Abstract
The development of HIV-1 protease inhibitors has been the historic paradigm of rational structure-based drug design, where structural and thermodynamic analyses have assisted in the discovery of novel inhibitors. While the total enthalpy and entropy change upon binding determine the affinity, often the thermodynamics are considered in terms of inhibitor properties only. In the current study, profound changes are observed in the binding thermodynamics of a drug resistant variant compared to wild-type HIV-1 protease, irrespective of the inhibitor bound. This variant (Flap+) has a combination of flap and active site mutations and exhibits extremely large entropy-enthalpy compensation compared to wild-type protease, 5–15 kcal/mol, while losing only 1–3 kcal/mol in total binding free energy for any of six FDA approved inhibitors. Although entropy-enthalpy compensation has been previously observed for a variety of systems, never have changes of this magnitude been reported. The co-crystal structures of Flap+ protease with four of the inhibitors were determined and compared with complexes of both the wildtype protease and another drug resistant variant that does not exhibit this energetic compensation. Structural changes conserved across the Flap+ complexes, which are more pronounced for the flaps covering the active site, likely contribute to the thermodynamic compensation. The finding that drug resistant mutations can profoundly modulate the relative thermodynamic properties of a therapeutic target independent of the inhibitor presents a new challenge for rational drug design.
INTRODUCTION
Development of potent inhibitors requires optimizing the binding affinity to the target, which is dictated by the binding free energy comprised of both enthalpic and entropic contributions. Structure-based drug design enormously benefits from thermodynamic profiles, which provide information on the driving forces for binding (1). HIV-1 protease inhibitors (PIs) were initially based on the substrate sequences as well as on the topology of the enzyme’s active site (2). The original structure-based drug design strategy was to optimize the entropy of binding by introducing conformational restraints into compounds so that they are pre-shaped to fit into the active site. In addition, these compounds are highly hydrophobic, resulting in an increase in solvation entropy upon binding. Thus, the first generation drugs bind with favorable entropy but with a corresponding loss in enthalpy (3). Some newer HIV-1 PIs (4–9) have favorable binding enthalpy and often higher affinity, as with Darunavir (DRV), leading to the hypothesis that favorable enthalpy may aid in attaining better inhibitors that are less susceptible to drug resistance. However, the binding of high affinity Tipranavir (TPV) is highly entropically driven (9). Hence, both entropy and enthalpy of binding can contribute significantly to the high affinity of potent inhibitors.
The interplay between entropy and enthalpy in attaining high affinity is not very well understood at the molecular level, and can be complex. In most cases, achieving higher affinity requires a saddle-point type of optimization, as enhancing the conformational entropy is balanced against the competing tendency to maximize intramolecular contacts and hence enthalpy (10). Entropy-enthalpy compensation has been observed in many biological systems after relatively minor perturbations to the system, including protein-metal interactions (11, 12), cAMP receptor protein variants and RNA polymerase binding (13), peptides binding to the Src Homology 2 domain of the Src kinase (14), as well as ligands binding to cyclodextrin variants (15, 16). This compensation comprises of nearly equal and opposite changes in TΔS and ΔH usually of 1–2 kcal/mol, resulting in only minimal differences in the overall ΔG when comparing the binding of different complexes (17). The consequence of entropy-enthalpy compensation makes it difficult to integrate the direct properties of enthalpy and entropy into rational drug design.
Drug resistant mutations in HIV protease throughout the enzyme can decrease the binding affinity with inhibitor molecules in a complex, interdependent and cooperative manner (18, 19). Combinations of thermodynamic and structural studies by many groups including our own, evaluated the consequences associated with drug-resistant mutations (6, 20–25). Our earlier thermodynamic study on DRV and the chemically similar inhibitor amprenavir (APV), hypothesized a structural rationale for their unprecedented highly favorable enthalpy even with drug resistant protease variants (6). The single-ringed tetrahydrofuran (THF) group of APV was replaced with a double-ringed bis-THF in DRV, which forms additional protease-inhibitor interactions (6) correlating with high affinity and highly favorable enthalpy. Such aspects of conformational changes in the bound structure may correlate with conserved thermodynamic changes, even though thermodynamics of binding is an equilibrium property between the liganded and unliganded forms of the enzymes.
In the present study, the crystal structures and thermodynamics are compared for the binding of inhibitors APV, atazanavir (ATV), DRV, indinavir (IDV), nelfinavir (NFV) and saquinavir (SQV) to the wild-type (WT) HIV-1 protease and two multi-drug-resistant (MDR) variants (Figure 1): (i) Act, with two active site mutations (V82T/I84V), and (ii) Flap+ (L10I/G48V/I54V/V82A) derived as a combination of mutations that simultaneously occur in patients’ sequences in flap and active site regions (26). Both these drug resistant protease variants lose similar amounts of inhibitor binding affinity relative to the WT protease. However the Flap+ variant exhibits extremely large entropy-enthalpy compensation, i.e. opposite changes in the entropy and enthalpy of interaction, for all the inhibitors studied, indicating that the drug resistant mutations in Flap+ are directly modulating the relative thermodynamics of binding.
Figure 1.
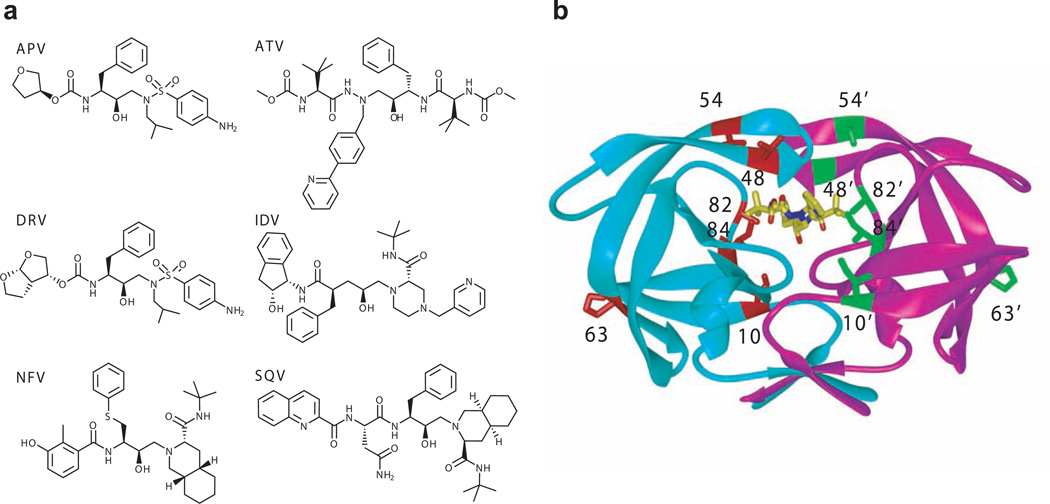
Structure of inhibitors and HIV-1 protease. (a) Chemical structures of inhibitors. (b) Overview of the mutation sites of Flap+ and Act mutants mapped on an HIV-1 protease dimer. The monomers are distinguished in cyan and magenta, while the inhibitor ATV is shown in yellow stick model. The mutation sites of Flap+ (L10I/G48V/I54V/V82A) and Act (V82T/I84V) along with the site of the natural polymorphism L63P are highlighted in red and green stick models.
RESULTS AND DISCUSSION
A major challenge in the treatment of HIV infections is the emergence of drug-resistant viruses. Mutations in the viral protease reduce the affinity of the inhibitors, thereby maintaining the viral replication and leading to therapy failure. Understanding the thermodynamics of binding to the prominent drug target HIV-1 protease is essential to aid the design and improvement of small molecule inhibitors to drug resistant variants.
Thermodynamics of inhibitor binding and entropy-enthalpy compensation in Flap+ protease
The affinity and binding thermodynamics of inhibitors with WT and MDR protease variants Act and Flap+ were determined by isothermal titration calorimetry (ITC) (Table 1). As expected, binding affinity of inhibitors to the mutant proteases is lower compared to WT. APV, followed by DRV, retains affinity to both MDR proteases more than the other inhibitors. In addition, the Flap+ mutations appear to have less effect on the binding of these two inhibitors when compared to those of the Act mutant. Relative to WT, the Kd for Act protease increases 5.9 and 14.7 fold for APV and DRV, respectively, while the same ratios for Flap+ protease binding are only 3.3 and 5.8. ATV binding affinity for Act and Flap+ protease decreases by 17.8 and 48.4 fold, respectively. IDV and NFV have a higher loss of affinity for the mutant proteases, in the range of 40–90 fold. SQV binding is the most compromised of all the inhibitors studied, with Kd ratios of 135 for Act and 353 for Flap+. Thus APV and DRV, which fit well within the substrate envelope (6, 21), are the most robust against the drug-resistant variants, consistent with results for other drug resistant HIV-1 protease variants (27).
