Abstract
Roses hold high symbolic value and great cultural importance in different societies throughout human history. They are widely used as garden ornamental plants, as cut flowers, and for the production of essential oils for the perfume and cosmetic industries. Domestication of roses has a long and complex history, and the rose species have been hybridized across vast geographic areas such as Europe, Asia, and the Middle East. The domestication processes selected several flower characters affecting floral quality, such as recurrent flowering, double flowers, petal colours, and fragrance. The molecular and genetic events that determine some of these flower characters cannot be studied using model species such as Arabidopsis thaliana, or at least only in a limited manner. In this review, we comment on the recent development of genetic, genomic, and transcriptomic tools for roses, and then focus on recent advances that have helped unravel the molecular mechanisms underlying several rose floral traits.
Key words: Flower, genomics, morphogenesis, rose.
Introduction
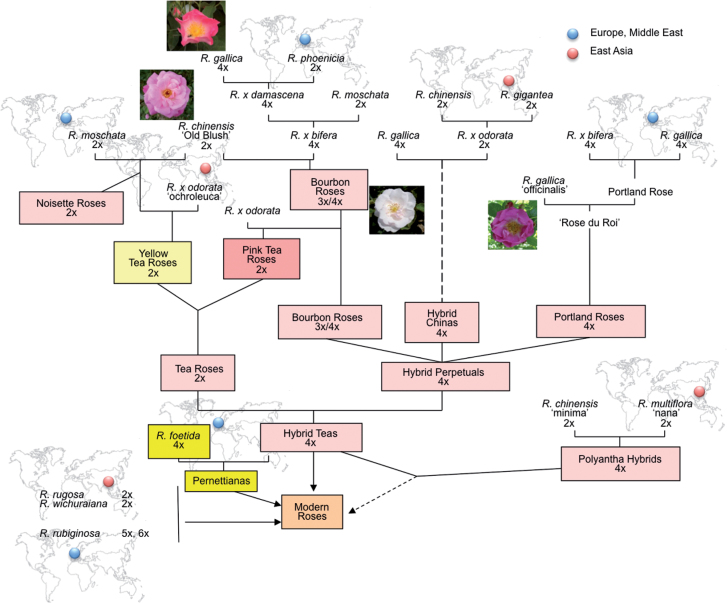
The genus Rosa belongs to the large family of the Rosaceae. Roses have been cultivated since antiquity, as early as 3000 BC in China, western Asia, and northern Africa. Wild roses were first domesticated and multiplied to use as an animal-proof fence. The Romans, Greeks, and Persians used domesticated roses as floral ornaments and/or as medicinal plants. Spontaneous interspecific hybrids, such as Rosa×damascena, have been stabilized by vegetative propagation. In the 14th century, missionaries introduced Chinese roses to Europe. The subsequent extensive hybridization amongst the Chinese, European, and Middle-Eastern roses formed the genetic basis of the ‘modern rose cultivars’ (Raymond, 1999) (Fig. 1). Nowadays, ~30 000–35 000 cultivated rose varieties are said to exist. They are usually referred to as Rosa×hybrida (Gudin, 2003). Today, roses are one of the most commonly cultivated ornamental plants in the world. They are highly popular as garden ornamental plants and cut flowers. They are also in enormous demand in the perfume and cosmetic industries. The ‘modern rose cultivars’ exhibit a huge variety of characteristics such as flower shape, colour, and fragrance that we now enjoy in gardens and parks.
Fig. 1.
Schematic representation of major steps of modern rose genealogy. Based on Raymond (1999).
Understanding the genetic and molecular mechanisms that regulate these traits is not just a fascinating question of basic rose biology, but will also help understand how these traits were selected during rose domestication and may also have important implications for the rational manipulation of rose quality.
Here we will review current knowledge about molecular and genetic approaches as well as the advances in understanding the genetic and molecular mechanisms controlling important rose traits, focusing mainly on flower initiation development and function.
The complex origin of cultivated roses
The genus Rosa comprises ~200 species, among which only 8–20 species have contributed to the genetic make-up of our present cultivars, namely the complex hybrid variety Rosa×hybrida (De Vries and Dubois, 1996; Reynders-Aloisi and Bollereau, 1996; Gudin, 2001). Each of these species may have contributed a specific trait. For example, Rosa gallica and other robust polyploid species lent the trait of cold hardiness, Rosa chinensis brought recurrent blooming, and Rosa foetida bestowed the yellow flower colour. However, despite such apparent mosaicity, the genome of modern rose varieties seems to bear the remains of a massive introgression of R. chinensis alleles (Martin et al., 2001). Therefore, although the genome of Rosa×hybrida is a patchy mix of several parental genomes, the origin of its genetic variation is relatively homogeneous because of the intensive backcrossing to R. chinensis cultivars during the recent processes of varietal creation. In fact, one of the major hindrances to the creation of novel rose varieties stems from such lack of allelic variation compounded by the difficulty in introgressing alleles of interest from wild diploid species due to the polyploidy barrier. More than half of the wild rose species are polyploid (Vamosi and Dickinson, 2006), ranging from 2n=2x=14 to 2n=8x=56 (Roberts et al., 2009), with permanent sexual pentaploids such as Rosa canina exhibiting unusual asymmetric meiosis (Lim et al., 2005; Kovarik et al., 2008). Recently, karyotyping of Rosa prealucens from the Sino-Himalayan region revealed decaploidy, the highest naturally occurring ploidy of the genus (Jian et al., 2010). A recent study suggests a possible evolutionary mechanism involving ploidy changes in response to adverse environmental conditions, such as exposure to high temperature (Pécrix et al., 2011). Therefore, manipulating temperature may be a strategy to overcome the ploidy barrier to introgress wild alleles of interest to modern rose cultivars.
Approaches and tools used to study important rose traits
From a commercial point of view, there are several very important rose traits. These traits include plant architecture, flower development, architecture, and senescence, scent biosynthesis and emission, ease of reproduction, and resistance to biotic and abiotic stresses. During the past decade, molecular and genetic approaches were used to identify and functionally characterize genes and gene networks associated with important rose traits and their inheritance among cultivated roses. Initial cloning relied on candidate gene approaches. However, until early 2000, very little information was available on gene expression in rose. This dearth of information hampered the discovery of genes associated with various traits.
In parallel, a number of laboratories created high-density linkage maps with the hope of locating or tag genes associated with important flowering traits. So far, linkage analysis in rose has been complicated by the quasi-impossibility of obtaining inbred lines, due to self-incompatibility and high heterozygosity. Many mapping populations exist and were used to unravel the genetic basis of monogenic or oligogenic characters such as the simple corolla, pink flower, recurrent blooming, and three volatile components of scent (Debener and Mattiesch, 1999; Crespel et al., 2002; Dugo et al., 2005; Yan et al., 2005; Linde et al., 2006; Spiller et al., 2011; Moghaddam et al., 2012). When it comes to identifying the genes controlling such characters, the mapping populations are usually of insufficient size (typically <100 individuals for the diploid maps), so that the observation of co-localization between one candidate gene and the locus of interest can be misleading. Not only is it difficult to carry out genetic analysis on quantitative and multigenic flower characteristics on a small mapping population, but also phenotyping roses can be demanding because many of the complex flowering traits are affected by environmental factors. Phenotyping has to be done on a pluri-annual basis, and such a task can be extremely labour intensive for complex traits such as inflorescence architecture. There is clearly a trade-off between the accuracy of phenotyping and size of the mapping population.
