Abstract
Escherichia coli and Proteus mirabilis are important urinary tract pathogens. The constant increase in the antibiotic resistance of clinical bacterial strains has become an important clinical problem. The aim of this study was to compare the antibiotic resistance of 141 clinical (Sweden and Poland) and 42 laboratory (Czech Republic) P. mirabilis strains and 129 clinical (Poland) uropathogenic E. coli strains. The proportion of unique versus diverse patterns in Swedish clinical and laboratory P. mirabilis strain collections was comparable. Notably, a similar proportion of unique versus diverse patterns was observed in Polish clinical P. mirabilis and E. coli strain collections. Mathematical models of the antibiotic resistance of E. coli and P. mirabilis strains based on Kohonen networks and association analysis are presented. In contrast to the three clinical strain collections, which revealed complex associations with the antibiotics tested, laboratory P. mirabilis strains provided simple antibiotic association diagrams. The monitoring of antibiotic resistance patterns of clinical E. coli and P. mirabilis strains plays an important role in the treatment procedures for urinary tract infections and is important in the context of the spreading drug resistance in uropathogenic strain populations. The adaptability and flexibility of the genomes of E. coli and P. mirabilis strains are discussed.
Keywords: Proteus mirabilis, Escherichia coli, UTI, Antibiotic resistance, Association analysis
Introduction
Human urinary tract infections (UTIs) are among the most common bacterial diseases [1, 2]. Persistent recurrences and asymptomatic infections are responsible for the difficult treatment of UTIs. This is mostly due to the presence of uropathogenic Escherichia coli and Proteus mirabilis strains in the urinary tract. E. coli accounts for 80 % of all bacteria isolated from the urine. P. mirabilis strains cause 10 % of uncomplicated urinary tract infections [3]. They are the fifth most common cause of nosocomial urinary tract infections and sepsis in hospitalized individuals [4, 5]. E. coli strains represent many different intestinal and extraintestinal pathotypes (IPEC and ExPEC, respectively) that are responsible for numerous diseases. Uropathogenic E. coli strains (UPECs) are ExPEC and constitute the most distinct phylogenetic pathotype among E. coli [6]. Importantly, uropathogenic P. mirabilis and E. coli strains may also manifest resistance to several antimicrobial agents, including extended-spectrum cephalosporins, fluoroquinolones, and aminoglycosides [7–9]. The increase in drug resistance has become a serious problem in effective antibiotic administration [10].
Although the molecular mechanisms of bacterial antibiotic resistance are known, they remain the object of studies worldwide. Several mechanisms explaining the emergence of drug resistance have been discovered in the past decades, the best known being beta-lactamase and quinolone resistance mechanisms. The presence of beta-lactamases and complex “efflux pumps” is considered to be the mechanism of beta-lactam resistance [7]. Beta-lactamases are enzymes that break the beta-lactam ring and deactivate this class of antimicrobial drugs [11]. Beta-lactamases are divided into four molecular classes (A, B, C, and D). Their molecular homology is diverse, and they do not seem to have one common ancestor. The presence of a serine residue in the active center is typical of A, C, and D classes, while in class B beta-lactamases, zinc ions are required. Beta-lactamases are encoded by either chromosomes or plasmids. The highly mobile nature of beta-lactamase genes remains an important problem in UTI treatment [8, 10].
The complex characteristics of bacterial antibiotic resistance may be analyzed by mathematical methods to model the dynamics of this process and anticipate its development [12, 13]. There is a need to create a tool for the development of a strategy against the spread of multi-drug-resistant (MDR) strains. In this work, the Kohonen network method was used to identify similar groups of antibiotics that were reactive against the investigated strains. To formulate a hypothesis about the dynamics of the resistance patterns, an association analysis of chosen antibiotics was performed for the investigated collections of strains [14].
The drug resistance profiles of E. coli strains and P. mirabilis strains were analyzed. One of the tasks was to compare the antibiotic resistance patterns of P. mirabilis and E. coli strains isolated from UTI patients with those of P. mirabilis strain collections stored for many years in the laboratory. Finally, a mathematical analysis of the antibiotic resistance patterns of P. mirabilis and E. coli strains was carried out to identify their correlation with virulence profiles.