Table 1.
Thermodynamic parameters with their standard errors for the binding of inhibitors to WT, Act and Flap+ variants of HIV-1 protease, at 20°C.
| protease variant |
ΔG (kcal mol−1) |
ΔH (kcal mol−1) |
−TΔS kcal mol−1) |
Kd (nM) |
Kd ratio |
ΔCp (cal K−1mol−1) |
|---|---|---|---|---|---|---|
| APV | ||||||
| WT | −12.4 ± 0.3 | −7.3 ± 0.9 | −5.3 ± 0.9 | 0.39 ± 0.20 | 1 | −430 ± 33 |
| Act | −11.4 ± 0.2 | −6.0 ± 2.0 | −5.6 ± 2.0 | 2.30 ± 0.79 | 5.9 | |
| Flap+ | −11.7 ± 0.0 | 3.3 ± 0.5 | −15.2 ± 0.5 | 1.30 ± 0.01 | 3.3 | −532 ± 65 |
| ATV | ||||||
| WT | −12.7 ± 0.3 | −1.1 ± 0.1 | −11.8 ± 0.3 | 0.23 ± 0.12 | 1 | |
| Act | −11.1 ± 0.0 | −0.3 ± 0.1 | −10.9 ± 0.1 | 4.00 ± 0.21 | 17.8 | |
| Flap+ | −10.5 ± 0.1 | 4.5 ± 0.1 | −15.2 ± 0.1 | 10.90 ± 1.60 | 48.4 | |
| DRV | ||||||
| WT | −15.0 ± 0.3 | −12.1 ± 0.9 | −3.1 ± 0.9 | 0.005 ± 0.00 | 1 | −373 ± 32 |
| Act | −13.4 ± 0.2 | −10.0 ± 0.1 | −3.7 ± 0.2 | 0.066 ± 0.02 | 14.7 | |
| Flap+ | −14.0 ± 0.1 | 2.0 ± 0.6 | −16.2 ± 0.6 | 0.026 ± 0.01 | 5.8 | −508 ± 16 |
| IDV | ||||||
| WT | −12.0 ± 0.1 | 1.7 ± 0.2 | −14.0 ± 0.2 | 0.74 ± 0.14 | 1 | |
| Act | −9.8 ± 0.2 | 7.4 ± 0.0 | −17.4 ± 0.2 | 36.0 ± 13.5 | 49.0 | |
| Flap+ | −9.6 ± 0.0 | 10.9 ± 0.1 | −20.6 ± 0.1 | 56.5 ± 22.9 | 76.9 | |
| NFV | ||||||
| WT | −12.4 ± 0.1 | 4.4 ± 0.1 | −17.0 ± 0.1 | 0.39 ± 0.07 | 1 | |
| Act | −10.2 ± 0.0 | 7.1 ± 0.1 | −17.4 ± 0.1 | 18.90 ± 0.25 | 48.2 | |
| Flap+ | −9.8 ± 0.0 | 10.8 ± 0.0 | −20.8 ± 0.0 | 34.1 ± 1.3 | 87.0 | |
| SQV | ||||||
| WT | −12.3 ± 0.0 | 3.6 ± 0.1 | −16.1 ± 0.1 | 0.50 ± 0.03 | 1 | |
| Act | −9.4 ± 0.0 | 9.5 ± 0.0 | −19.1 ± 0.0 | 67.4 ± 0.3 | 135.1 | |
| Flap+ | −8.9 ± 0.0 | 12.9 ± 0.1 | −21.9 ± 0.1 | 176.0 ± 4.2 | 352.7 | |
A closer examination of the specific enthalpic and entropic contributions to the free energy (Table 1) reveals that the binding of the first generation inhibitors IDV, NFV, and SQV to all protease variants is entropically driven (negative −TΔS). In contrast, binding of APV, ATV, and DRV to WT and Act proteases is both enthalpically (negative ΔH) and entropically driven (Table 1). However, the binding of these same compounds (APV, ATV, and DRV) becomes endothermic (positive ΔH) with the Flap+ protease. In this case, inhibitor binding is entropically driven, suggesting considerable solvation effects and/or enhanced protease/inhibitor flexibility upon complex formation.
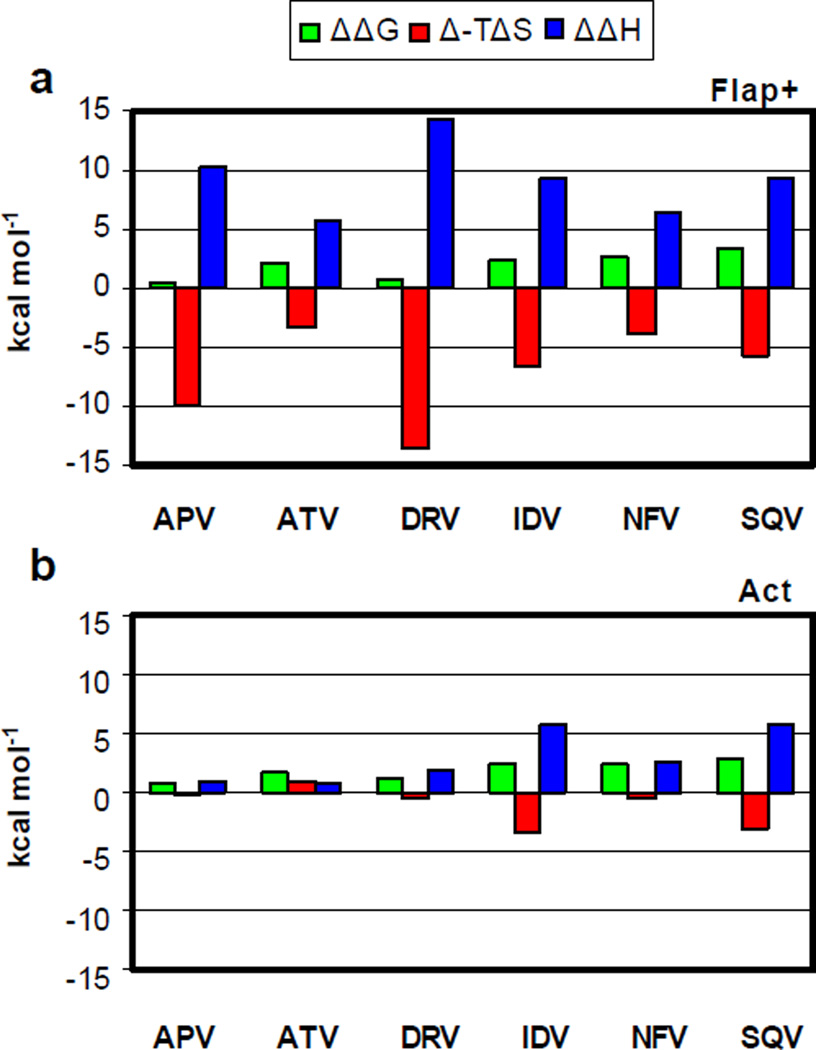
To better evaluate the changes in binding thermodynamics due to drug resistance mutations, the difference in entropic and enthalpic contributions to the binding free energy with respect to the WT protease were calculated (Figure 2). Strikingly, there is a significant entropy-enthalpy compensation in the Flap+ protease for all the inhibitors: difference in entropy (Δ(−TΔS) and enthalpy (ΔΔH) of binding have opposite sign and similar magnitude, canceling out to yield relatively small changes in the overall binding free energy (ΔΔG). APV and DRV bind to Flap+ protease with an entropy-enthalpy compensation in the order of 10 kcal mol−1. Even though ΔΔG values for Act protease are comparable to those for Flap+, the entropy-enthalpy compensation is either minimal or nonexistent (Figure 2b).
Figure 2.
Thermodynamics of inhibitor binding. Differences in binding energetics between (a) WT and Flap+ and (b) WT and Act variants. The differences in ΔG, TΔS and ΔH are shown in green, red and blue, respectively.