Because each of the above approaches has limitations, combinations of genetic mapping, quantitative trait locus (QTL) analyses, and candidate gene approaches are being undertaken by a number of research groups in order to identify genes and genetic pathways associated with important rose traits. In the late 2000s, researchers started to develop advanced molecular tools and used them to study Rosa sp. The first rose expressed sequence tag (EST) sequencing (Channeliere et al., 2002; Guterman et al., 2002; Foucher et al., 2008) and two sets of microarrays, although containing only 350 (Guterman et al., 2002) and 5000 genes (Dubois et al., 2011), respectively, were instrumental in identifying genes associated with flower initiation, flower organ morphogenesis and senescence, as well as scent biosynthesis (Guterman et al., 2002, 2006; Lavid et al., 2002; Scalliet et al., 2002, 2006, 2008; Dubois et al., 2011). Very recently, Dubois et al. (2012) used next-generation Illumina and 454 sequencing technologies to cover most of the rose transcriptome. This study permitted the authors to obtain (i) information on transcripts representing ~21 000 unisequences or putative genes expressed in various rose tissues and organs (i.e. roots, flowers, leaves, stems, and cynorhodon) and in response to biotic and abiotic stresses (http://iant.toulouse.inra.fr/R.chinensis); (ii) digital expression of each of these transcripts in plant organs, some at different development stages or under different stress conditions; and (iii) information on the putative peptides and putative enzymatic pathways (http://pathway-tools.toulouse.inra.fr/ROSACYC). These recently developed rose databases represent comprehensive resources for transcript detection and accumulation, and a valuable prerequisite to identify genes and gene pathways associated with various rose traits.
In the following sections, we will provide an updated summary of recent findings regarding the molecular and genetic mechanisms that control floral initiation, development, and function, as well as the available molecular and genetic tools.
Recurrent flowering and flowering time
Roses are perennial shrubs with axillary buds that undergo floral transition in late autumn, remain dormant in winter, and bloom in spring when temperatures are permissive. Short-day cultivars of roses flower once a year in spring; most of the ever-blooming long-day cultivars flower recurrently until autumn or even until the first frost. The flowers occurring in spring originate from buds that have undergone floral transition in autumn.
The flower burst of rose buds needs light (Maas and Bakx, 1995). White, far-red, or blue light can trigger bud burst (Girault et al., 2008). Bud blooming also requires sucrose, but experiments on bud explants showed that sucrose alone in the dark fails to trigger bud burst. Both light and sucrose function as signals that are necessary for bud burst and vacuolar invertase activity that permits sucrose catabolism (Rabot et al., 2012). The phytohormones gibberellins (GAs) are also implicated in the bud burst process of recurrent flowering in hybrid roses, as Choubane et al. (2012) showed that the GA inhibitors paclobutrazol or ancymidol (at 50 µM) can inhibit bud burst, and GA biosynthesis genes are up-regulated during the bud burst process.
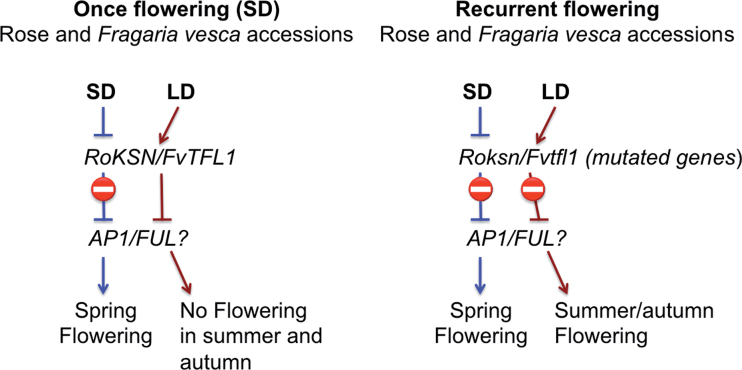
In non-recurrent cultivars, floral transition occurs within the main and axillary buds during short photoperiods in autumn and early spring. In recurrent flowering cultivars, floral transition also occurs during long photoperiods such as in late spring and summer. Recurrent or continuous flowering (CF) (i.e. the ability of the plant to flower many times a year) appears to be under the control of a single locus, ‘Recurrent Blooming’ or RB. Crespel et al. (2002) mapped the RB locus and demonstrated that RB co-localizes with the rose homologue of SPINDLY (RoSPINDLY), an Arabidopsis gene involved in GA signalling (Silverstone et al., 2007; Remay et al., 2009). However, in a recent study using more mapping populations, the same authors failed to confirm the co-localization of RB and RoSPINDLY on IM-LG3 (Integrated Map-Linkage Group3; Spiller et al., 2011). In 2012, Iwata et al. discovered a tight map linkage between RB and RoKSN in 670 diploid F1 progeny. This observation, combined with sequence comparison results from once-flowering versus CF roses, suggests that RoKSN is responsible for the CF phenotype. RoKSN encodes an orthologue of Arabidopsis TERMINAL FLOWER 1 (TFL1). In Arabis alpina, Populus, and Malus domestica, TFL1 can repress flowering through the regulation of APETALA1 and FRUITFUL, and control the length of the juvenile phase (Kotoda et al., 2006; Mohamed et al., 2010; R.H. Wang et al., 2011). In agreement with these data, a transposon insertion and a point mutation in the TFL1 orthologue correlate with the CF trait in roses and in Fragaria vesca, respectively (Iwata et al., 2012). In a recent study in F. vesca, FvTFL1 was shown to act as a photoperiod-regulated floral repressor. In the short-day (non-recurrent) flowering F. vesca, FvTFL1 is repressed by short photoperiods in winter and early spring, so that floral identity genes such as APETALA1 and FRUITFUL can be expressed. Consequently, floral buds are then formed and are ready to burst in spring. After blooming, TFL1 expression is activated by a long photoperiod and represses further flower formation in summer. In strawberry accessions that were selected for recurrent flowering (i.e. long-day accessions), TFL1 is mutated and prevents long-day suppression of flowering (Fig. 2) (Koskela et al., 2012). A similar mechanism may occur in roses where a mutation in the TFL1 orthologue enables flowering in long days (Iwata et al., 2012; Wang et al., 2012). Interestingly, a study in a different genetic background using distinct mapping populations identified two QTLs for flowering time, suggesting that the CF trait may be under the control of multiple regulators (Dugo et al., 2005).
