental and genetic studies described above generally support one or the other hypothesis.
Developmental and genetic information seem to support selection over neutral mutation as an explanation for eye loss. First, the genes involved in eye development that have been identified thus far do not appear to have mutated to a degree in which they have lost function. For example, the restoration of eyes by lens transplantation suggests that genes operating downstream of lens function are present and potentially active in cavefish (23, 66). Second, also supportive of selection is that most genes with modified expression patterns, such as those in the Shh signaling pathway and hsp90α, increase rather than decrease their activity in cavefish (18, 67). However, until the genes and mutations that are directly involved in eye degeneration are identified and fully characterized, we must be cautious about concluding that neutral mutations have not occurred in eye forming genes. Third, probably the strongest evidence currently available supporting a role for natural selection in eye loss is the polarity of the many eye QTL, all of which decrease the extent of eye development (41). If neutral mutation and genetic drift were indeed operating, one might expect some eye QTL to define increases as well as decreases in eye formation.
The benefits of eye loss in the cave environment are also still a mystery but some reasonable ideas have been suggested. First, a benefit of eye loss may be energy conservation, as it takes considerable effort to sustain an eye, in particular the high cost of maintaining a retina (41). Thus, there might be an advantage in casting aside this nonessential burden during life in darkness. Another idea is that the genes involved in eye development are pleiotropic and have selectable effects on constructive traits. For example, shh is known to have possible effects on taste bud and forebrain development, which are enhanced in cavefish, as well as negative effects on eye development (32, 46, 67, 69). Therefore, enhancement of these constructive traits might only be possible at the expense of eye development. Also relevant to the pleiotropy hypothesis, many QTL involved in constructive and regressive features co-map in genetic linkage analysis, particularly some of the eye and taste bud QTL (42). The role of pleiotropy in eye regression may be clarified when we understand more about the genes directly involved in eye degeneration and their roles in development.
Our current understanding of the evolutionary mechanisms causing pigment regression suggests that neutral mutations are more likely than natural selection to control this regressive trait. Recent genetic analysis shows that individual QTL governing the extent of pigmentation can either increase or decrease melanophore abundance, supporting the role of neutral mutation and genetic drift (41). However, we could also entertain a model in which selection could operate to control changes in pigmentation. For example, in the case of albinism it might be advantageous to block melanin formation at the level of L-tyrosine conversion to L-DOPA because this could shunt more L- tyrosine substrate into the branch pathway in which specialized neural cells produce the neural transmitter L-dopamine (Figure 10a), a possible prerequisite for behavioral changes in cavefish. The relationships between neutral mutation and selection are very poorly understood in evolutionary biology, and further research on Astyanax cavefish may clarify them.
CONCLUSIONS
At present, a single model system, the cavefish Astyanax mexicanus, has been developed to study the evolution of cave-related phenotypes. Based on the evidence described above, the following sequence of events may have occured during Astyanax cavefish evolution. First, surface fish colonized (or were entrapped in) caves, and their differential survival led to the formation of cave-adapted populations in which regressive evolution could have been initiated. Second, the established cavefish populations spread horizontally and vertically underground, in some cases for relatively long distances, whilst continuing regressive evolution. Third, the dispersed cavefish populations were isolated from each other in the caves in which they are now located, and further regressive evolution occurred. As a result of these events, many individual cavefish populations evolved. Some of these cavefish populations share genes and mutations responsible for regressive evolution with other populations, whereas other populations probably contain unique genes responsible for regressive evolution. The final phenotypes are similar (although not identical), suggesting that there are many different ways to produce the same phenotypes. The next frontier in cavefish research will be to identify more of the genes and mutations involved in regressive evolution, especially those controlling the loss of eyes and reduction of melanophores, and from this information learn more about the forces that drove the evolution of these regressive phenotypes during adaptation of cavefish to perpetual darkness.
SUMMARY POINTS.
Astyanax cavefish have originated independently several different times from a surface fish ancestor. As expected, the founder cavefish populations have undergone eye and pigmentation regression independently. In addition, it is possible that multiple episodes of regressive evolution occurred after a single colonization event.
Developmental analysis indicates that lens apoptosis plays a major role in eye regression via discarding its normal protective effect on the retina. Other defects in the cavefish eye may collaborate with the lens to cause eye degeneration.