These dramatic changes in binding thermodynamics of APV and DRV to Flap+ relative to WT protease were further evaluated by comparing the heat capacity changes upon binding (ΔCp in Table 1). The mutations in the Flap+ protease substantially change the overall heat capacity change associated with binding of these two inhibitors, by more than 100 cal mol−1K−1. This large and negative change in ΔCp suggests altered hydrophobic effects in inhibitor binding to Flap+, and may be due to increased nonpolar interactions with the inhibitor, and release of solvation water upon complex formation. The favorable solvation entropy indicated by more negative ΔCp (28, 29) is also consistent with entropy-driven binding to Flap+ protease.
Structural comparison of protease complexes
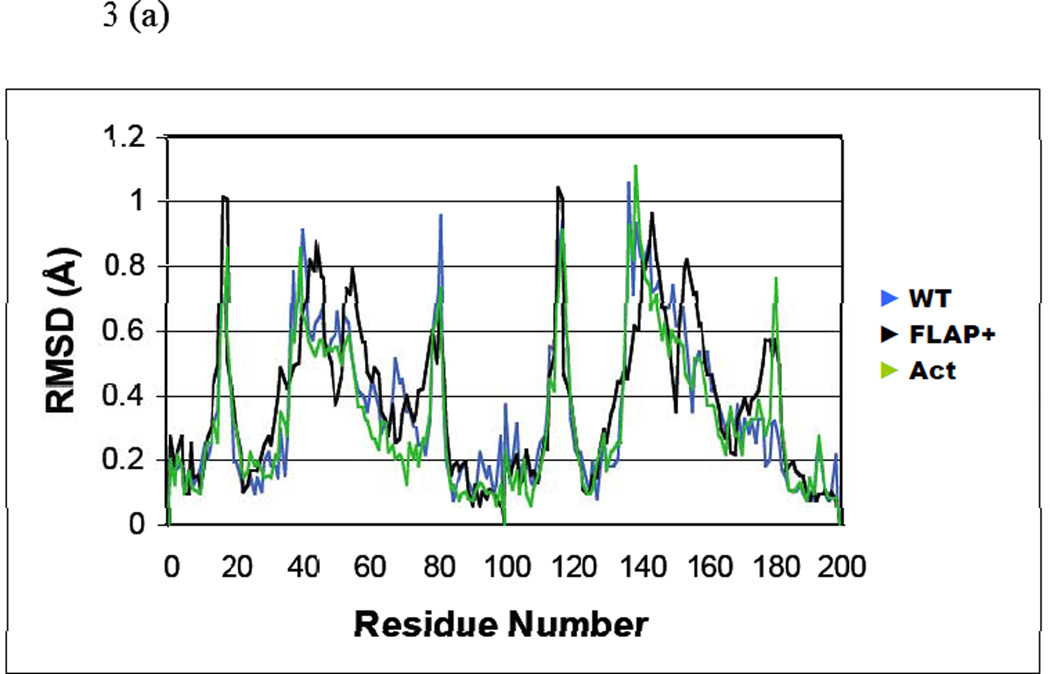
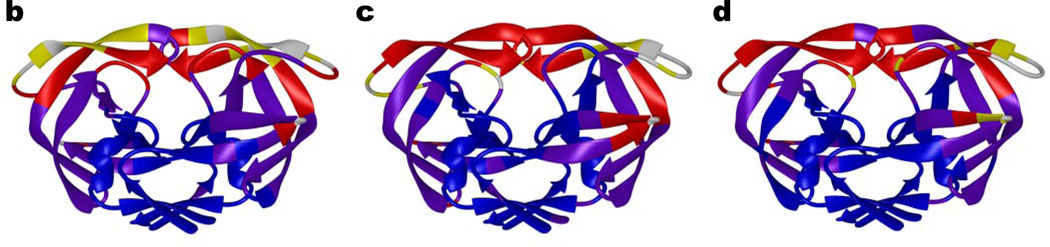
Crystal structures of the six inhibitors in complex with WT, Act and Flap+ protease variants were determined and compared (Table 2). The twelve inhibitor complexes chosen for this structural analysis were superimposed onto the capsid-p2 substrate complex (PDB code 1F7A), as described in Methods. The root mean squared deviations (RMSD) of Cα atoms with respect to the substrate complex were calculated for each protease variant (Figure 3a). Although the distributions of RMSD values for the WT and Act complexes follow the same pattern, the distribution observed in the Flap+ complexes is considerably different. Interestingly, the Flap+ complex structures exhibit the largest difference around the flap region (residues 44–57). The RMSD in coordinates near Lys41 and Lys57 for both monomers of Flap+ is the highest (> 0.8 Å), while for the tip of the flap (Ile50) the RMSD value drops to < 0.25 Å. For the WT and Act complexes, the RMSD values for the entire flap region remain relatively uniform at 0.5–0.65 Å. Thus, the relative changes in the backbone suggest that the flap region of Flap+ complexes is conformationally distinct from WT and Act proteases.
Table 2.
Crystallographic and refinement statistics.
| Flap+ (L10I/G48V/I54V/V82A) |
WT | Act (V82T/I84V) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | APV | ATV | DRV | SQV | APV | ATV | DRVa,b | SQVc | APVRT,a | ATV | DRVa,b | SQVRT |
| PDB code | 3EKP | 3EKW | 3EKT | 3EKQ | 3EKV | 3EKY | 1T3R | 1HXB | 1T7J | 3EL9 | 1T7I | 3EL4 |
| Resolution (Å) | 2.15 | 1.6 | 1.97 | 2.2 | 1.8 | 1.7 | 1.2 | 2.3 | 2.2 | 1.6 | 1.35 | 2.0 |
| Space group | P61 | P212121 | P61 | P212121 | P212121 | P212121 | P212121 | P61 | P212121 | P212121 | P212121 | P212121 |
| Unit Cell a (Å) | 92.0 | 51.2 | 92.0 | 51.2 | 50.7 | 51.0 | 54.9 | 63.3 | 51.1 | 50.7 | 50.9 | 51.3 |
| b (Å) | - | 58.5 | - | 59.3 | 57.4 | 58.8 | 57.8 | 59.3 | 58.1 | 58.0 | 59.3 | |
| c (Å) | 106.2 | 61.3 | 106.2 | 61.4 | 61.7 | 61.3 | 62.0 | 83.5 | 61.8 | 61.4 | 61.7 | 62.1 |
| Total reflections | 161411 | 175086 | 202358 | 37340 | 49469 | 68018 | 302022 | 49950 | 87982 | 256671 | 81540 | |
| Unique reflections | 27531 | 24185 | 34578 | 9789 | 14987 | 19646 | 55056 | 9705 | 23103 | 39998 | 12975 | |
| Completeness (%) | 99.5 | 96.6 | 95.8 | 98.3 | 79.7 | 93.5 | 95.5 | 96.8 | 93.6 | 98.0 | 97.2 | |
| Rmerge (%) | 5.4 | 3.3 | 5.0 | 4.6 | 2.9 | 5.0 | 3.8 | 8.7 | 4.8 | 4.4 | 7.0 | |
| I/σI | 11.1 | 17.1 | 15.4 | 11.4 | 19.0 | 11.3 | 25.0 | 5.9 | 11.7 | 13.5 | 6.0 | |
| REFINEMENT | CCP4 | CCP4 | CCP4 | CCP4 | CCP4 | CCP4 | CCP4 | X-PLOR | CNS | CCP4 | CCP4 | CNS |
| R factor (%) | 19.6 | 18.7 | 20.9 | 19.5 | 19.0 | 17.3 | 14.1 | 16.1 | 20.3 | 19.2 | 16.3 | 19.0 |
| Rfree (%) | 25.5 | 22.2 | 25.6 | 24.6 | 22.6 | 20.5 | 17.9 | NA | 24.4 | 21.7 | 20.0 | 22.3 |
| RMSD in | ||||||||||||
| bond length (Å) | 0.009 | 0.006 | 0.007 | 0.008 | 0.007 | 0.007 | 0.004 | 0.013 | 0.007 | 0.006 | 0.005 | 0.007 |
| bond angle (°) | 1.4 | 1.4 | 1.1 | 1.3 | 1.4 | 1.2 | 1.5 | 2.9 | 1.4 | 1.3 | 1.4 | 1.3 |
Figure 3.