Fig. 2.
Models showing the control of flowering for once- and recurrent-flowering Rosa sp. and Fragaria vesca cultivars. Adapted from Iwata et al. (2012) and Koskela et al. (2012).
Determination of rose flower organ identity and morphogenesis
Like most core eudicots, the flower of wild-type roses consists of four organ types: five sepals, five petals, and a large number of stamens and carpels are arranged in concentric whorls, with the sepals occupying the outermost whorl (Fig. 3). The number of stamens and carpels varies among rose species. Through mutant analysis, molecular cloning, and functional characterization of the corresponding genes in model species such as A. thaliana and Antirrhinum majus, several groups have helped establish a model which shows that the combinatory actions of four classes of homeotic genes (A, B, C, and E) determine flower organ identity and trigger the developmental programmes required for flower organogenesis (Theissen and Saedler, 2001; Krizek and Fletcher, 2005; Irish, 2010). These homeotic genes have globally conserved functions among Eudicots. In Rosa sp., orthologues of the MADS-box genes involved in flower organ identity determination have been cloned and, for some, functionally characterized (Kitahara and Matsumoto, 2000; Kitahara et al., 2001, 2004; Hibino et al., 2006; Remay et al., 2009; Dubois et al., 2010). Organ identity determination genes such as RhAPETALA3, RhPISTILLATA, RhAGAMOUS, and RhSHATTERPROOF were cloned in Rosa sp. Expression analysis in Rosa sp. and overexpression experiments of these rose MADS-encoding cDNAs in Arabidopsis confirmed that the functions of these B and C function genes are conserved (Kitahara and Matsumoto, 2000; Kitahara et al., 2001, 2004; Hibino et al., 2006).
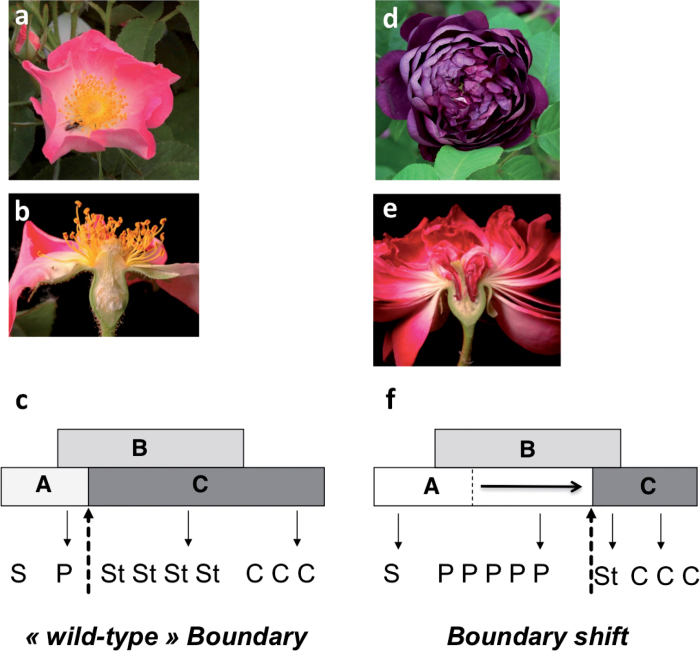
Fig. 3.
Double flower phenotype in Rosa. (a) Simple flower Rosa gallica. (b) Longitudinal section of a R. gallica flower. (d) Double flower of R. gallica cv Cardinal de Richelieu. (e) Longitudinal section of a double flower rose of R. gallica cv Cardinal de Richelieu. (c and f) Model showing the origin of the double flower phenotype. (c) ABC model in wild-type simple flower roses. (f) In double flower roses, a shift of the boundary between A and C functions towards the centre of the flower leads to homeotic conversion of stamens into petals. Adapted from Dubois et al. (2010).
In Rosa sp., the number of flower organs, especially that of petals, greatly influences flower architecture. While the wild roses have a ‘simple flower’ with typically five petals per flower, the modern roses have double flowers consisting of >10 petals. In a number of species, researchers have uncovered some morphological, developmental, and genetic aspects (i.e. QTLs) of the double flower phenotype (Innes et al., 1989; Lehmann and Sattler, 1993; Maclntyre and Lacroix, 1996; Y.Q. Wang et al., 2011), and it was not until 2010 that the underlying molecular mechanisms were first unravelled in Rosa (Dubois et al., 2010). Dubois et al. (2010) demonstrated that in Rosa sp. the orthologue of AGAMOUS (RhAG) was differentially expressed in double flowers as compared with simple flowers. Using in situ hybridization, they showed that in double flower roses, RhAG expression is down-regulated and its domain of expression is restricted towards the centre of the floral meristem, hence allowing more petals and fewer stamens to form. The authors also demonstrated that the restriction of the RhAG (C-function) expression domain towards the centre of the floral meristem was selected independently in two major regions of rose domestication (China and the peri-Mediterranean areas) (Fig. 3). Recently, misexpression of the AG orthologue in ranunculids was also proposed to be responsible for the double flower phenotype (Galimba et al., 2012). Similarly, in Prunus lannesiana, a deletion of C-terminal motifs in AGAMOUS was proposed to be responsible for the double flower phenotype (Liu et al., 2012). Therefore, in all analysed species the double flower phenotype was always associated with alterations in the C-function gene AGAMOUS.
Using genetic mapping studies, rose researchers showed that the simple versus double corolla phenotype is linked to a single dominant locus. This locus, referred to as Blfo or d6, was mapped on IM-LG3 on the integrated map of diploid roses (Debener and Mattiesch, 1999; Crespel et al., 2002; Spiller et al., 2011). Crosses between a simple corolla and double corolla rose typically produce a double corolla in half of the F1 progeny. In these double corolla progeny, the petal number is variable and can be scored quantitatively for QTL identification. However, as petal number is influenced by various environmental factors (Debener, 1999), the accuracy of categorizing the double corolla phenotype can be compromised. Therefore, when scoring the double corolla phenotype, one must compute the average number of petals per flower and take seasonal effects into account. Despite these limitations, a major QTL associated with several flower traits was identified on IM-LG3 in the close vicinity of Blfo. It accounts for almost 30% of phenotypic variation (Hibrand Saint Oyant et al., 2008; Remay et al., 2009). Interestingly, when researchers crossed two double corolla progenitors and applied single marker QTL mapping on a tetraploid F1 progeny, they identified an IM-LG3 marker that correlates with 12% of petal number variance (Koning-Boucoiran et al., 2012).