Genetic analysis indicates that cavefish eye degeneration is a multigenic trait, and consistent with the conclusion of independent regressive events, some of the genes involved are different in individual cavefish populations.
Candidate gene analysis has earmarked several genes for a role in eye degeneration, including the proapoptotic factor Hsp90α, the antiapoptotic factor αA-crystallin, and the midline signal Shh. These genes induce lens apoptosis when their expression is modified in cavefish.
Genetic linkage analysis has identified 12 QTL that are directly involved in eye degeneration. The responsible genes in these QTL are just beginning to be identified.
The reduction or loss of pigmentation is caused by defects in melanophore development downstream of the point in which neural crest-derived progenitors diverge into different types of pigment cells.
Genetic analysis indicates that melanophore reduction is a polygenic trait but includes individual genes that are inherited by a simple Mendelian basis. The albinism gene is oca2. MC1r is a gene involved in the reduction of melanophore number and melanin synthesis.
Current evidence suggests that natural selection may be involved in regressive evolution of eye development, whereas neutral mutation and drift is the best explanation for regressive evolution of pigmentation.
Acknowledgments
While writing this review, I was supported by grants from the US National Institutes of Health (EYO14619) and the National Science Foundation (IBN-0611529). I thank Boris Sket, Maja Zagmajster, Katie Schneider, and Jim Krest for generously allowing me to use some of the photographs in Figure 1.
Glossary
- Regressive evolution
the loss of an ancestral trait during evolution
- Parallel evolution
independent evolution of similar traits in two or more species or genetically diverging populations of the same species descended from a common ancestor
- RAPD
DNA sequence amplified by polymerase chain reaction using random sets of primers
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- RPE
retinal pigment epithelium
- CMZ
ciliary marginal zone of the retina
- Quantitative trait locus (QTL)
a chromosomal region defined by genetic linkage that usually controls a small portion of an additive phenotypic trait
- Candidate gene
gene likely to be involved in a process based on characteristics such as temporal and spatial expression pattern or broad function
- Convergent evolution
independent evolution of the same or similar traits in two or more species lacking a recent common ancestor
- Pleiotropy
the circumstance in which a single gene controls two or more phenotypic traits that may seem unrelated
- Multigenic trait
a phenotypic trait controlled by more than one gene and usually many genes
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alunni A, Menuet A, Candal E, Pénigault J-B, Jeffery WR, Rétaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Comp Neurol. 2007;505:221–33. doi: 10.1002/cne.21488. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez J. Revisión del generero Anoptichthys con descriptión de una especies nueva (Pisc. Characidae) An Esc Nac Cien Biol Mexico. 1946;4:263–82. [Google Scholar]
- 3.Alvarez J. Descriptión de Anoptichthys hubbsi caracinído ciego de La Cueva de Los Sabinos, S. L. P. Rev Soc Mexicana Hist Nat. 1947;8:215–19. [Google Scholar]
- 4.Behrens M, Wilkens H, Schmale H. Cloning of the αA-crystallin genes of the blind cave form and the epigean form of Astyanax fasciatus: a comparative analysis of structure, expression and evolutionary conservation. Gene. 1998;216:319–26. doi: 10.1016/s0378-1119(98)00346-1. [DOI] [PubMed] [Google Scholar]
- 5.Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 6.Borowsky R, Wilkens H. Mapping a cavefish genome: polygenic systems and regressive evolution. J Hered. 2002;93:19–21. doi: 10.1093/jhered/93.1.19. [DOI] [PubMed] [Google Scholar]
- 7.Cahn PH. Comparative optic development in Astyanax mexicanus and two of its blind cave derivatives. Bull Am Mus Nat Hist. 1958;115:75–112. [Google Scholar]
- 8.Culver DC, Pipan T. The Biology of Caves and Other Subterranean Habitats. Oxford UK: Oxford Univ. Press; 2009. [Google Scholar]