Structural deviation between different inhibitor complexes. (a) Distribution of root mean squared deviations (RMSD) in Cα coordinates between the four inhibitor complexes of Flap+ (black), WT (blue) and Act (green) variants. The RMSD in Cα coordinates in (b) Flap+, (c) WT and (d) Act mapped on an HIV-1 protease dimer model. The color code for distinguishing the RMSD values are: blue, 0–0.25 Å; purple, 0.25–0.5 Å; red, 0.5–0.65 Å; yellow, 0.65–0.8 Å; white, 0.8 Å and above.
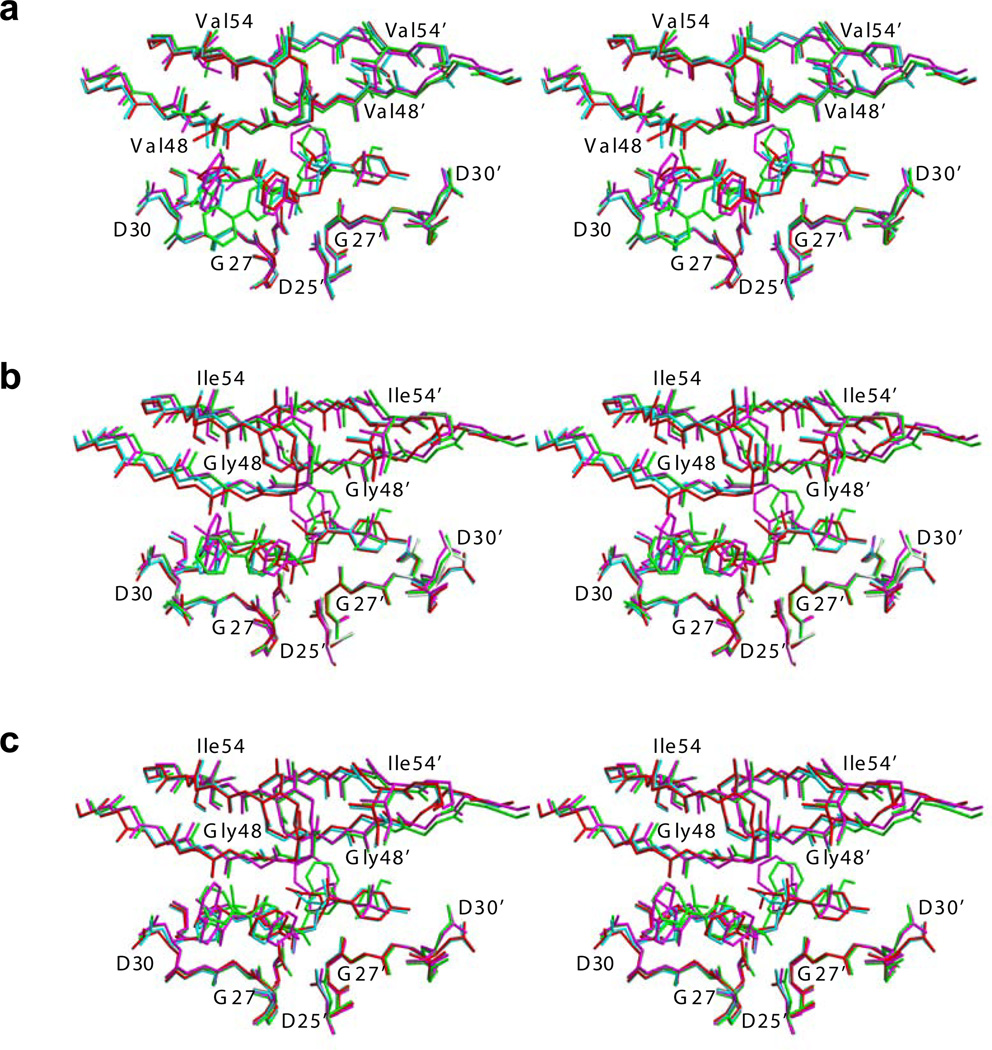
The RMSD values mapped onto the HIV-1 protease structure further illustrate that Flap+ inhibitor complexes are different from the WT and Act complexes, especially at the flaps (Figure 3). Figure 3b demonstrates the high-degree of structural conservation at the tips of flaps (Ile50–Gly51) in Flap+ complexes, along with significant variability in the β-strands flanking the tip. In contrast, the entire flap region in the WT and Act complexes exhibits uniform structural variation (Figure 3 c, d). This distinct behavior of flaps in the Flap+ protease is evident from the structural comparison of Flap+, WT and Act complexes (Figure 4). While the structure of the flap tips for the WT and Act complexes are conformationally variable, the Flap+ complexes appear to converge to a single conformation regardless of the inhibitor bound (Figure 4a). Thus, the flap tips of Flap+ protease may not have much conformational freedom but rather the rest of the flap region adapts to accommodate the binding of various inhibitors.
Figure 4.
Binding of inhibitors in HIV-1 protease complex structures. The active site region (Asp25–Asp30), flaps (Lys45–Lys55) and the inhibitors of (a) Flap+, (b) WT and (c) Act are superposed and illustrated as stereo pairs. The complexes involving APV, ATV, DRV and SQV are distinguished in red, green, cyan and magenta, respectively.
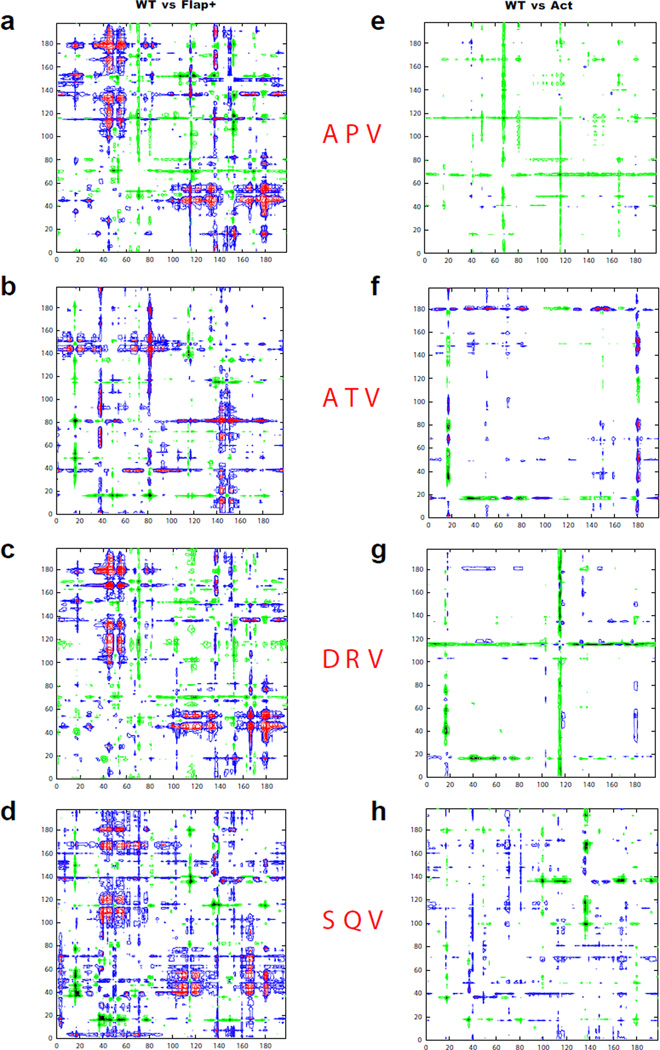
Additionally, pair-wise structural comparisons (WT vs Flap+ and WT vs Act) were carried out for complexes of each inhibitor (Figure 5) (30, 31). The double-difference plots comparing WT and Flap+ complexes reveal structural differences mainly concentrated around the flaps (Pro44–Lys57) and the P1-loop regions (Pro79–Ile84), which are asymmetric in that they occur in one monomer only. The flap of that particular monomer (Pro44–Lys57) in the Flap+ variant, compared to WT complexes, is closer to non-flap regions (1’–43’ and 58’–99’) of the opposite monomer by over 1 Å (Figure 5a–d). In contrast, the plots for WT-Act pairs exhibit fewer peaks for all four inhibitors (Figures 5e–h), indicating that the structures of WT and Act, when bound to the same inhibitor, are similar to each other.
Figure 5.