However, mapping also showed that RhAG did not co-localize with Blfo or the QTL for petal number (Spiller et al., 2011). This is in agreement with Dubois et al. (2010) who proposed that RhAG, whose expression strongly correlates with petal number, is under the control (direct or indirect) of a master regulatory gene likely to be localized at the Blfo locus. Moreover, candidate genes known to regulate AG expression in Arabidospis were systematically rejected as master determinants of the petal number phenotype for the following two reasons: (i) they did not co-localize with Blfo or the QTL for petal number; and (ii) their expression was not affected in the double flower roses (Spiller et al., 2011; unpublished data). Further studies are required to elucidate the molecular mechanisms controlling RhAG expression in rose double flowers. Furthermore, in Rosa sp. flower organ morphogenesis, post-organ identity determination remains completely unknown. Even in model species such as Arabidopsis, the link between gene regulation/function and organ morphogenesis remains largely unknown (Dornelas et al., 2011).
Until recently, studies on rose floral organ differentiation focused on stamens and petals, especially petal differentiation, because the ornamental and commercial value of the rose depends heavily on petal characteristics. At the cellular level, rose petal expansion is a result of cell expansion rather than cell division (Yamada et al., 2009). Furthermore, cell expansion is inhibited by transient down-regulation of RhPIP2;1, a gene encoding a plasma membrane aquaporin that is strongly down-regulated by ethylene treatment in the cultivar R. hybrida ‘Samantha’ (Ma et al., 2008). The final petal expansion process and rose flower opening process rely heavily on turgor pressure, and, thus, water availability for cut flowers is the factor that allows correct opening. It is at this crucial moment of flower opening that rose petals become coloured and start emitting scent compounds.
Flower colour
Petals in roses exhibit such a wide variety of colours that the only colour absent from this genus is blue. The absence of the blue colour is a result of roses lacking the flavonol 3’,5’-hydroxylase (F3′5′H) activity to generate dihydromyrcetin, a precursor of delphinidin. Delphinidin is at the origin of the blue colour. However, roses can be forced to harbour the blue colour. In 2007, Katsumuto et al. first knocked down endogenous rose dihydroflavonol 4-reductase (DFR) to reduce the production of red anthocyanin precursors, and overexpressed the viola DFR that uses dihydromyrcetin to make delphinidin from Iris. The petals of these roses exclusively accumulated delphinidin and showed blue hues (Katsumoto et al., 2007). However, because of the numerous parameters controlling the final petal colour, such as vacuolar pH or genetic background, these delphinidin-enriched flowers exhibited blue–purple-like petal colours.
Yellow and orange rose petals mostly contain carotenoid pigments. The pink and red colours are due to anthocyanins and in particular to 3,5-diglycosyl anthocyanidins in association with 3-glycosylated flavonols (Biolley and Jay, 1993; Ogata et al., 2005). Three enzymes encoding flavonoid 3-glycosyltransferases (RhGT1–RhGT3) have been characterized. RhGT1 was shown to be expressed in petals of cultivars that synthesize cyanidine 3-glucoside from cyanidins. RhGT2 and RhGT3 are co-expressed in rose petals with the flavonol synthase gene and catalyse 3-glycosylation of flavonols (Fukuchi-Mizutani et al., 2011)
The pathways leading to petal colour in angiosperms are well characterized (Koes et al., 2005). Genes involved in the anthocyanin biosynthesis pathway are fairly well described in the rose (Tanaka et al., 1995; Suzuki et al., 2000). However, only one locus (Blfa) associated with pink flower colour was mapped on IM-LG2 (Debener and Mattiesch, 1999; Yan et al., 2005; Spiller et al., 2011). It should be noted that genes well known to be involved in the anthocyanin biosynthesis pathway in rose were not mapped on the IM-LG2 map.
The anthocyanidin biosynthesis enzymes are regulated at the transcriptional level by complexes such as the basic helix–loop–helix transcription factors interacting with R2R3 MYB, and WD40 proteins (Koes et al., 2005; Hichri et al., 2011). However, how the metabolic fluxes towards specific pathways are regulated at the cell and tissue levels remains a very challenging question. The regulation of these pathways may vary between species, depending on their specific metabolism. For example, in a rose hybrid cultivar, overexpression of the PRODUCTION OF ANTHOCYANIN PIGMENT 1 gene from Arabidopsis led to increased levels of phenylpropanoid-derived colour and scent compounds (Ben Zvi et al., 2012). Moreover, these transformed roses also produced significantly higher levels of terpenoid scent compounds, suggesting an enhanced metabolic flux not only towards the phenylpropanoid pathway, but also towards the isoprenoid pathway. Colour and scent compounds of the rose petals thus appear intricately linked through regulation of secondary metabolite fluxes. Thus, the study of secondary metabolism pathways in the rose can provide new information on the regulation and cross-talk between the metabolic pathways.
Rose flower as machinery for scent production
In Rosa sp., the petal is the major organ for scent biosynthesis. Rose probably manufactures the most diverse scent compounds. Roses produce not only the typical rose flower fragrance, but also scents described as resembling tea, various fruits, myrrh, and even aniseed. However, not all roses currently on the market are heavily scented; roses bred for cut flowers often lack scent. The cause of the lack of fragrance in the cut flowers of these cultivars is unknown. Rose scent is a complex trait based on the emission of hundreds of volatile molecules. Variations in the composition of the volatile molecules, in both quality and quantity, lead to different rose scent profiles (Joichi et al., 2005). There are three major scent molecule classes in roses: the terpenes, the benzenoids/phenylpropanoids, and the fatty acid derivatives (Schnepp and Dudareva, 2007). For the typical rose-scented flowers, the most abundant molecules are monoterpene alcohols and 2-phenylethanol. The tea fragrance is mostly due to the presence of aromatic compounds such as 3,5-dimethoxytoluene (DMT) and 1,3,5-trimethoxybenzene (TMB) (Lavid et al., 2002; Scalliet et al., 2002, 2006, 2008; Joichi et al., 2005).
During the past decade, a number of studies have illustrated the molecular mechanisms controlling the biosynthesis and emission of rose scent compounds. The pathways leading to the biosynthesis of DMT and TMB have been described. Phloroglucinol O-methyltransferase (POMT) was shown to catalyse the methylation of phloroglucinol to 3,5-dihydroxyanisole, a precursor of TMB (Scalliet et al., 2002, 2006; Wu et al., 2004). The petal-specific enzymes, orcinol-O-methyl transferases (RcOOMT1 and RcOOMT2), were shown to be involved in DMT biosynthesis (Scalliet et al., 2006). RcOOMT1 and RcOOMT2 originate from recent gene duplication and exhibit high identity at the nucleotide and the amino acid levels (Scalliet et al., 2002, 2008). Functional characterization of the rose OOMTs revealed that the tea scent has been introgressed into modern rose cultivars from a unique origin in China (Scalliet et al., 2008). The carotenoid cleavage oxygenases CCD1 and CCD4 are highly expressed in flowers and participate in the biosynthesis of terpenes, such as β-ionone (Huang et al., 2009a, b). 2-Phenylethyl alcohol is another volatile organic compound responsible for the typical rose scent. Its synthesis occurs in two steps. The first step is catalysed by phenylacetaldehyde synthase (PAAS) that converts phenylalanine to phenylacetaldehyde (Kaminaga et al., 2006). Phenylacetaldehyde reductase (PAR), whose expression is restricted to flowers, catalyses the second step that reduces phenylacetaldehyde to 2-phenylethyl alcohol (Sakai et al., 2007). Little is known about the genes acting in the terpene pathway in Rosa sp. Only two genes (alcohol acetyltransferase RhAAT1- and sesquiterpene synthase-encoding genes) have been characterized so far, and no monoterpene synthase-encoding gene has been isolated (Guterman et al., 2002; Shalit et al., 2003). We are still lacking a detailed picture of the various pathways leading to the enormous diversity of scent compounds synthesized by rose flowers.