- 9.Culver DC, Wilkens H. Critical review of the relevant theories of the evolution of subterranean animals. In: Wilkens H, Culver DC, Humphreys WF, editors. Ecosystems of the World. Vol. 30. Amsterdam: Elsevier; 2000. pp. 381–98. [Google Scholar]
- 10.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. 6. London: John Murray; 1872. [PMC free article] [PubMed] [Google Scholar]
- 11.Dowling TE, Martasian DP, Jeffery WR. Evidence for multiple genetic lineages with similar eyeless phenotypes in the blind cavefish, Astyanax mexicanus. Mol Biol Evol. 2002;19:446–55. doi: 10.1093/oxfordjournals.molbev.a004100. [DOI] [PubMed] [Google Scholar]
- 12.Ekker SC, Ungar AR, von Greenstein P, Porter JA, Moon RT, Beachy P. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr Biol. 1995;5:944–55. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- 13.Espinasa L, Borowsky RB. Origin and relationships of cave populations of the blind Mexican tetra, Astyanax fasciatus, in the Sierra de El Abra. Environ Biol Fishes. 2001;62:233–37. [Google Scholar]
- 14.Espinasa L, Rivas-Manzano P, Espinosa Pérez H. A new blind cave fish population of the genus Astyanax: geography, morphology and behavior. Environ Biol Fishes. 2001;62:329–44. [Google Scholar]
- 15.Fairbank PD, Lee C, Ellis A, Hildebrand JD, Gross JM, Wallingford JB. Shroom2 (APXL) regulates melanosome biogenesis and localization in the retinal epithelium. Development. 2006;133:4109–18. doi: 10.1242/dev.02563. [DOI] [PubMed] [Google Scholar]
- 16.Gross JB, Protas M, Conrad M, Scheid PE, Vidal O, et al. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc Natl Acad Sci USA. 2008;105:20106–11. doi: 10.1073/pnas.0806238105. Illustrates the linkage between the Astyanax genetic map and the zebrafish physical genome map. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5(1):e1000326. doi: 10.1371/journal.pgen.1000326. Identifies MC1r as the gene underlying the brown phenotype, which reduces number of melanophores and amount of melanin in several cavefish populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooven TA, Yamamoto Y, Jeffery WR. Blind cavefish and heat shock protein chaperones: a novel role for hsp90α in lens apoptosis. Int J Dev Biol. 2004;48:731–38. doi: 10.1387/ijdb.041874th. [DOI] [PubMed] [Google Scholar]
- 19.Hubbs CL, Innis WT. The first known blind fish of the family Characidae: a new genus from Mexico. Occas Papers Mus Zool Univ Michigan. 1936;342:1–7. [Google Scholar]
- 20.Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- 21.Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: apoptosis and the Pax-6 gene. Amer Zool. 1998;38:685–96. [Google Scholar]
- 22.Jeffery WR, Strickler AG, Guiney S, Heyser D, Tomarev SI. Prox1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000;210:223–30. doi: 10.1007/s004270050308. [DOI] [PubMed] [Google Scholar]
- 23.Jeffery WR, Strickler AG, Yamamoto Y. To see or not to see: evolution of eye degeneration in Mexican blind cavefish. Comp Int Biol. 2003;43:531–41. doi: 10.1093/icb/43.4.531. [DOI] [PubMed] [Google Scholar]
- 24.Johns P. Growth of the adult goldfish eye III. Source of the new retinal cells. J Comp Neurol. 1977;176:348–58. doi: 10.1002/cne.901760304. [DOI] [PubMed] [Google Scholar]
- 25.King RA, Pietsch J, Fryer JP, Savage S, Brott MJ, et al. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): definition of the phenotype. Hum Genet. 2003;113:502–13. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- 26.King RA, Willaert RK, Schmidt RM, Pietsch J, Savage S, et al. MC1R mutations modify the class phenotypes of oculocutaneous albinism type 2 (OCA2) Am J Hum Genet. 2003;73:638–45. doi: 10.1086/377569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurita R, Sagara H, Aoki Y, Link BA, Arai K, Watanabe S. Suppression of lens growth by αA-crystallin promoter driven diptheria toxin results in disruption of retinal cell organization in zebrafish. Dev Biol. 2003;255:113–27. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- 28.Kusakabe T, Swalla BJ, Satoh N, Jeffery WR. Mechanism of an evolutionary change in muscle cell differentiation in ascidians with different modes of development. Dev Biol. 1996;174:379–92. doi: 10.1006/dbio.1996.0082. [DOI] [PubMed] [Google Scholar]