Comparison of mutant complex structures with WT HIV-1 protease. Double difference plots (see Methods) illustrating the WT vs Flap+ structural changes in (a) APV, (b) ATV, (c) DRV and (d) SQV, and WT vs Act structural changes in (e) APV, (f) ATV, (g) DRV, (h) SQV. The key for contours: (i) black −5.0 to −1.0 Å and green −1.0 to −0.5Å (Corresponding residue distances in the mutant structures have increased); (ii) blue 0.5 to 1.0 Å and magenta 1.0 to 5.0 Å (Corresponding distances in the mutant structures have decreased).
Hence, flap-specific changes with respect to WT protease are observed in the Flap+ variant complexes and not in Act, as revealed by double-difference plots (Figure 5) and RMSD values (Figure 4). The symmetry of the HIV-1 protease homodimer is broken upon binding either a substrate or inhibitor ligand (32–34). The structural analysis reveals that the asymmetry induced by inhibitor binding to Flap+ variant is more pronounced compared to WT and Act protease. This conserved asymmetric structural change observed in the Flap+ variant inhibitor complexes and altered flap behavior may contribute to the unique thermodynamic characteristics.
Inhibitor-protease hydrogen bonds
The number of hydrogen bonds between the inhibitor and the protease atoms is 9, 13, 11, and 12 for APV, ATV, DRV and SQV, respectively, for each protease variant. None of these bonds are with the flaps in APV and DRV complexes. The APVWT and APVAct complexes form two hydrogen bonds between Asp29 N and Asp30 N and the inhibitor THF ring O, which are absent in the Flap+ complex. However, these two interactions are maintained in all the complexes of DRV. One new hydrogen bond is observed in both the APV and DRV Flap+ complexes between Asp30’ OD1 and APVFlap+ (N3) or DRVFlap+ (N1), respectively. ATV and SQV form 4 and 2 hydrogen bonds, respectively, to the backbone of residue 48 in the flap. However, the N to Gly48 O hydrogen bond in the SQVWT and SQVAct does not exist in SQVFlap+. Thus, there are specific changes in the hydrogen bonds for each of the inhibitors in the Flap+ complexes, rendering the hydrogen bonding pattern distinct from those in the corresponding inhibitor complexes of WT and Act protease variants.
Rearrangement of packing around the inhibitor in drug resistant proteases
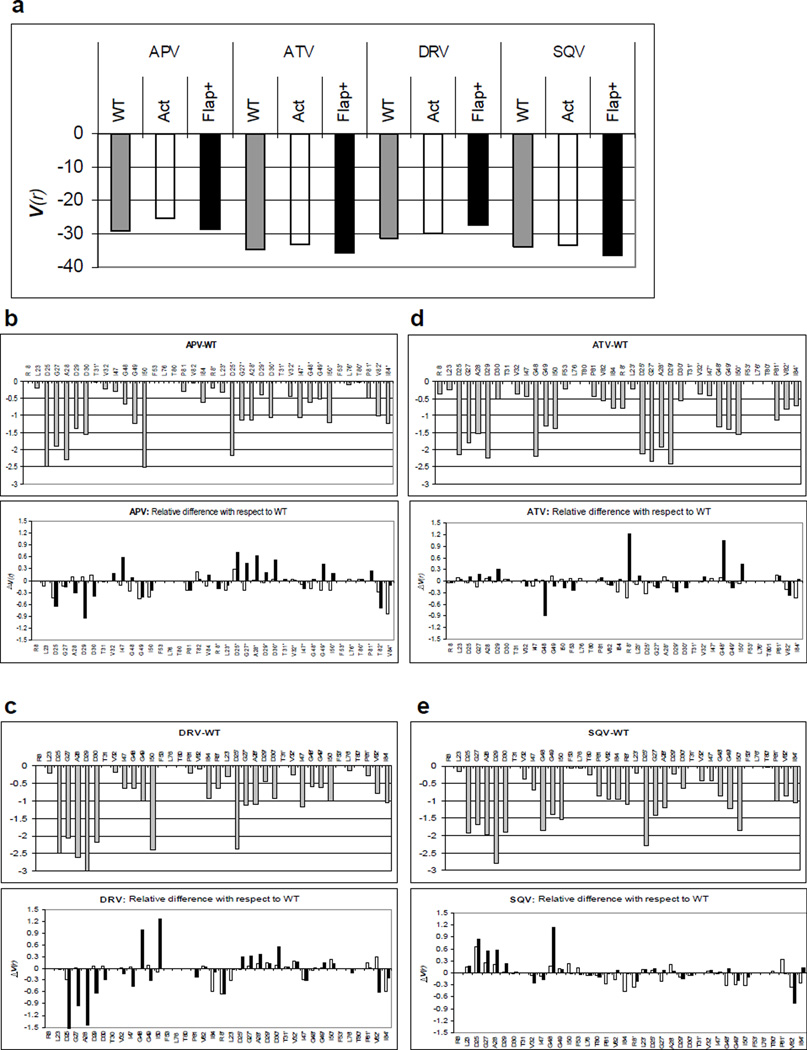
Details of binding and packing around the bound inhibitor in WT and MDR proteases were assessed by van der Waals (vdW) interactions between the inhibitor and the protein active site (Figure 6a). The difference in vdW interaction energy for each protease active site residue in Flap+ and Act complexes (ΔV(r)) with respect to WT protease (V(r)WT) were calculated (Figure 6b–e). Increase in vdW contacts by one inhibitor-protease residue pair is usually compensated by a decrease in contacts between another pair, rendering the net change in van der Waals contacts (ΣΔV(r)) (panel a) relatively small. These changes in residue contacts are due to the rearrangement of packing around the inhibitors in the MDR protease variants, and are reflected in the total absolute value of changes in vdW contacts (Σ|(ΔV(r)|) relative to WT complex structures. Repacking around the inhibitors is most pronounced for APVFlap+ and DRVFlap+ complexes, while the changes are smaller in APVAct and DRVAct (Figures 6b, d). Thus, the inhibitor-protease packing in the APV and DRV complexes of the Flap+ variant, which display severe entropy-enthalpy compensation, are significantly different compared to WT protease.
Figure 6.
Packing around the bound inhibitor in WT and MDR protease variants. (a) Total inhibitor-protease van der Waals interaction energies for WT (gray), Act (white) and Flap+ (black). Residue-wise distribution of interaction energy is shown in (b) APV, (c) ATV, (d) DRV and (e) SQV. The distribution of energies in the WT complexes are shown in the upper panels while the lower panels illustrate the WT vs Flap+ (black) and WT vs Act (white) differences in energy distribution.
A closer look at vdW interactions of the catalytic site and flap regions reveals the details of the active site rearrangement in Flap+ complex structures (Figures 6b–e). In APVFlap+ and DRVFlap+ complexes, inhibitor contacts increase for catalytic site residues Asp25–Asp30 in one monomer, but decrease in the other monomer (Asp25’–Asp30’). Concurrent with the large rearrangement of flap regions (Figure 3 and 4), these changes define the asymmetric conformation assumed by inhibitor complexes of Flap+ protease, which display extreme entropy-enthalpy compensation. Changes in contacts of flaps with the inhibitors in Flap+ variant are more subtle but distinct from Act and WT complexes. Interestingly, in all Flap+ complexes, nonpolar P1’ moiety of the inhibitor is in close proximity to flap tips, possibly stabilizing the unvaried conformation of the flaps (Figure 4a).
To assess the direct effect of resistance mutations, van der Waals contacts of mutation sites (L10I/G48V/I54V/V82A in Flap+ and V82T/I84V in Act) with the inhibitors were examined in detail. Only Val48 and Ala82 are involved in direct contacts with the inhibitors in Flap+ complexes. The sidechain of Val48 forms interactions with the inhibitor in SQVFlap+, while the inhibitor in all the other Flap+ complexes interact with the backbone of Val48. Residue 82 is a Thr in Act protease and Ala in Flap+. The V82T mutation in the Act complexes results in minor changes in van der Waals contacts. Compared to Thr82 in the Act complexes, the Flap+ complexes exhibit a loss in contacts by Ala82. Despite this loss in contacts due to the valine to alanine substitution, the three-dimensional arrangement of this region among the Flap+ complexes is highly conserved. Similar to the V82A mutation in the Flap+ complexes, the I84V mutation in the Act complexes results in a reduction of van der Waals contacts. Hence, the impact of the mutations appears not to be a direct change in overall van der Waals contacts, but rather an indirect change subtly rearranging the active site and therefore the energetics of inhibitor binding.