The genetic control of the key components of rose scent was examined by metabolic profiling coupled to marker analysis in two mapping populations (Spiller et al., 2011). Loci associated with nerol and neryl acetate biosynthesis were mapped on IM-LG3 and IM-LG4. Inheritance of the production of geranyl acetate was under the control of two loci, one of which co-segregates with a known scent gene (Alcohol acyltranferase1) on IM-LG2. Six QTLs for three other volatiles (geraniol, citronellol, and 2-phenylethanol) were also found (Spiller et al., 2011). Four additional markers, derived from known scent genes (Germacrene D-synthase, Orcinol O-methyltransferase, Eugenol O-methyltransferasE, and Caffeic acid O-methyltransferase 3), were mapped.
Despite this knowledge, the mechanisms of scent production and regulation are complex, as every single rose cultivar has a very specific and highly complex scent composition. One of the major challenges in the field is to decipher how these pathways are finely tuned depending on the genetic background and/or on environmental conditions.
Flower senescence
The molecular bases of petal senescence and abscission are of great research interest as senescence and abscission processes influence the vase life of cut flowers (Rogers, 2012). Abscisic acid (ABA) is usually involved in petal senescence in both ethylene-sensitive and ethylene-insensitive senescence processes. ABA levels were reported to be higher in senescent rose petals concomitant with reduced flower water potential and reduced water uptake (Kumar et al., 2008a). However, the action of ABA in rose senescing petals remains uncharacterized. Similar to other senescent organs, senescent rose petals have increased endogenous H2O2 levels and they exhibit decreased activities of antioxidant enzymes (Kumar et al., 2008b). So far, there is no report on autophagy, nucleic acid degradation, protein turnover, or nutrient remobilization processes in senescing rose flowers.
In roses, ethylene was shown to promote petal abscission through cell wall modifications. Ethylene levels dramatically increase during rose petal senescence, probably through the activity of the rose 1-aminocyclopropane-1-carboxylate (ACC) synthase in petals (Wang et al., 2004). The R. bourboniana genes Expansin 1A (RbEXPA1) and pectate lyase1 (RbPEL1) are specifically induced during petal abscission. The expression of RbEXPA1 and RbPEL1 increases in abscission zones of petals during the course of natural abscission and post-ethylene treatment (Sane et al., 2007; Singh et al., 2011a). Two other genes, RbXTH1 and RbXTH2, encoding the xyloglucan endotransglucosylase/hydrolase, were shown to be induced in senescing petals and their expression is also induced by ethylene (Singh et al., 2011b). Further work is needed to identify and functionally characterize the genetic and molecular determinants associated with rose vase life. Recent transcriptomic data using high throughput sequencing approaches are providing novel information on putative molecular markers for rose flower senescence (Dubois et al., 2011, 2012). Moreover, in many model species, the understanding of regulators of senescence is improving and is providing us with insights into possible signalling pathways associated with flower senescence (Rogers, 2012). Future prospects will be to use this information from model species, namely to study the molecular and genetic control of vase life in ornamental species.
Discussion and conclusions
The past decade has witnessed the development of a number of molecular and genetic tools for the study of rose. These tools were instrumental for the discovery of the molecular and genetic basis of flower initiation and development in Rosa sp.
First, many rose mapping populations were used to unravel the genetic basis of the monogenic or oligogenic characters such as simple corolla, pink flower, recurrent blooming, and scent based on three volatile components. However, because these mapping populations are usually small, co-localization between a given candidate gene and the locus of interest was unsuccessful.
In parallel, candidate gene approaches brought novel information to many aspects of rose floral development. Tools for gene functional validation have been developed and applied to rose. For instance, in situ RNA hybridization was adapted to Rosa in both floral meristems and petals (Ma et al., 2008; Dubois et al., 2010; Nakamura et al., 2011). Protein subcellular localization was made possible in rose petal epidermal cells using a green fluorescent protein (GFP) fusion for rose OOMTs (Scalliet et al., 2006). However, most of these experiments are still classically carried out in heterologous systems such as Arabidopsis for RhPIP2;1 after mesophyll protoplast polyethylene glycol (PEG)-mediated transformation (Ma et al., 2008) or tobacco for RoDELLA after leaf agroinfiltration (Hamama et al., 2012).
Therefore, two important roadblocks hamper the in-depth understanding of the molecular control of flower development in Rosa sp. First, because of the difficulty in generating stable genetic rose transformants, gene function validation is limited. Although several transformation protocols were developed, no universal efficient method is currently available. Agrobacterium-mediated or biolistic methods were used, but the critical bottleneck for transformed plant establishment remains regeneration of transformed plants (Marchant et al., 1998; Li et al., 2002; Kim et al., 2004; Katsumoto et al., 2007; Vergne et al., 2010). Most genetic transformation protocols have been applied to tetraploid roses, and only one report showed the successful transformation of diploid roses (Vergne et al., 2010). Overall, it is difficult to generate new mutant rose populations by insertion or ethyl methanesulphonate (EMS) mutagenesis due to the difficulties related to genetic transformation of the rose, the relatively high heterozygosity of the rose genome, and the self-incompatibility issue. To circumvent these issues, most functional validations of rose genes were performed in heterologous systems such as in Petunia, Arabidopsis, Torenia, tobacco, pelargonium, Escherichia coli, or yeast (Tanaka et al., 1995; Guterman et al., 2002, 2006; Shalit et al., 2003; Kitahara et al., 2004; Hibino et al., 2006; Scalliet et al., 2006; Hamama et al., 2012). Although such strategies provided some information, functional validation in roses will be required. A promising approach for gene knockdown would be the use of virus-induced gene silencing (VIGS). In 2008, a study demonstrated that aquaporin controls petal expansion by knocking down RhPIP2;1 with VIGS in rose (Ma et al., 2008). However the efficiency of VIGS in roses remains very low compared with that initially reported in tobacco and tomato.