- 29.Langecker TG, Schmale H, Wilkens H. Transcription of the opsin gene in degenerate eyes of cave dwelling Astyanax fasciatus (Teleostei, Characidae) and its conspecific ancestor during early ontogeny. Cell Tiss Res. 1993;273:183–92. [Google Scholar]
- 30.Macdonald R, Anukampa Barth K, Xu Q, Holder N, Mikkola I, Wilson S. Midline signaling is required for Pax6 gene regulation and patterning of the eyes. Development. 1995;121:3267–78. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- 31.McCauley DW, Hixon E, Jeffery WR. Evolution of pigment cell regression in the cavefish Astyanax: a late step in melanogenesis. Evol Dev. 2004;6:209–18. doi: 10.1111/j.1525-142X.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 32.Menuet A, Alunni A, Joly J-S, Jeffery WR, Rétaux S. Shh overexpression in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development. 2006;134:845–55. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell RW, Russell WH, Elliot WR. Mexican eyeless characin fishes, genus Astyanax: environment, distribution, and evolution. Spec Publ Mus Texas Tech Univ. 1977;12:1–89. [Google Scholar]
- 34.Mogil JS, Ritchie J, Smith SB, Staaburg K, Kaplan L, et al. Melanocortin-1 receptor gene variants affect pain and muopiod analgesia in mice and humans. J Med Genet. 2005;42:583–87. doi: 10.1136/jmg.2004.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moritz OL, Molday RS. Molecular cloning, membrane topology, and localization of bovine rom-1 in rod and cone photoreceptor cells. Invest Ophthalmol Vis Sci. 1996;37:352–62. [PubMed] [Google Scholar]
- 36.Oetting WS, Garrett SS, Brott M, King RA. P gene mutations associated with oculocutaneous albinism type II (OCA2) Hum Mut. 2005;25:323. doi: 10.1002/humu.9318. [DOI] [PubMed] [Google Scholar]
- 37.Otteson DC, D’Costa AR, Hitchcock RF. Putative stem cells and the lineage of rod photoreceptors in the mature retina of the goldfish. Dev Biol. 2001;232:62–76. doi: 10.1006/dbio.2001.0163. [DOI] [PubMed] [Google Scholar]
- 38.Poulson TL, White WB. The cave environment. Science. 1969;165:971–81. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]
- 39.Protas M, Patel NH. Evolution of color patterns. Annu Rev Cell Dev Biol. 2008;24:425–46. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- 40.Protas M, Hersey E, Kochanek C, Zhou Y, Wilkens H, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–11. doi: 10.1038/ng1700. Identifies oca2 as the single gene responsible for the cavefish albinism phenotype. [DOI] [PubMed] [Google Scholar]
- 41.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–54. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Protas M, Tabansky I, Conrad M, Gross JB, Vidal O, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- 43.Proudlove G. Subterranean Fishes of the World. Moulis, France: International Society for Subterranean Biology; 2006. [Google Scholar]
- 44.Rasquin P. Progressive pigmentary regression in fishes associated with cave environments. Zoologica. 1947;32:35–44. [PubMed] [Google Scholar]
- 45.Rees JL. Genetics of hair and skin color. Annu Rev Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- 46.Rétaux S, Pottin K, Alunni A. Shh and forebrain evolution in the blind cavefish Astyanax mexicanus. Biol Cell. 2008;100:139–47. doi: 10.1042/BC20070084. [DOI] [PubMed] [Google Scholar]
- 47.Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, et al. A gene for mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Proc Natl Acad Sci USA. 1993;91:12071–75. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 48.Sadoglu P. Mendelian inheritance in hybrids between the Mexican blind fish and their overground ancestors. Verh Dtsch Zool Ges. 1957;1957:432–39. [Google Scholar]
- 49.Sadoglu P. A Mendelian gene for albinism in natural cave fish. Experientia. 1957;13:394. [Google Scholar]
- 50.Sadoglu P, McKee A. A second gene that affects eye and body color in Mexican blind cavefish. J Hered. 1969;60:10–14. doi: 10.1093/oxfordjournals.jhered.a107917. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz M, Cecconi F, Berneir G, Andrejewski N, Kammandel B, et al. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–34. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- 52.Soares D, Yamamoto Y, Strickler AG, Jeffery WR. The lens has a specific influence on optic nerve and tectum development in the blind cavefish Astyanax. Dev Neurosci. 2004;26:308–17. doi: 10.1159/000082272. [DOI] [PubMed] [Google Scholar]