Molecular basis and implications of entropy-enthalpy compensation
Majority of thermodynamic studies on drug-resistant variants of HIV-1 protease have revealed a loss in the binding enthalpy, decreasing the overall free energy and hence affinity with respect to WT (6, 7, 19, 22, 35–37). The multi-drug resistant variant Flap+ displays unique thermodynamics of inhibitor binding, with an extreme entropy-enthalpy compensation and significant reduction in heat capacity change compared to the WT enzyme. This entropy-enthalpy compensation is observed with all the six different HIV-1 protease inhibitors studied here, especially for APV and DRV. The loss in the binding enthalpy with respect to WT is very high, on the order of 10 kcal mol−1, and this loss is compensated by favorable entropy changes of similar magnitude. This compensation leads to relatively low but still significant losses in the overall free energy, and hence binding affinity. The other multi-drug resistant protease variant, Act, has comparable reductions in inhibitor binding affinity as the Flap+ protease, but Act does not display any significant entropy-enthalpy compensation.
Comparing Flap+ to WT and Act protease complexes, rearrangement of packing around the bound inhibitor revealed by changes in vdW interactions, and distinct hydrogen bonding patterns were observed. The most significant structural changes in the inhibitor complexes of Flap+ compared to WT and Act are in the conformation of the flap regions (Figures 3–5). The Flap+ variant of HIV-1 protease has two mutations in the flaps at G48V, I54V, as well as an active site mutation V82A and a surface mutation just outside the active site, L10I. The flaps of HIV-1 protease are critical to enzyme function, and they are known to be highly mobile in the WT apo enzyme (38–40). The flaps of the protease must open up to allow substrate or inhibitor binding. When bound, the flaps close over the inhibitor in the active site. Different than the WT and Act variants, the tips of the Flap+ protease adopt a conserved conformation among all inhibitor complexes (Figure 4), and the rest of the flaps rearrange to accommodate various inhibitors. The two mutations in Flap+ flanking the flap tips (G48V/I54V) and the positioning of the nonpolar P1’ group of the inhibitors perhaps restricts the conformational freedom of this region, resulting in the apparent rigidity when bound to inhibitors. The conformationally restricted flap tips may act as hinges and cause the distal parts of the flaps (including the 40s loop) and the inhibitor to be more flexible in Flap+ complexes. Such an enhancement in conformational entropy would be consistent with the entropy-driven binding of inhibitors to Flap+ protease.
Another possible explanation for the entropy-driven binding of inhibitors to Flap+ could be the release of substantial amounts of solvation water from the active site upon inhibitor binding. Changes in solvent accessibility (41) and water structure (42) have been previously implicated as potential reasons for entropy-enthalpy compensation. There are no significant changes in the solvent accessibility and crystallographic water structure (see Supporting Information) of Flap+ complexes compared to WT protease. Hence, any possible role of water structure in the entropy-enthalpy compensation is not apparent from the crystal structures.
While implicating changes in the flap behavior, the unique entropy-enthalpy compensation phenomenon in the Flap+ variant cannot be inferred solely from the inhibitor-bound structures. The changes in binding thermodynamics are likely caused by highly interdependent, subtle but significant alterations in the structure and dynamics, involving both the inhibited and free states of the enzyme. However, deducing changes in the dynamics of a system from the static crystal structures is very challenging, if not impossible. Protein dynamics is related to the conformational entropy, and changes therein could potentially be significant enough to impact entropy and hence free energy of inhibitor binding (43). Flexibility and dynamics of Flap+ protease may be substantially altered in both apo and inhibitor-bound states. Molecular dynamics simulations comparing WT HIV-1 protease with a G48V mutant indicated a marked difference in the flexibility of the flap tips (44), reducing the frequency of trans-cis isomerization of the ω-bond for Val48 relative to Gly48. More recently, extensive MD simulations and detailed NMR relaxation experiments indeed indicated differential flap dynamics in the Flap+ variant compared to WT protease (45). Together with future analysis of inhibitor-bound structure, the evaluation of changes in dynamics and flexibility will contribute to a better understanding of the molecular mechanisms underpinning entropy-enthalpy compensation in this drug-resistant protease variant. Additionally, investigating the effect of individual mutations in contributing to the observed thermodynamic behavior will be informative (unpublished data). Future studies may also address the structural basis of resistance in variants with fully diminished susceptibility to APV and DRV to gain insight into the binding mode of these inhibitors.
The challenge in rational drug design is to truly integrate interdependent sequence, structure, energetic and dynamic data in a productive manner to minimize the emergence of drug resistance. To completely integrate thermodynamics in structure-based drug design, a comprehensive approach is necessary involving structures and dynamic information of both the free and the bound states of the inhibitor and the therapeutic target. The binding thermodynamic properties can be profoundly modulated not only by the inhibitor, but also by alteration of the target, such as in the evolution of drug resistance. In drug resistance, the target mutates to avoid drug binding, but still needs to maintain substrate binding and processing. The interplay between these two processes relies on a balance determined by the dynamics and kinetics of the system, and the target can evolve in many different ways to maintain this essential balance. The extreme entropy-enthalpy compensation observed here is a manifestation of subtle but significant changes that lead to drug resistance, and emerges as an additional challenge to rational drug design.
METHODS
Protease gene construction
The WT protease gene was constructed using synthetic oligonucleotides optimized for Escherichia coli codon usage, and included the Q7K substitution to prevent autoproteolysis (46). Additionally, all the constructs included the natural polymorphism L63P. Mutations were introduced using the Quick Change site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing.
Protein expression and purification
HIV-1 protease was overexpressed in E. coli Tap106 cells using heat induction, as previously described (20). The protease was extracted from inclusion bodies using 50% acetic acid (47). High molecular weight proteins were separated from the desired protease by size exclusion chromatography on a 2.1-L Sephadex G-75 superfine (Sigma Chemical) column equilibrated with 50% acetic acid. The protein was then refolded by rapid dilution into a 10-fold volume of 50 mM sodium acetate buffer at pH 5.5, also containing 10% glycerol, 5% ethylene glycol and 5 mM dithiothreitol (refolding buffer). The refolded protein was concentrated using an Amicon ultrafiltration cell, followed by dialysis to remove any residual acetic acid (6, 20, 48). The protein was further concentrated to approximately 1 to 2 mg/mL and stored at −80°C. Protease used for crystallization was further purified on a Pharmacia Superdex 75 fast-performance liquid chromatography column equilibrated with refolding buffer.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were carried out at 20°C using a VP-ITC microcalorimeter (MicroCal). All solutions were prepared in a buffer consisting of 10 mM sodium acetate (pH 5.0), 2% v/v dimethyl sulfoxide, and 2 mM Tris (2-carboxyethyl) phosphine as final concentrations. The protease underwent buffer exchange using PD10 gel filtration columns (Amersham Biosciences). Due to the high affinity of WT protease to all inhibitors as well as the sharp transition to saturation, the thermodynamic parameters for complex formation were obtained by the displacement titration method (4, 6, 7, 49). Likewise, affinities for ATV binding to Act protease, and APV and DRV binding to both mutant enzymes were determined using competition experiments. With one exception, acetyl-pepstatin (Bachem Bioscience) was used as the competing ligand at concentrations ranging from 250 to 400 µM. For the Flap+ displacement experiments, acetyl-pepstatin concentration was 300 µM with 250 µM APV, while 900 µM acetyl-pepstatin was required for titrations with 66 µM DRV. IDV was used as the weaker binder in displacement experiments with Act protease and DRV, as described (6). Protease concentrations of approximately 6 to 20 µM were used in displacement experiments. The protease in the calorimetric cell was initially titrated with the weaker binding inhibitor, followed by titration with the higher affinity ligand for displacement of acetylpepstatin (or IDV). APV, ATV, DRV, IDV, NFV and SQV were used at concentrations of 150 to 250 µM. The thermodynamic parameters for APV and DRV with the WT protease were obtained as previously described (6).