The second issue is related to the limited information on Rosa-expressed genes. The recent study in which next-generation Illumina and 454 sequencing technologies were used to identify most of the rose transcriptome has provided information on transcripts representing ~21 000 unisequences expressed in various tissues and organs and in response to biotic and abiotic stresses (http://iant.toulouse.inra.fr/R.chinensis; Dubois et al., 2012). However, a major task in the near future is the rose whole-genome sequencing. Despite the relatively high heterozygosity, a complete rose genome sequence is now conceivable. Recently, the ‘Rose Genome Sequencing Initiative’ (RGSI) international consortium has initiated the rose genome sequencing. Access to the whole-genome information is necessary to improve genomics research in rose and in the field of woody ornamentals. Besides its economic importance, the rose is well suited as a model for woody ornamental species as it has a relatively small genome size (~560 Mbp) and it has a short life cycle for a perennial woody plant (~1 year; seedlings can flower the same spring). The rose genome sequence will also be useful to help understand the molecular bases for important ornamental traits, which can in turn facilitate and accelerate rose breeding by marker-assisted selection or genomic selection, and to study genetic diversity and genome evolution. Altogether, these arguments prove that the rose can be an excellent model for ornamental species and especially woody ornamentals.
Acknowledgements
We thank Dr Dali Ma and Judit Szecsi for helpful discussion and for critical reading of this review. Work in our laboratory is funded by the ‘Biologie Végétale’ Department of the French ‘Institut National de la Recherche Agronomique’, by the ‘Ecole Normale Supérieure de Lyon’, and by the ‘Région Rhone-Alpes-France’.
References
- Ben Zvi MM, Shklarman E, Masci T, Kalev H, Debener T, Shafir S, Ovadis M, Vainstein A. 2012. PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytologist 195, 335–345 [DOI] [PubMed] [Google Scholar]
- Biolley JP, Jay M. 1993. Anthocyanins in modern roses—chemical and colorimetric features in relation to the color range. Journal of Experimental Botany 44, 1725–1734 [Google Scholar]
- Channeliere S, Riviere S, Scalliet G, et al. 2002. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Letters 515, 35–38 [DOI] [PubMed] [Google Scholar]
- Choubane D, Rabot A, Mortreau E, et al. 2012. Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp . Journal of Plant Physiology 169, 1271–1280 [DOI] [PubMed] [Google Scholar]
- Crespel L, Chirollet M, Durel CE, Zhang D, Meynet J, Gudin S. 2002. Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theoretical and Applied Genetics 105, 1207–1214 [DOI] [PubMed] [Google Scholar]
- Debener T. 1999. Genetic analysis of horticulturally important morphological and physiological characters in diploid roses. Gartenbauwissenschaft 64, 14–20 [Google Scholar]
- Debener T, Mattiesch L. 1999. Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theoretical and Applied Genetics 99, 891–899 [Google Scholar]
- De Vries DP, Dubois L. 1996. Rose breeding: past, present, prospects. Acta Horticulturae 424, 241–248 [Google Scholar]
- Dornelas MC, Patreze CM, Angenent GC, Immink RG. 2011. MADS: the missing link between identity and growth?. Trends in Plant Science 16, 89–97 [DOI] [PubMed] [Google Scholar]
- Dubois A, Raymond O, Maene M, Baudino S, Langlade NB, Boltz V, Vergne P, Bendahmane M. 2010. Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One 5, e9288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A, Remay A, Raymond O, et al. 2011. Genomic approach to study floral development genes in Rosa sp . PLoS One 6, e28455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A, Carrere S, Raymond O, et al. 2012. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics 13, 638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugo ML, Satovic Z, Millan T, Cubero JI, Rubiales D, Cabrera A, Torres AM. 2005. Genetic mapping of QTLs controlling horticultural traits in diploid roses. Theoretical and Applied Genetics 111, 511–520 [DOI] [PubMed] [Google Scholar]
- Foucher F, Chevalier M, Corre C, Soufflet-Freslon V, Legeai F, Hibrand-Saint Oyant L. 2008. New resources for studying the rose flowering process. Genome 51, 827–837 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Akagi M, Ishiguro K, Katsumoto Y, Fukui Y, Togami J, Nakamura N, Tanaka Y. 2011. Biochemical and molecular characterization of anthocyanidin/flavonol 3-glucosylation pathways in Rosa×hybrida . Plant Biotechnology 28, 239–244 [Google Scholar]
- Galimba KD, Tolkin TR, Sullivan AM, Melzer R, Theissen G, Di Stilio VS. 2012. Loss of deeply conserved C-class floral homeotic gene function and C- and E-class protein interaction in a double-flowered ranunculid mutant. Proceedings of the National Academy of Sciences, USA 109, E2267–E2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault T, Bergougnoux V, Combes D, Viemont JD, Leduc N. 2008. Light controls shoot meristem organogenic activity and leaf primordia growth during bud burst in Rosa sp. Plant, Cell and Environment 31, 1534–1544 [DOI] [PubMed] [Google Scholar]
- Gudin S. 2001. Rose breeding technologies. Acta Horticulturae 547, 23–26 [Google Scholar]
- Gudin S. 2003. Breeding. In: Roberts AV, Debener T, Gudin S, eds. Encyclopedia of rose science Oxford: Academic Press, 25–30 [Google Scholar]
- Guterman I, Masci T, Chen X, Negre F, Pichersky E, Dudareva N, Weiss D, Vainstein A. 2006. Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Molecular Biology 60, 555–563 [DOI] [PubMed] [Google Scholar]
- Guterman I, Shalit M, Menda N, et al. 2002. Rose scent: genomics approach to discovering novel floral fragrance-related genes. The Plant Cell 14, 2325–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamama L, Naouar A, Gala R, et al. 2012. Overexpression of RoDELLA impacts the height, branching, and flowering behaviour of Pelargonium×domesticum transgenic plants. Plant Cell Reports 31, 2015–2029 [DOI] [PubMed] [Google Scholar]
- Hibino Y, Kitahara K, Hirai S, Matsumoto S. 2006. Structural and functional analysis of rose class B MADS-box genes ‘MASAKO BP, euB3, and B3’: paleo-type AP3 homologue ‘MASAKO B3’ association with petal development. Plant Science 170, 778–785 [Google Scholar]
- Hibrand Saint Oyant L, Crespel L, Rajapakse S, Zhang L, Foucher F. 2008. Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genetics and Genomes 4, 11–23 [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. 2011. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany 62, 2465–2483 [DOI] [PubMed] [Google Scholar]