Materials and methods
Bacterial strain collections
In this study, the following bacterial strains of E. coli and P. mirabilis were used:
A collection of 129 clinical E. coli strains isolated from the urine of patients in different wards of Military Teaching Hospital No. 2, Medical University of Lodz, Poland, in 2005–2007.
A collection of 43 clinical P. mirabilis strains isolated from the urine of patients of the Swietokrzyskie Oncology Center in Kielce, Poland, in 2002.
A collection of 99 clinical isolates of P. mirabilis strains collected at the Department of Clinical Microbiology of the Karolinska Hospital in Stockholm, Sweden, between October 1999 and January 2000. These strains were isolated from UTI patients and sub-cultured four times prior to the study.
A collection of 42 laboratory P. mirabilis strains from the Czech National Collection of Type Cultures from the Institute of Epidemiology and Microbiology, Prague, Czech Republic. These strains were stored for more than 20 years and sub-cultured at least 20 times.
For everyday work, the clinical and laboratory strains were stored at 4 °C in bacterial media, and for longer storage, they were stored in glycerol stock solutions at −70 °C.
Bacterial identification and cultivation media
Escherichia coli and P. mirabilis strains were identified based on their differential growth on CPS3 medium (bioMerieux). UTI cases were confirmed by the presence of >104 cfu/ml of bacteria in a urine sample. All of the strains were grown at 37 °C on LB plates for further tests.
Antibiotic susceptibility testing
Proteus mirabilis strains were subjected to an in vitro antimicrobial susceptibility disc diffusion test according to the guidelines of the National Committee for Clinical Laboratory Standards [15]. A standardized inoculum of bacteria (0.5 McFarland standard, 1.5 × 108 cfu/ml) was swabbed onto the surface of Mueller–Hinton agar (MHA) plates (Difco Laboratories, Detroit MI, USA). Filter paper discs impregnated with antimicrobial agents were placed on the agar surface. After 16–18 h of incubation at 35 °C, the diameter of the inhibition zone around each disc was measured, and these measurements were compared with the NCCLS disc diffusion tables [15].
The drug resistance of E. coli strains was determined for enterobacteria of urinary origin by a susceptibility test (ATB UR5, bioMerieux) at the Faculty of Laboratory Diagnostics and Clinical Biochemistry, Military Teaching Hospital No. 2, Medical University of Lodz, Poland.
Bacterial isolates were determined to be sensitive (S), moderately sensitive (M), or resistant (R) to the antimicrobial agents tested.
Antimicrobial agents
Antimicrobial discs with ampicillin (AP) 10 μg, cotrimoxazole (trimethoprim/sulfamethoxazole) (TS) 1.25/27.75 μg, nitrofurantoin (NI) 30 μg, norfloxacin 10 μg, carbenicillin (PY) 100 μg, ofloxacin (OFX) 5 μg, tetracycline (T) 30 μg, amoxicillin/clavulanate (AUG) 20/10 μg, ciprofloxacin (CIP) 5 μg, amikacin (AK) 30 μg, aztreonam (ATM) 30 μg, cefuroxime (CXM) 30 μg, imipenem (IMI) 10 μg, polymyxin B (PB) 300 μg, and colistin sulfate (CO) 100 μg (Mast Diagnostics, Mast Group Ltd., Merseyside, UK) were used on the studied collections of P. mirabilis strains.
Escherichia coli strains was screened for their susceptibility to amoxicillin (A), AUG, piperacillin (PIP), cefalotin (CF), cefoxitin (CFX), cefotaxime (CFT), ceftazidime (CFZ), IMI, tobramycin (TB), AK, gentamicin (Gm), netilmicin (NT), nalidixic acid (Na), NOR, CIP, NI, TS and fosfomycin (F). The list of antibiotics used in the study is presented in Table 1.
Table 1.