- 53.Strecker U, Bernachez L, Wilkens H. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei) Mol Ecol. 2003;12:699–710. doi: 10.1046/j.1365-294x.2003.01753.x. Provides evidence that cavefish were derived from surface fish ancestors several different times in northeastern Mexico. [DOI] [PubMed] [Google Scholar]
- 54.Strecker U, Faúndez VH, Wilkens H. Phylogeography of surface and cave Astyanax (Teleostei) from Central and North America based on cytochrome b sequence data. Mol Phylogenet Evol. 2004;33:469–81. doi: 10.1016/j.ympev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Strickler AG, Jeffery WR. Differentially expressed genes identified by cross-species microarray in the blind cavefish Astyanax. Int Zool. 2009;4:31–40. doi: 10.1111/j.1749-4877.2008.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strickler AG, Yamamoto Y, Jeffery WR. Early and late changes in Pax6 expression accompany eye degeneration during cavefish development. Dev Genes Evol. 2001;211:138–44. doi: 10.1007/s004270000123. [DOI] [PubMed] [Google Scholar]
- 57.Strickler AG, Famuditimi K, Jeffery WR. Retinal homeobox genes and the role of cell proliferation in cavefish eye degeneration. Int J Dev Biol. 2002;46:285–94. [PubMed] [Google Scholar]
- 58.Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina: evidence from the blind cavefish. Astyanax Dev Biol. 2007;311:512–23. doi: 10.1016/j.ydbio.2007.08.050. Establishes protective effect of lens over the retina and shows that its loss is a reason for eye degeneration in cavefish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone αA-crystallin during cavefish eye degeneration. Dev Genes Evol. 2007;217:771–82. doi: 10.1007/s00427-007-0190-z. [DOI] [PubMed] [Google Scholar]
- 60.Wilkens H. Genetic interpretation of regressive evolutionary processes: studies of hybrid eyes of two Astyanax cave populations (Characidae, Pisces) Evolution. 1971;25:530–44. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 61.Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces) Evol Biol. 1988;23:271–367. [Google Scholar]
- 62.Wilkens H. Regressive evolution: ontogeny and genetics of cavefish eye rudimentation. Biol J Linn Soc. 2007;92:287–96. [Google Scholar]
- 63.Wilkens H, Burns RL. A new Anoptichthys cave population (Characidae, Pisces) Ann Spéléol. 1972;27:263–70. [Google Scholar]
- 64.Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Charcidae, Telesotei): genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc. 2003;80:545–54. Describes complementation tests showing whether genes underlying eye and pigment regressive are the same or different in various cavefish populations. [Google Scholar]
- 65.Yamamoto Y. Cavefish. Curr Biol. 2004;14:R943. doi: 10.1016/j.cub.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto Y, Jeffery WR. Central role for the lens in cavefish eye degeneration. Science. 2000;289:631–33. doi: 10.1126/science.289.5479.631. Establishes a major role for lens apoptosis in cavefish eye degeneration using embryonic lens transplantation. [DOI] [PubMed] [Google Scholar]
- 67.Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–47. doi: 10.1038/nature02864. Illustrates that overexpression of Shh along the cavefish midline causes eye degeneration by inducing lens apoptosis. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye-dependent and -independent processes in the cavefish Astyanax. Evol Dev. 2003;5:435–46. doi: 10.1046/j.1525-142x.2003.03050.x. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009;330:200–11. doi: 10.1016/j.ydbio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi Z, Garrison N, Cohen-Barak O, Karafet TM, King RA, et al. A 122.5 kilobase deletion of the P gene underlies the high prevalence of occulocutaneous albinism type 2 in the Navajo population. Am J Hum Genet. 2003;72:62–72. doi: 10.1086/345380. [DOI] [PMC free article] [PubMed] [Google Scholar]