- Huang FC, Horvath G, Molnar P, Turcsi E, Deli J, Schrader J, Sandmann G, Schmidt H, Schwab W. 2009a. Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from Rosa damascena . Phytochemistry 70, 457–464 [DOI] [PubMed] [Google Scholar]
- Huang FC, Molnar P, Schwab W. 2009b. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. Journal of Experimental Botany 60, 3011–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RL, Remphrey WR, Lenz LM. 1989. An analysis of the development of single and double flowers in Potentilla fruticosa . Canadian Journal of Botany 67, 1071–1079 [Google Scholar]
- Irish VF. 2010. The flowering of Arabidopsis flower development. The Plant Journal 61, 1014–1028 [DOI] [PubMed] [Google Scholar]
- Iwata H, Gaston A, Remay A, Thouroude T, Jeauffre J, Kawamura K, Oyant LH, Araki T, Denoyes B, Foucher F. 2012. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. The Plant Journal 69, 116–125 [DOI] [PubMed] [Google Scholar]
- Jian H, Zhang H, Tang K, Li S, Wang Q, Zhang T, Qiu X, Yan H. 2010. Decaploidy in Rosa praelucens Byhouwer (Rosaceae) endemic to Zhongdian Plateau, Yunnan, China. Caryologia 63, 162–167 [Google Scholar]
- Joichi A, Yomogida K, Awano K–i, Ueda Y. 2005. Volatile components of tea-scented modern roses and ancient Chinese roses. Flavour and Fragrance Journal 20, 152–157 [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, et al. 2006. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. Journal of Biological Chemistry 281, 23357–23366 [DOI] [PubMed] [Google Scholar]
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, et al. 2007. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant and Cell Physiology 48, 1589–1600 [DOI] [PubMed] [Google Scholar]
- Kim CK, Chung JD, Park SH, Burrell AM, Kamo KK, Byrne DH. 2004. Agrobacterium tumefaciens-mediated transformation of Rosa hybrida using the green fluorescent protein (GFP) gene. Plant Cell, Tissue and Organ Culture 78, 107–111 [Google Scholar]
- Kitahara K, Hibino Y, Aida R, Matsumoto S. 2004. Ectopic expression of the rose AGAMOUS-like MADS-box genes ‘MASAKO C1 and D1’ causes similar homeotic transformation of sepal and petal in Arabidopsis and sepal in Torenia . Plant Science 166, 1245–1252 [Google Scholar]
- Kitahara K, Hirai S, Fukui H, Matsumoto S. 2001. Rose MADS-box genes ‘MASAKO BP and B3’ homologous to class B floral identity genes. Plant Science 161, 549–557 [DOI] [PubMed] [Google Scholar]
- Kitahara K, Matsumoto S. 2000. Rose MADS-box genes ‘MASAKO C1 and D1’ homologous to class C floral identity genes. Plant Science 151, 121–134 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242 [DOI] [PubMed] [Google Scholar]
- Koning-Boucoiran CFS, Gitonga VW, Yan Z, et al. 2012. The mode of inheritance in tetraploid cut roses. Theoretical and Applied Genetics 125, 591–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela EA, Mouhu K, Albani MC, Kurokura T, Rantanen M, Sargent DJ, Battey NH, Coupland G, Elomaa P, Hytonen T. 2012. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca . Plant Physiology 159, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoda N, Iwanami H, Takahashi S, Abe K. 2006. Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. Journal of the American Society for Horticultural Science 131, 74–81 [Google Scholar]
- Kovarik A, Werlemark G, Leitch AR, Souckova-Skalicka K, Lim YK, Khaitová L, Koukalova B, Nybom H. 2008. The asymmetric meiosis in pentaploid dogroses (Rosa sect. Caninae) is associated with a skewed distribution of rRNA gene families in the gametes. Heredity 101, 359–367 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. 2005. Molecular mechanisms of flower development: an armchair guide. Nature Reviews Genetics 6, 688–698 [DOI] [PubMed] [Google Scholar]
- Kumar N, Srivastava GC, Dixit K. 2008a. Hormonal regulation of flower senescence in roses (Rosa hybrida L.). Plant Growth Regulation 55, 65–71 [Google Scholar]
- Kumar N, Srivastava GC, Dixit K. 2008b. Senescence in rose (Rosa hybrida L.): role of the endogenous anti-oxidant system. Journal of Horticultural Science & Biotechnology 83, 125–131 [Google Scholar]
- Lavid N, Wang J, Shalit M, et al. 2002. O-Methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiology 129, 1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann NL, Sattler R. 1993. Homeosis in floral development of Sanguinaria canadensis and S. canadensis ‘Multiplex’ (Papaveraceae). American Journal of Botany 80, 1323–1335 [Google Scholar]
- Li X, Krasnyanski SF, Korban SS. 2002. Optimization of the uidA gene transfer into somatic embryos of rose via Agrobacterium tumefaciens . Plant Physiology and Biochemistry 40, 453–459 [Google Scholar]
- Lim KY, Werlemark G, Matyasek R, Bringloe JB, Sieber V, Mokadem HE, Meynet J, Hemming J, Leitch AR, Roberts AV. 2005. Evolutionary implications of permanent odd polyploidy in the stable sexual, pentaploid of Rosa canina L. Heredity 94, 501–506 [DOI] [PubMed] [Google Scholar]
- Linde M, Hattendorf A, Kaufmann H, Debener T. 2006. Powdery mildew resistance in roses: QTL mapping in different environments using selective genotyping. Theoretical and Applied Genetics 113, 1081–1092 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang D, Liu D, Li F, Lu H. 2012. Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiana (Rosaceae). Plant Cell Reports (in press) [DOI] [PubMed] [Google Scholar]
- Ma N, Xue JQ, Li YH, Liu XJ, Dai FW, Jia WS, Luo YB, Gao JP. 2008. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiology 148, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas FM, Bakx EJ. 1995. Effects of light on growth and flowering of Rosa hybrids ‘Mercedes’. Journal of the American Society for Horticultural Science 120, 571–576 [Google Scholar]
- Maclntyre JP, Lacroix CR. 1996. Comparative development of perianth and androecial primordia of the single flower and the homeotic double-flowered mutant in Hibiscus rosa-sinensis (Malvaceae). Canadian Journal of Botany 74, 1871–1882 [Google Scholar]
- Marchant R, Power JB, Lucas JA, Davey MR. 1998. Biolistic transformation of rose (Rosa hybrida L.). Annals of Botany 81, 109–114 [Google Scholar]
- Martin M, Piola F, Chessel D, Jay M, Heizmann P. 2001. The domestication process of the Modern Rose: genetic structure and allelic composition of the rose complex. Theoretical and Applied Genetics 102, 398–404 [Google Scholar]
- Moghaddam HH, Leus L, De Riek J, Van Huylenbroeck J, Van Bockstaele E. 2012. Construction of a genetic linkage map with SSR, AFLP and morphological markers to locate QTLs controlling pathotype-specific powdery mildew resistance in diploid roses. Euphytica 184, 413–427 [Google Scholar]
- Mohamed R, Wang C-T, Ma C, et al. 2010. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus . The Plant Journal 62, 674–688 [DOI] [PubMed] [Google Scholar]