List of antibiotics used in the study
| Antibiotics used against | Antibiotics by class | |
|---|---|---|
| E. coli strains | P. mirabilis strains | |
| I. β-lactam antibiotics | ||
| Amoxicillin (A) | Carbenicillin (PY) | Penicillins |
| Piperacillin (PIP) | Ampicillin (AP) | |
| Amoxicillin/Clavulanate (AUG) | ||
| Cefalotin (1st G) (CF) | Cephalosporins | |
| Cefoxitin (2nd G) (CFX) | Cefuroxime (2nd G) (CXM) | |
| Cefotaxime (3rd G) (CFT) | ||
| Ceftazidime (3rd G) (CFZ) | ||
| Imipenem (IMI) | Carbapenems | |
| Aztreonam (ATM) | Monobactams | |
| Amikacin (AK) | II. Aminoglycosides | |
| Tobramycin (TB) | ||
| Gentamicin (Gm) | ||
| Netilmicin (NT) | ||
| Nalidixic acid (Na) | III. Quinolones | |
| Norfloxacin (NOR) | ||
| Ciprofloxacin (CIP) | ||
| Ofloxacin (OFX) | ||
| Tetracycline (T) | IV. Tetracyclines | |
| Polymyxin B (PB) | V. Polypeptides | |
|
Colistin (Polymyxin E) (CO) |
||
| Nitrofurantoin (NI) | VI. Nitrofurans | |
| Cotrimoxazole (TS) | VII. Sulfonamides | |
| Fosfomycin (F) | VIII. Folic acid derivatives | |
Mathematical and statistical analysis
The following methods were used in mathematical and statistical analysis: Kohonen networks and association analysis using SAS® Data Miner tools.
Results
Antibiotic resistance of clinical and laboratory collections of P. mirabilis strains
A comparative analysis (Table 2) showed that two clinical P. mirabilis strain collections from Sweden and Poland had much more diverse patterns of resistance than the third collection (laboratory). The number of diverse resistance patterns was as follows: 66 % of all strains (44) in the Polish collection, 36 % of all strains (99) in the Swedish collection and 21 % of strains (42) in the laboratory collection. Among the Polish collection strains, there were 23 unique patterns of resistance, representing 79 % of all patterns of resistance (29). In the Swedish collection, 15 unique patterns of drug resistance were identified, representing 42 % of all patterns of resistance (36). In the laboratory P. mirabilis collection, six unique patterns of drug resistance were identified, accounting for 67 % of all patterns of resistance (9) (Table 2).
Table 2.
Comparison of antibiotic resistance patterns of E. coli and P. mirabilis collections
| Strain collections | Antimicrobial resistance patterns | |
|---|---|---|
| No. of unique patterns | No. of diverse patterns | |
| P. mirabilis | 15 | 36 |
| 99 Swedish clinical strains | ||
| P. mirabilis | 6 | 9 |
| 42 Czech laboratory strains | ||
| P. mirabilis | 23 | 29 |
| 44 Polish clinical strains | ||
| E. coli 129 Polish clinical strains | 63 | 76 |
The bacterial strains revealed a high resistance to some of the applied antibiotics (Table 3). Over 80 % of all P. mirabilis strains were resistant to T, NI, and polypeptides. The majority of bacterial strains in the Polish collection were resistant to beta-lactam antibiotics: 60 % of Polish P. mirabilis strains were resistant to CXM, and almost 50 % of Polish strains were resistant to PY, AP, IMI, and ATM. No single antibiotic was found to be an effective bactericidal agent against all P. mirabilis strains in this collection. In the other two collections (laboratory and Swedish), the strains displayed a much higher sensitivity. IMI, ATM and AK were found to be effective bactericidal agents against all P. mirabilis strains in these collections. Additionally, in the laboratory P. mirabilis collection, all strains were susceptible to AUG, PY, norfloxacin, CIP, OFX and cotrimoxazole.
Table 3.