Due to the decrease in the binding affinity of ATV, IDV, NFV and SQV with Flap+ protease, the thermodynamic parameters for these complexes were obtained by direct titrations. The same was true for experiments done on Act protease with IDV, NFV, and SQV. In each case, 200 µM inhibitor stock solution was titrated directly into the calorimetric cell containing protease at concentrations ranging from 16 to 26 µM. For experiments carried out using the displacement method, direct titrations were also performed in order to confirm the enthalpy changes acquired through competition experiments. All experiments were carried out at least twice and their mean values are reported. Heats of dilution obtained after saturation were subtracted from the heats of reaction in order to obtain the heat due solely to the inhibitor binding to the enzyme. Data were analyzed using the Origin 7 software package provided by MicroCal. Concentrations of active, folded protease reported here were determined during curve fitting, by adjusting the protease concentration to the value which results in a stoichiometric ratio of inhibitor to enzyme at half saturation.
The heat capacity change (ΔCp) associated with binding of APV and DRV to WT and Flap+ variants was determined by measuring the binding enthalpy over a temperature range of 10 to 42°C, titrating 200 µM APV or 84 µM DRV into 13 to 50 µM protein in the calorimetric cell.
Crystal structures
The following nomenclature will be followed to refer to each crystal structure: Inhibitorprotease variant. For example, APVWT, APVAct and APVFlap+ represent the WT, Act and Flap+ complexes, respectively, of APV. Crystal structures of the six inhibitors in complex with WT, Act and Flap+ protease variants were determined. The IDV complex did not form diffraction quality crystals and the flaps in NFVFlap+ exhibit an unusual conformation (30) and therefore, structural comparisons with those two inhibitor complexes are not included. In addition, crystals for the SQVWT complex could not be grown and therefore, the complex structure from the PDB was used (PDB code 1HXB (50)). The inhibitor in all of the complexes, except ATVWT and SQVWT, binds in a unique conformation. In the ATVWT and SQVWT complexes, the inhibitor binds in two orientations with nearly 50% occupancy for each orientation. In the ATVWT structure, the second phenyl group at P1' does not have electron density, and therefore, was not included during any of the subsequent structural analyses. Altogether, this analysis includes eight new HIV-1 protease-inhibitor complexes, three previously reported from our laboratory and one from the database, allowing for a detailed structural comparison of drug resistant variants with the WT enzyme.
Crystallization and data collection
Crystal screens were set up with a three- to five-fold molar excess of inhibitor to protease to ensure ubiquitous binding. The final protein concentration ranged from 0.5–2.5 mg mL−1 in refolding buffer. The hanging drop vapor diffusion method was used for crystallization as previously described (6). With two exceptions, crystal screens were set up at ambient temperature using reservoir solutions consisting of 126 mM phosphate buffer at pH 6.2, 63 mM sodium citrate and ammonium sulfate at a range of 25 to 33 % w/v. Crystals for the APVFlap+ and DRVFlap+ complexes however, were grown at 4°C using 10:1 molar ratios of inhibitor to protein. The buffer used in each case consisted of 50 mM citrate phosphate at pH 5.0 with 7% v/v DMSO, and ammonium sulfate at concentrations of 38% and 28% w/v for APV and DRV respectively.
Crystallographic data for all the complexes was collected on an RAXIS IV. The raw frames were indexed and integrated using DENZO and subsequently scaled using ScalePack (51, 52). All of the complexes, except two, crystallized in the usual orthorhombic crystal form with isomorphous cell dimensions. APVFlap+ and DRVFlap+ crystallized in an unusual hexagonal space group with 12 HIV-1 protease dimers per unit cell. The data collection statistics are listed in Table 2.
Structure solution and crystallographic refinement
The methods used for structure solution is detailed in Supporting Information. The refinement statistics are provided in Table 2.
Structural analysis
(i) Structural superimpositions: The inhibitor complexes were superimposed on the substrate capsid-p2 complex with WT protease (PDB code 1F7A(34)) using the protease terminal domain (Pro1–Pro9 and Arg87–Phe99). This substrate complex was chosen to preserve consistency and enable comparisons with our previous analyses (6, 21).
(ii) Double difference plots: Double-difference plots, computed using Cα–Cα distances, reveal structural differences between similar structures without the bias due to superimposition (30, 31). Distances between all the Cα atoms within each dimer were computed (dij, where d is the distance and i and j are residue numbers). This was repeated for each of the structures. Double differences (D) were then calculated as the difference of the distances between two structures n and m (Dij = ndij − mdij). The (I x j) matrix was then displayed as a contour diagram using GnuPlot (31).
(iii) Estimation of van der Waals potential: Inhibitor-protease van der Waals contacts were estimated by a simplified Lennard-Jones potential V(r) using the relation 4ε[(σ/r)12 − (σ/r)6]; where r is the inter-atomic distance, and ε and σ are the well depth and hard sphere diameter, respectively, for each protease-inhibitor atom pair (53). V(r) is computed for all possible protease-inhibitor atom pairs within 5 Å, and equated to ε for non-bonded pairs separated less than a distance corresponding to the minimum of the potential. Using this simplified potential for each non-bonded protease-inhibitor atom pair, ΣV(r) was then computed for each protease residue.
Supplementary Material
Acknowledgements
The authors thank D. Cooper, C. Ng, V. Chou, C. McRell, S. Kathuria, M. Kolli for help with experiments, Y. Cai for the force field, and B. Bhyravbhatla and L. Leone for technical assistance. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: amprenavir, atazanavir, indinavir, nelfinavir and saquinavir. Darunavir was kindly provided by Tibotec, Inc. This work has been funded by National Institutes of Health (P01-GM66524).
Footnotes
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Chaires JB. Annual Review of Biophysics. Palo Alto: Annual Reviews; 2008. Calorimetry and thermodynamics in drug design; pp. 135–151. [DOI] [PubMed] [Google Scholar]
- 2.Wlodawer A, Erickson JW. Structure-based inhibitors of HIV-1 protease. Ann. Rev. of Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 3.Todd MJ, Luque I, Velazquez-Campoy A, Freire E. Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry. 2000;39:11876–11883. doi: 10.1021/bi001013s. [DOI] [PubMed] [Google Scholar]
- 4.Valzaquez-Campoy A, Luque I, Todd MJ, Milutinovich M, Kiso Y, Freire E. Thermodynamic dissection of the binding energetics of KNI-272, a potent HIV-1 protease inhibitor. Protein Sci. 2000;9:1801–1809. doi: 10.1110/ps.9.9.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valzaquez-Campoy A, Todd MJ, Freire E. HIV-1 protease inhibitors: enthalpic versus entropic optimization of the binding affinity. Biochemistry. 2000;39:2201–2207. doi: 10.1021/bi992399d. [DOI] [PubMed] [Google Scholar]
- 6.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune M-P, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type-1 protease inhibitor. J. Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtaka H, Velazquez-Campoy A, Xie D, Freire E. Overcoming drug resistance in HIV-1 chemotherapy: the binding thermodynamics of Amprenavir and TMC-126 to wild-type and drug-resistant mutants of the HIV-1 protease. Protein Sci. 2002;11:1908–1916. doi: 10.1110/ps.0206402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surleraux DL, de Kock HA, Verschueren WG, Pille GM, Maes LJ, Peeters A, Vendeville S, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. Design of HIV-1 protease inhibitors active on multidrug-resistant virus. J. Med. Chem. 2005;48:1965–1973. doi: 10.1021/jm049454n. [DOI] [PubMed] [Google Scholar]
- 9.Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, Amzel LM, Freire E. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J. Virol. 2007;81:5144–5154. doi: 10.1128/JVI.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez A, Fraser C, Scott LR. Purposely engineered drug-target mismatches for entropy-based drug optimization. Trends Biotechnol. 2012;30:1–7. doi: 10.1016/j.tibtech.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Kuroki R, Nitta K, Yutani K. Thermodynamic changes in the binding of Ca2+ to a mutant human lysozyme (D86/92). Enthalpy-entropy compensation observed upon Ca2+ binding to proteins. J. Biol. Chem. 1992;267:24297–24301. [PubMed] [Google Scholar]