- Nakamura N, Fukuchi-Mizutani M, Katsumoto Y, et al. 2011. Environmental risk assessment and field performance of rose (Rosa×hybrida) genetically modified for delphinidin production. Plant Biotechnology 28, 251–261 [Google Scholar]
- Ogata J, Kanno Y, Itoh Y, Tsugawa H, Suzuki M. 2005. Plant biochemistry: anthocyanin biosynthesis in roses. Nature 435, 757–758 [DOI] [PubMed] [Google Scholar]
- Pécrix Y, Rallo G, Folzer H, Cigna M, Gudin S, Le Bris M. 2011. Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. Journal of Experimental Botany 62, 3587–3597 [DOI] [PubMed] [Google Scholar]
- Rabot A, Henry C, Ben Baaziz K, et al. 2012. Insight into the role of sugars in bud burst under light in the rose. Plant and Cell Physiology 53, 1068–1082 [DOI] [PubMed] [Google Scholar]
- Raymond O. 1999. Domestication et sélection dirigée chez le rosier: analyse historique via les phénotypes morphologique, chimique et biochimique. PhD Thesis Université Claude Bernard-Lyon1, Lyon, France: [Google Scholar]
- Remay A, Lalanne D, Thouroude T, Le Couviour F, Hibrand-Saint Oyant L, Foucher F. 2009. A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theoretical and Applied Genetics 119, 767–781 [DOI] [PubMed] [Google Scholar]
- Reynders-Aloisi S, Bollereau P. 1996. Characterisation of genetic diversity in genus Rosa by randomly amplified polymorphic DNA. Acta Horticulturae 424, 253–259 [Google Scholar]
- Roberts AV, Gladis T, Brumme H. 2009. DNA amounts of roses (Rosa L.) and their use in attributing ploidy levels. Plant Cell Reports 28, 61–71 [DOI] [PubMed] [Google Scholar]
- Rogers HJ. 2012. From models to ornamentals: how is flower senescence regulated?. Plant Molecular Biology (in press) [DOI] [PubMed] [Google Scholar]
- Sakai M, Hirata H, Sayama H, Sekiguchi K, Itano H, Asai T, Dohra H, Hara M, Watanabe N. 2007. Production of 2-phenylethanol in roses as the dominant floral scent compound from l-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Bioscience, Biotechnology and Biochemistry 71, 2408–2419 [DOI] [PubMed] [Google Scholar]
- Sane AP, Tripathi SK, Nath P. 2007. Petal abscission in rose (Rosa bourboniana var Gruss an Teplitz) is associated with the enhanced expression of an alpha expansin gene, RbEXPA1. Plant Science 172, 481–487 [Google Scholar]
- Scalliet G, Journot N, Jullien F, et al. 2002. Biosynthesis of the major scent components 3,5-dimethoxytoluene and 1,3,5-trimethoxybenzene by novel rose O-methyltransferases. FEBS Letters 523, 113–118 [DOI] [PubMed] [Google Scholar]
- Scalliet G, Lionnet C, Le Bechec M, et al. 2006. Role of petal-specific orcinol O-methyltransferases in the evolution of rose scent. Plant Physiology 140, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalliet G, Piola F, Douady CJ, et al. 2008. Scent evolution in Chinese roses. Proceedings of the National Academy of Sciences, USA 105, 5927–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp J, Dudareva N. 2007. Floral scent: biosynthesis, regulation and genetic modifications. In: Ainsworth C, ed. Annual Plant Reviews Volume 20: Flowering and its Manipulation Oxford: Blackwell Publishing, 240–257 [Google Scholar]
- Shalit M, Guterman I, Volpin H, et al. 2003. Volatile ester formation in roses. Identification of an acetyl-coenzyme A:geraniol/citronellol acetyltransferase in developing rose petals. Plant Physiology 131, 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng T-S, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun T-p. 2007. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiology 143, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Pandey SP, Rajluxmi, Pandey S, Nath P, Sane AP. 2011a. Transcriptional activation of a pectate lyase gene, RbPel1, during petal abscission in rose. Postharvest Biology and Technology 60, 143–148 [Google Scholar]
- Singh AP, Tripathi SK, Nath P, Sane AP. 2011b. Petal abscission in rose is associated with the differential expression of two ethylene-responsive xyloglucan endotransglucosylase/hydrolase genes, RbXTH1 and RbXTH2 . Journal of Experimental Botany 62, 5091–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller M, Linde M, Hibrand-Saint Oyant L, Tsai CJ, Byrne DH, Smulders MJ, Foucher F, Debener T. 2011. Towards a unified genetic map for diploid roses. Theoretical and Applied Genetics 122, 489–500 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsuda S, Fukui Y, Fukuchi-Mizutani M, Yonekura-Sakakibara K, Tanaka Y, Kusumi T. 2000. Molecular characterization of rose flavonoid biosynthesis genes and their application in Petunia . Biotechnology & Biotechnological Equipment 14, 56–62 [Google Scholar]
- Tanaka Y, Fukui Y, Fukuchi–Mizutani M, Holton TA, Higgins E, Kusumi T. 1995. Molecular cloning and characterization of Rosa hybrida dihydroflavonol 4-reductase gene. Plant and Cell Physiology 36, 1023–1031 [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. 2001. Plant biology. Floral quartets. Nature 409, 469–471 [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Dickinson TA. 2006. Polyploidy and diversification: a phylogenetic investigation in Rosaceae . International Journal of Plant Sciences 167, 349–358 [Google Scholar]
- Vergne P, Maene M, Gabant G, Chauvet A, Debener T, Bendahmane M. 2010. Somatic embryogenesis and transformation of the diploid Rosa chinensis cv Old Blush. Plant Cell, Tissue and Organ Culture 100, 73–81 [Google Scholar]
- Wang D, Fan J, Ranu RS. 2004. Cloning and expression of 1-aminocyclopropane-1-carboxylate synthase cDNA from Rosa (Rosa×hybrida). Plant Cell Reports 22, 422–429 [DOI] [PubMed] [Google Scholar]
- Wang LN, Liu YF, Zhang YM, Fang RX, Liu QL. 2012. The expression level of Rosa Terminal Flower 1 (RTFL1) is related with recurrent flowering in roses. Molecular Biology Reports 39, 3737–3746 [DOI] [PubMed] [Google Scholar]
- Wang RH, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G. 2011. Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina . The Plant Cell 23, 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Melzer R, Theiben G. 2011. A double-flowered variety of lesser periwinkle (Vinca minor fl. pl.) that has persisted in the wild for more than 160 years. Annals of Botany 107, 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Watanabe N, Mita S, Dohra H, Ueda Y, Shibuya M, Ebizuka Y. 2004. The key role of phloroglucinol O-methyltransferase in the biosynthesis of Rosa chinensis volatile 1,3,5-trimethoxybenzene. Plant Physiology 135, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Norikoshi R, Suzuki K, Nishijima T, Imanishi H, Ichimura K. 2009. Cell division and expansion growth during rose petal development. Journal of the Japanese Society for Horticultural Science 78, 356–362 [Google Scholar]
- Yan Z, Denneboom C, Hattendorf A, Dolstra O, Debener T, Stam P, Visser PB. 2005. Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theoretical and Applied Genetics 110, 766–777 [DOI] [PubMed] [Google Scholar]