Number (percentage) of bacterial strains resistant to the antibiotics used
| Antibiotics | No. (%) of resistant strains | |||
|---|---|---|---|---|
| P. mirabilis collections | E. coli | |||
| Laboratory | Swedish | Polish | ||
| AUG | 0 | 2 (2.0) | 5 (12.0) | 8 (6.0) |
| A | – | – | – | 73 (57.0) |
| PIP | – | – | – | 27 (21.0) |
| PY | 0 | 13 (13.0) | 19 (44.0) | – |
| AP | 2 (9.0) | 15 (15.0) | 21 (48.0) | – |
| CF | – | – | – | 38 (30.0) |
| CFX | – | – | – | 12 (9.0) |
| CXM | 2 (9.0) | 3 (3.0) | 26 (60.0) | – |
| CFT | – | – | – | 7 (5.5) |
| CFZ | – | – | – | 8 (6.0) |
| IMI | 0 | 0 | 20 (46.0) | 0 |
| ATM | 0 | 0 | 21 (49.0) | – |
| AK | 0 | 0 | 10 (23.0) | 1 (0.7) |
| TB | – | – | – | 12 (9.0) |
| Gm | – | – | – | 20 (16.0) |
| NT | – | – | – | 4 (3.0) |
| Na | – | – | – | 65 (51.0) |
| NOR | 0 | 3 (3.0) | 14 (32.0) | 38 (30.0) |
| CIP | 0 | 3 (3.0) | 12 (28.0) | 38 (30.0) |
| OFX | 0 | 2 (2.0) | 18 (42.0) | – |
| T | 38 (90.0) | 94 (94.0) | 42 (98.0) | – |
| PB | 42 (100) | 97 (97.0) | 36 (84.0) | – |
| CO | 42 (100) | 99 (100) | 39 (91.0) | – |
| NI | 42 (100) | 91 (91.0) | 39 (91.0) | 24 (19.0) |
| TS | 0 | 19 (19.0) | 18 (42.0) | 42 (33.0) |
| F | – | – | – | 2 (1.0) |
Antibiotics: AUG amoxicillin/clavulanate, A amoxicillin, PIP piperacillin, PY carbenicillin, AP ampicillin, CF cefalotin, CFX cefoxitin, CXM cefuroxime, CFT cefotaxime, CFZ ceftazidime, IMI imipenem, ATM aztreonam, AK amikacin, TB tobramycin, Gm gentamicin, NT netilmicin, Na nalidixic acid, NOR norfloxacin, CIP ciprofloxacin, OFX ofloxacin, T tetracycline, PB polymyxin B, CO colistin sulfate, NI nitrofurantoin, TS cotrimoxazole, F fosfomycin; bold over 45 % of resistant strains;- not studied