- 12.Blasie CA, Berg JM. Entropy-enthalpy compensation in ionic interactions probed in a zinc finger peptide. Biochemistry. 2004;43:10600–10604. doi: 10.1021/bi0363230. [DOI] [PubMed] [Google Scholar]
- 13.Krueger S, Gregurick S, Shi Y, Wang S, Wladkowski BD, Schwarz FP. Entropic Nature of the Interaction between Promoter Bound CRP Mutants and RNA Polymerase. Biochemistry. 2003;42:1958–1968. doi: 10.1021/bi026755v. [DOI] [PubMed] [Google Scholar]
- 14.Davidson JP, Lubman O, Rose T, Waksman G, Martin SF. Calorimetric and Structural Studies of 1,2,3-Trisubstituted Cyclopropanes as Conformationally Constrained Peptide Inhibitors of Src SH2 Domain Binding. J. Am. Chem. Soc. 2002;124:205–215. doi: 10.1021/ja011746f. [DOI] [PubMed] [Google Scholar]
- 15.Houk KN, Leach AG, Kim SP, Zhang X. Binding Affinities of Host-Guest, Protein-Ligand, and Protein-Transition-State Complexes. Angewandte Chemie International Edition. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 16.Rekharsky MV, Inoue Y. Complexation Thermodynamics of Cyclodextrins. Chemical reviews. 1998;98:1875–1918. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- 17.Sharp K. Entropy—enthalpy compensation: Fact or artifact? Prot. Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen DB, Stahlhut MW, Rutkowski CA, Schock HB, vanOlden AL, Kuo LC. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J. Biol. Chem. 1999;274:23699–23701. doi: 10.1074/jbc.274.34.23699. [DOI] [PubMed] [Google Scholar]
- 19.Ohtaka H, Schon A, Freire E. Multidrug Resistance to HIV-1 Protease Inhibition Requires Cooperative Coupling between Distal Mutations. Biochemistry. 2003;42:13659–13666. doi: 10.1021/bi0350405. [DOI] [PubMed] [Google Scholar]
- 20.King NM, Melnick L, Prabu-Jeyabalan M, Nalivaika EA, Yang S-S, Gao Y, Nie X, Zepp C, Heefner DL, Schiffer CA. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 2002;11:418–429. doi: 10.1110/ps.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King NM, Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Combating Susceptibility to Drug Resistance: Lessons from HIV-1 Protease. Chem. Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Lafont V, Armstrong AA, Ohtaka H, Kiso Y, Mario Amzel L, Freire E. Compensating enthalpic and entropic changes hinder binding affinity optimization. Chem. Biol. Drug Des. 2007;69:413–422. doi: 10.1111/j.1747-0285.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 23.Ohtaka H, Muzammil S, Schön A, Velazquez-Campoy A, Vega S, Freire E. Thermodynamic rules for the design of high affinity HIV-1 protease inhibitors with adaptability to mutations and high selectivity towards unwanted targets. Int. J Biochem. Cell Biol. 2004;36:1787–1799. doi: 10.1016/j.biocel.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Steuber H, Heine A, Klebe G. Structural and thermodynamic study on aldose reductase: nitro-substituted inhibitors with strong enthalpic binding contribution. Journal of molecular biology. 2007:368. doi: 10.1016/j.jmb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Bottcher J, Blum A, Heine A, Diederich WE, Klebe G. Structural and kinetic analysis of pyrrolidine-based inhibitors of the drug-resistant Ile84Val mutant of HIV-1 protease. Journal of molecular biology. 2008;383:347–357. doi: 10.1016/j.jmb.2008.07.062. [DOI] [PubMed] [Google Scholar]
- 26.Shafer RW, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eftink MR, Anusiem AC, Biltonen RL. Enthalpy-entropy compensation and heat capacity changes for protein-ligand interactions: general thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry. 1983;22:3884–3896. doi: 10.1021/bi00285a025. [DOI] [PubMed] [Google Scholar]
- 29.Guan R, Ho MC, Brenowitz M, Tyler PC, Evans GB, Almo SC, Schramm VL. Entropy-Driven Binding of Picomolar Transition State Analogue Inhibitors to Human 5 '-Methylthioadenosine Phosphorylase. Biochemistry. 2011;50:10408–10417. doi: 10.1021/bi201321x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabu-Jeyabalan M, King NM, Nalivaika E, Heilek-Snyder G, Cammack N, Schiffer CA. Substrate Envelope and Drug Resistance: Crystal Structure of RO1 in Complex with Wild-Type Human Immunodeficiency Virus Type 1 Protease. Antimicrob. Agents Chemother. 2006;50:1518–1521. doi: 10.1128/AAC.50.4.1518-1521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams T, Kelley C. GNUPLOT. 1998 [Google Scholar]
- 32.Freedberg DI, Wang YX, Stahl SS, Kaufman JD, Wingfield PT, Kiso Y, Torchia DA. Flexibility and Function in HIV Protease: Dynamics of the HIV-1 Protease Bound to the Asymmetric Inhibitor Kynostatin 272. J. Am. Chem. Soc. 1998;120:7916–7923. [Google Scholar]
- 33.Jhoti H, Singh OM, Weir MP, Cooke R, Murray-Rust P, Wonacott A. X-ray crystallographic studies of a series of penicillin-derived asymmetric inhibitors of HIV-1 protease. Biochemistry. 1994;33:8417–8427. doi: 10.1021/bi00194a005. [DOI] [PubMed] [Google Scholar]
- 34.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 2000;301:1207–1220. doi: 10.1006/jmbi.2000.4018. [DOI] [PubMed] [Google Scholar]
- 35.Ohtaka H, Freire E. Adaptive inhibitors of the HIV-1 protease. Prog Biophys Mol Biol. 2005;88:193–208. doi: 10.1016/j.pbiomolbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Muzammil S, Ross P, Freire E. A Major Role for a Set of Non-Active Site Mutations in the Development of HIV-1 Protease Drug Resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 37.Stoica I, Sadiq SK, Coveney PV. Rapid and accurate prediction of binding free energies for saquinavir-bound HIV-1 proteases. J. Am. Chem. Soc. 2008;130:2639–2648. doi: 10.1021/ja0779250. [DOI] [PubMed] [Google Scholar]
- 38.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure with Folding and Design. 1999;7:1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 40.Scott WRP, Schiffer CA. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 41.Ladbury JE, Chowdhry BZ, editors. Biocalorimetry. Applications of Calorimetry in the Biological Sciences. Chichester: John Wiley & Sons Ltd; 1998. [Google Scholar]
- 42.Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem. Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 43.Diehl C, Engstrom O, Delaine T, Hakansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, Akke M. Protein Flexibility and Conformational Entropy in Ligand Design Targeting the Carbohydrate Recognition Domain of Galectin-3. J. Am. Chem. Soc. 2010;132:14577–14589. doi: 10.1021/ja105852y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamelberg D, McCammon JA. Fast Peptidyl cis-trans Isomerization within the Flexible Gly-Rich Flaps of HIV-1 Protease. J. Am. Chem. Soc. 2005;127:13778–13779. doi: 10.1021/ja054338a. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y, Kurt Yilmaz N, Myint W, Ishima R, Schiffer C. Differential Flap Dynamics in Wild-type and a Drug Resistant Variant of HIV-1 Protease Revealed by Molecular Dynamics and NMR Relaxation. Journal of Chemical Theory and Computation. 2012 doi: 10.1021/ct300076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose JR, Salto R, Craik CS. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J. Biol. Chem. 1993;268:11939–11945. [PubMed] [Google Scholar]
- 47.Hui JO, AG T, Reardon IM, Lull JM, Brunner DP, Tomich CC, Heinrikson RL. Large scale purification and refolding of HIV-1 protease from Escherichia coli inclusion bodies. J. Prot. Chem. 1993;12:323–327. doi: 10.1007/BF01028194. [DOI] [PubMed] [Google Scholar]
- 48.Prabu-Jeyabalan M, Nalivaika EA, King NM, Schiffer CA. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 2004;78:12446–12454. doi: 10.1128/JVI.78.22.12446-12454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sigurskjold B. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 50.Krohn A, Redshaw S, Ritchie JC, Graves BJ, Hatada MH. Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere. J. Med. Chem. 1991;34:3340–3342. doi: 10.1021/jm00115a028. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. CCP4 Study weekend: Data collection and processing, 29–30, Jan 1993. England: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 52.Minor W. XDISPLAYF program. West Lafayette, Indiana: Purdue University; 1993. [Google Scholar]
- 53.Engh RA, Huber R. Accurate Bond and Angle Parameters for X-ray Protein Structure Refinement. Acta Crystallogr. Sect. A. 1991;47:392–400. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.