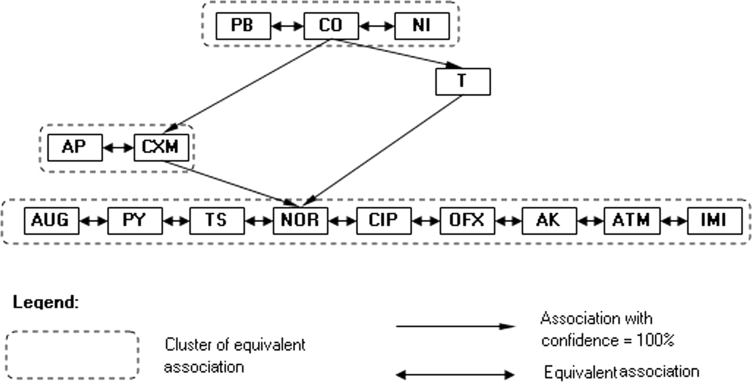
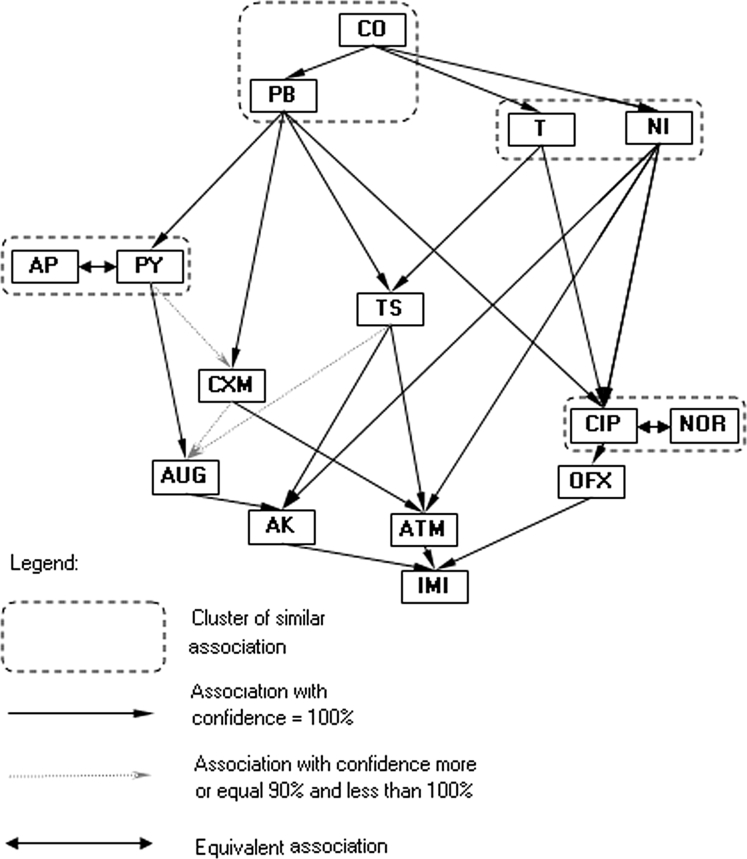
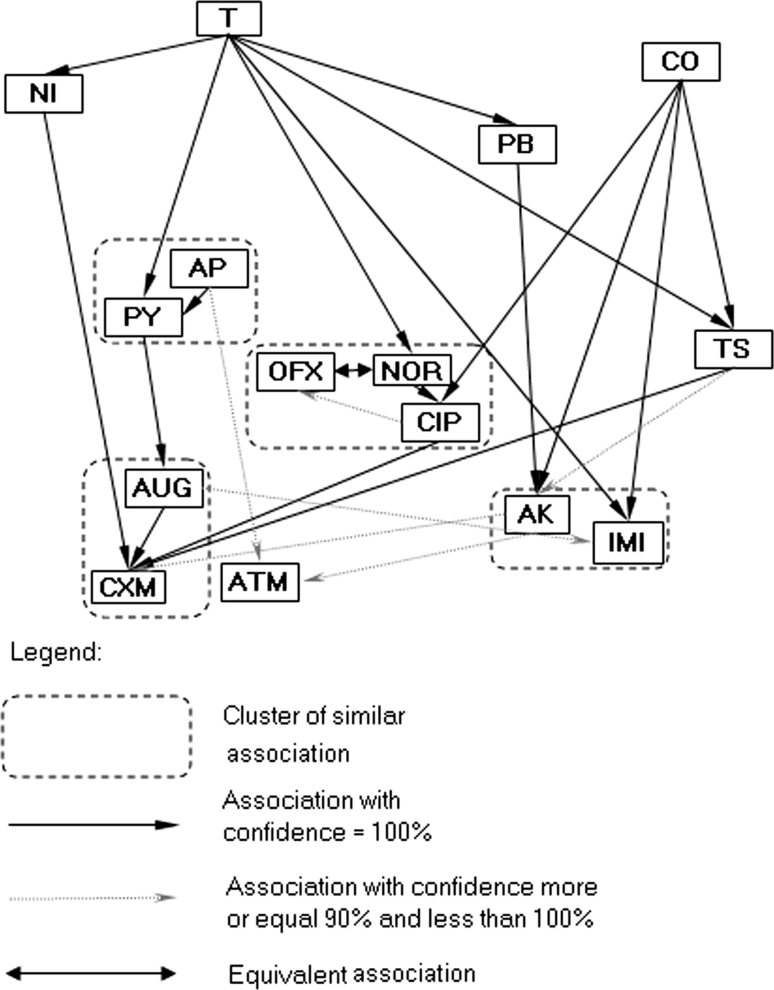
The Kohonen network method and association analysis based on the antibiotic resistance patterns of laboratory P. mirabilis strains resulted in a simple association diagram of antibiotic reaction with only three major similarity clusters (Fig. 1). The arrow (e.g. A → B) in the association diagram should be interpreted as follows: “if a strain is sensitive to antibiotic A, then it is sensitive to B” or “if a strain is not sensitive to antibiotic B, then it is not sensitive to A”. For example, resistance to one of the antibiotics from the first cluster (CO; NT or T) was accompanied by a lack of sensitivity to CXM, AP, and T. The analysis did not indicate diverse patterns of antibiotic resistance. This result is in contrast to two the clinical collections, in which much more diverse correlation patterns were observed (Figs. 2, 3). However, similar associations of CO, T, and NOR were observed in the Swedish laboratory collection (Fig. 2). The cluster patterns and association diagrams of the two clinical strain collections differed significantly (compare Figs. 2, 3). Interestingly, antibiotics with similar chemical structures formed one cluster. The diagrams of associations made it possible to present the correlation between antibiotic action patterns and might help to identify hidden relationships in antibiotic resistance mechanisms.
Fig. 1.
Kohonen map of drug resistance patterns of the P. mirabilis laboratory strain collection
Fig. 2.
Kohonen map of drug resistance patterns of the P. mirabilis swedish strain collection
Fig. 3.
Kohonen map of drug resistance patterns of the P. mirabilis polish strain collection
Antibiotic resistance of the clinical collection of E. coli strains
The results demonstrated significantly diverse drug resistance patterns among 129 E. coli strains. Sixty-three unique resistance patterns were found, which consisted of 83 % of all diverse drug resistance profiles (76) in these E. coli strains. Diverse resistance patterns were found for 59 % of all E. coli strains (Table 2).
This collection of strains revealed simultaneous resistance to some of the applied antibiotics (Table 3). E. coli strains that were resistant to penicillins were also resistant to quinolones, which was in contrast to cephalosporins and aminoglycosides, which were effective. All of the strains were also susceptible to IMI and AK. More than 70 % of bacterial strains were susceptible to many of the applied antibiotics, with the exception of A (57 % resistant strains), Na (51 % resistant strains), TS (33 % resistant strains), and CF (30 % resistant strains). A total of 60 % of bacterial strains were resistant to at least one beta-lactam antibiotic (A). Moreover, 15 % of the strains (20 isolates) were resistant to at least one aminoglycoside. All bacterial isolates that were resistant to TB were also resistant to Gm. Resistance to quinolones was revealed in 50 % of the strains, and 80 % of these strains were also resistant to A. In conclusion, as many as 50 % of the studied E. coli strains displayed an MDR phenotype due to their resistance to at least two antibiotics of two different classes.
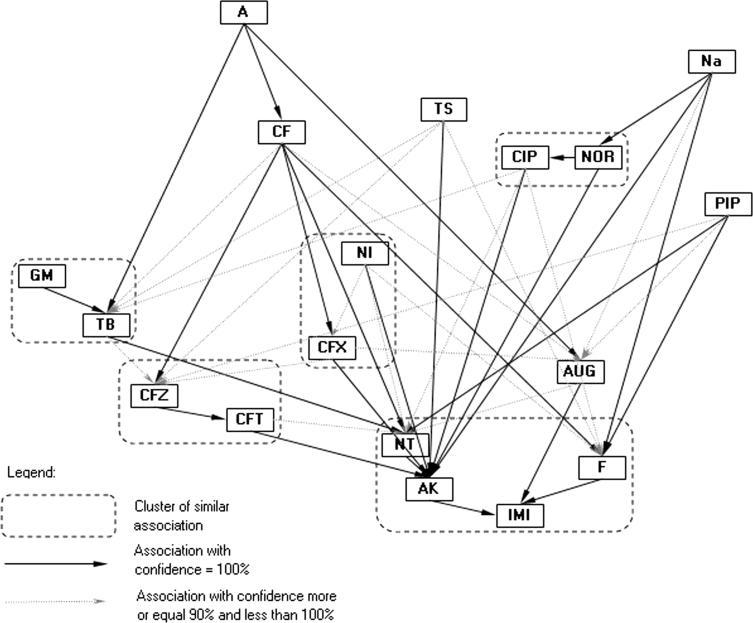
Similar to P. mirabilis clinical strains, E. coli strains presented a complex pattern of resistance/susceptibility associations (Fig. 4). The mathematical analysis of the resistance of E. coli strains showed a much more diverse correlation pattern in contrast to P. mirabilis strains. Resistance to A was accompanied by a lack of sensitivity to TB, CF, and AUG. Additionally, resistance to Na was accompanied by a lack of sensitivity to NOR, CIP, AK, fosfomycin (F) and AUG. A weaker correlation was observed for the other antibiotics (TS and PIP). Antibiotics with a similar degree of correlation formed one association cluster. Interestingly, this model indicated that the antibiotics NT, AK, IMI and F rarely led to resistant strains. In contrast, other antibiotics, including A, Na and TS, frequently induced resistance. This finding is consistent with the data shown in Table 3. There were substantial differences in resistance associations among four P. mirabilis and E. coli strain collections with respect to clustering patterns.
Fig. 4.
Kohonen map of drug resistance patterns of the E. coli polish strain collection
Discussion
Escherichia coli and P. mirabilis are the most important etiological factors of UTIs. The pathogenicity of these bacteria is specific to uropathogenic strains due to the presence of virulence factors, such as fimbrial adhesins (S and P in UPEC and MR/P and PMF in P. mirabilis) [3, 6]. Toxins, such as α-hemolysin, cnf1, and bacteriocin usp, are some of the typical pathogenic factors of E. coli, while urease, protease, and hemolysins are characteristic of P. mirabilis. It has been shown that two major groups of E. coli strains can invade human urinary tracts. The first group is characterized by a statistically limited presence of virulence factors and a multi-drug resistance pattern (including resistance to quinolones). The second E. coli group encodes many virulence factors but is susceptible to quinolones and many other antibiotics [16–18]. This finding may imply that the latter group consists of uropathogenic E. coli.
A comparison of the antibiotic resistance patterns of one E. coli and three P. mirabilis collections (Table 2) revealed that the percentage of unique versus diverse patterns in the Swedish clinical and laboratory P. mirabilis strain collections remained at the same level. Interestingly, a similar proportion was observed for Polish clinical E. coli and P. mirabilis collections. This diversity may result from the types of antibiotics and the frequency of their use in Poland and Sweden. In addition, it was shown that there are some groups of antibiotics to which bacterial strains were rarely resistant. If a strain was resistant to an antibiotic in the group, that strain was usually resistant to the majority of antibiotics within the same group (see Figs. 1, 2, 3, 4). The association graphs demonstrated a high probability of co-existing resistance toward antibiotics in particular strain collections. Resistance often concerned antibiotics with different chemical groups. This finding may be a manifestation of general mechanisms for the acquisition of resistance. Therefore, this association may show some new tendencies of the emergence of drug resistance. Observing the structure of the graphs, there were similarities in associations depending on antibiotic types used.
MDR Proteus and Escherichia strains pose a serious hazard for patients hospitalized as a result of UTIs [19]. Therefore, monitoring changes in the increase in drug resistance and anticipating these changes seem to be an important medical issue. The mathematical analysis revealed much more complex antibiotic resistance patterns in E. coli strains than P. mirabilis strains [12]. This finding may suggest that the former are characterized by greater genome plasticity. This work offers a complex analysis of bacterial populations of E. coli and P. mirabilis strains responsible for UTIs, including antibiotic resistance patterns, a mathematical analysis of those patterns, multiplex PCR for the detection of virulence factors, and the correlation of drug resistance patterns with virulence factors. Such a complex approach may allow us to trace the evolution of changes in the most important UTI pathogens, E. coli and P. mirabilis. The prediction of the emergence of future strain resistance is a prerequisite for the rational planning of medical treatment.
Acknowledgments
This study was supported by Grant 2011/01/D/NZ7/00107 from National Science Centre, Poland and co-financed by the European Regional Development Fund under the Operational Programme Innovative Economy, Grant POIG.01.01.02-10-107/09.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Abbreviations
- UTI
Urinary tract infection
- UPEC
Uropathogenic E. coli
- IPEC
Intestinal pathogenic E. coli
- ExPEC
Extraintestinal pathogenic E. coli
- MDR
Multi-drug resistant
References
- 1.Mazzulli T. Resistance trends in urinary tract pathogens and impact on management. J Urol. 2002;168:1720–17222. doi: 10.1016/S0022-5347(05)64397-2. [DOI] [PubMed] [Google Scholar]
- 2.Schollum JB, Walker RJ. Adult urinary tract infection. Br J Hosp Med. 2012;73:218–223. doi: 10.12968/hmed.2012.73.4.218. [DOI] [PubMed] [Google Scholar]
- 3.Rozalski A, Sidorczyk Z, Kotelko K. Potential virulence factors of Proteus bacilli. Microbiol Mol Biol Rev. 1997;61:65–89. doi: 10.1128/mmbr.61.1.65-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daza R, Gutierrez J, Piedrola G. Antibiotic susceptibility of bacterial strains isolated from patients with community-acquired urinary tract infections. Int J Antimicrob Agents. 2001;18:211–215. doi: 10.1016/S0924-8579(01)00389-2. [DOI] [PubMed] [Google Scholar]
- 5.Lindsay E, Nicolle MD. Resistant pathogens in urinary tract infections. J Am Geriatr Soc. 2002;50:230–235. doi: 10.1046/j.1532-5415.50.7s.3.x. [DOI] [PubMed] [Google Scholar]
- 6.Silveira WD, Benetti F, Lancellotti M, Ferreira A, Solferini VN, Brocchi M. Biological and genetic characteristics of uropathogenic Escherichia coli strains. Rev Inst Med Trop. 2001;43:303–310. doi: 10.1590/S0036-46652001000600001. [DOI] [PubMed] [Google Scholar]
- 7.Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Sahm DF. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob Agents Chemother. 2002;46:2540–2545. doi: 10.1128/AAC.46.8.2540-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagani L, Migliavacca R, Pallecchi L, Matti C, Giacobone E, Amicosante G, Romero E. Emerging extended-spectrum beta-lactamases in Proteus mirabilis. J Clin Microbiol. 2002;40:1549–1552. doi: 10.1128/JCM.40.4.1549-1552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan AU, Musharraf A. Plasmid-mediated multiple antibiotic resistance in Proteus mirabilis isolated from patients with urinary tract infection. Med Sci Monit. 2004;10:598–602. [PubMed] [Google Scholar]
- 10.Garcia-Rodriguez JA, Jones RN. Antimicrobial resistance in gram-negative isolates from European intensive care units: data from the Meropenem yearly susceptibility test information collection (MYSTIC) programme. J Chemother. 2002;14:25–32. doi: 10.1179/joc.2002.14.1.25. [DOI] [PubMed] [Google Scholar]
- 11.Empel J, Baraniak A, Literacka E, Mrówka A, Fiett J, Sadowy E, Hryniewicz W, Gniadkowski M. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother. 2008;52:2449–2454. doi: 10.1128/AAC.00043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonten MJM, Austin DJ, Lipsitch M. Understanding the spread of antibiotic resistant pathogens in hospitals: mathematical models as tools for control. Clin Infect Dis. 2001;33:1739–1746. doi: 10.1086/323761. [DOI] [PubMed] [Google Scholar]
- 13.Bootsma MCJ. (2005) Mathematical studies of the dynamics of antibiotic resistance. Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht
- 14.Tavare S (2001) Ancestral Inference in Population Genetics. In: Proceedings of saint flour summer school in probability and statistics, Saint Flour, Springer lecture notes in mathematics
- 15.National Committee for Clinical Laboratory Standards (NCCLS) (1997) Performance standards for antimicrobial disc susceptibility test, 6th edn. Approved Standard: M2-A6 and Supplemental Tables M100-S7
- 16.Adamus-Bialek W, Wojtasik A, Majchrzak M, Sosnowski M, Parniewski P. (CGG)4-based PCR as a novel tool for discrimination of uropathogenic Escherichia coli strains: comparison with enterobacterial repetitive intergenic consensus-PCR. J Clin Microbiol. 2009;47:3937–3944. doi: 10.1128/JCM.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horcajada JP, Soto S, Gajewski A, Smithson A, Jimenez de Anta MT, Mensa J, Vila J, Johnson JR. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J Clin Microbiol. 2005;43:2962–2964. doi: 10.1128/JCM.43.6.2962-2964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno E, Prats G, Sabate M, Perez T, Johnson JR, Andreu A. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother. 2006;57:204–211. doi: 10.1093/jac/dki468. [DOI] [PubMed] [Google Scholar]
- 19.Lipsitch M, Bergstrom CT, Levin BR. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc Natl Acad Sci USA. 2000;97:1938–1943. doi: 10.1073/pnas.97.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]