Abstract
The ciliated protozoan Tetrahymena thermophila is a useful unicellular model organism for studies of eukaryotic cellular and molecular biology. Researches on T. thermophila have contributed to a series of remarkable basic biological principles. After the macronuclear genome was sequenced, substantial progress has been made in functional genomics research on T. thermophila, including genome-wide microarray analysis of the T. thermophila life cycle, a T. thermophila gene network analysis based on the microarray data and transcriptome analysis by deep RNA sequencing. To meet the growing demands for the Tetrahymena research community, we integrated these data to provide a public access database: Tetrahymena functional genomics database (TetraFGD). TetraFGD contains three major resources, including the RNA-Seq transcriptome, microarray and gene networks. The RNA-Seq data define gene structures and transcriptome, with special emphasis on exon–intron boundaries; the microarray data describe gene expression of 20 time points during three major stages of the T. thermophila life cycle; the gene network data identify potential gene–gene interactions of 15 049 genes. The TetraFGD provides user-friendly search functions that assist researchers in accessing gene models, transcripts, gene expression data and gene–gene relationships. In conclusion, the TetraFGD is an important functional genomic resource for researchers who focus on the Tetrahymena or other ciliates.
Database URL: http://tfgd.ihb.ac.cn/
Introduction
Tetrahymena thermophila is a free-living ciliated protozoan that normally has two types of functionally distinct nuclei (1), the silent germ line micronucleus (MIC) and the actively transcribed somatic macronucleus (MAC) in each cell. Its typical eukaryotic biology and many molecular genetic tools have enabled Tetrahymena researchers to contribute to landmark discoveries of fundamental eukaryotic cellular mechanisms, such as the first cytoskeletal motor (2), catalytic RNA (3), telomere structure (4) and telomerase (5) and the role of small RNAs in programmed somatic genome rearrangement (6). Although its genome has been sequenced and its genetics and molecular biology have been extensively studied, research on T. thermophila has been limited by the lack of some basic genomic resources, in particular, functional genomics data.
In 2006, the MAC genome sequence project of T. thermophila was completed (7), which provides the first ciliate genome sequence. After this, the Tetrahymena genome database (TGD and TGD Wiki, http://ciliate.org), containing the genome sequence and the predicted gene models, was established (8–9). The first insights into the Tetrahymena transcriptome came from expressed sequence tag (EST) sequences generated by the initial genome sequence project and the protist EST program, and they could be accessed through the NCBI EST database and the PEP database (TBestDB, http://www.bch.umontreal.ca/pepdb/pepdb.html), respectively. Miao et al. (10) initiated Tetrahymena functional genomics by establishing the first T. thermophila microarray platform performing a genome-wide investigation of gene expression during the three major physiological and developmental stages of the T. thermophila life cycle. These microarray data could be accessed via the Tetrahymena gene expression database (TGED, http://tged.ihb.ac.cn/) (11). Recently, the microarray data were extended to infer a T. thermophila gene network (12), providing the first insight of gene–gene relationships in T. thermophila. Finally, an analysis using deep RNA sequencing (RNA-Seq) provided more comprehensive and detailed analysis of the transcriptome and greatly improved the gene models (13). Given these recent genomic studies, an integrated database is needed to provide easy access to functional genomics data for members of the scientific research community interested in T. thermophila.
In this study, we describe the Tetrahymena functional genomics database (TetraFGD), which provides user-friendly search functions for accessing gene models, transcripts, gene expression data and gene–gene relationships in Tetrahymena.
Content
The TetraFGD is an online resource containing three major functional genomic data sets of T. thermophila: (i) RNA-Seq data; (ii) microarray data; and (iii) gene network.
RNA-Seq data
These data were obtained from six RNA samples in three major physiological and developmental stages of T. thermophila, including one in growth (∼3.5 × 105 cells/ml), three in starvation (mating type V and VI in 3 h, mating type VI in 15 h) and two in conjugation (2 and 8 h after mixing of two mating types) by using Illumina deep RNA sequencing (13). To ensure that the RNA-Seq data could be compared with the microarray data, time points were selected a subset of those in the previous microarray expression studies that covered 20 states of the three stages of the T. thermophila life cycle. More than 96% of the predicted genes have detectable reads after mapping the RNA-Seq data to the genome. More than 30 000 transcripts were assembled, including >1000 new transcripts, which were not found by gene scanning. These transcripts were used to improve the previous gene models. Over 7000 predicted gene models showed errors when the RNA-Seq data and gene annotation (13) were compared, greatly improving identification of coding sequences, untranslated regions and exon–intron boundaries. Although the RNA-Seq technology is powerful, some assembled transcripts do not contain complete open reading frames because of limited coverage and assembly. Compared with the RNA-Seq, gene scanning may be relatively less accurate when it was used to predict the transcription information, whereas it supplies the complete open reading frames. Thus, we shared the RNA-Seq data with the Tetrahymena team at the Broad Institute, who are using them to update gene prediction and genome annotation.
Microarray data
The TetraFGD now contains the microarray gene expression data containing 20 time points during the three major physiological and developmental stages of the T. thermophila life cycle, including 3 points in growth, 7 points in starvation and 10 points in conjugation. Because the microarray expression values might be wrong if a gene was mis-predicted, and the RNA-Seq assembled transcripts provide more correct transcription information. Therefore, the TetraFGD now integrates two types of microarray expression values: (i) normalization based on the predicted genes (gene model may be incorrectly predicted); and (ii) normalization based on the RNA-Seq assembled transcripts (some of them are transcription fragments). These two types of microarray expression values were both normalized using the microarrays reported previously (10).
There are two kinds of gene expression values normalized by the predicted genes. Take the expression values of the gene TTHERM_00257230 (http://tfgd.ihb.ac.cn/search/detail/gene/TTHERM_00257230) for example, two expression profile are showed when you search the database and are represented by the blue and red line. The blue line represents the expression values from the Tetrahymena gene expression database (TGED) (11), and the red line represents the expression values that were normalized by Prof. Ronald Pearlman laboratory using a different method (most of the raw data are the same as those used in the blue line, and 10 new microarrays in the Tetrahymena conjugation stage were added).
For the RNA-Seq assembled transcripts (each transcript was regarded as a gene model, although it may be only a transcription fragment), all previous designed microarray probes (10) were re-mapped to the RNA-Seq assembled transcripts, and the microarray expression values for these transcripts were re-normalized as in the study by Miao et al. (10) and provided in the TetraFGD.
Gene network
Network analysis can be used to identify genes in the same biological processes or pathways, to infer interactions of bio-molecules, such as their physical association, metabolite flow, regulatory relationships and co-expression relationships and so forth. An important resource in the TetraFGD is the Tetrahymena gene network (TGN). The TGN was constructed using the context likelihood of relatedness algorithm (CLR, mutual information-based method, which is an extension of the relevance networks) (14) based on 67 Roche NimbleGen single-channel microarray expression data (12), which means that the connected genes in the TGN have similar expression profiles. After gene filtering strategies (12), 15 049 genes were used to infer the gene network. By determining an appropriate threshold with a CLR Z-score threshold 3.49, 1 958 477 gene–gene interactions were included in the TGN (12). The larger the Z-score value between the two genes in the TGN, the more reliable interaction (more similar expression profile) between them. Several experimentally verified cases showed that the TGN-predicted gene connections were likely to have related functions, such as the proteasome complex (12), the adenosine triphosphate synthase complex (15) and genes involved in DNA rearrangement during Tetrahymena MAC development (6). Thus, the TGN presents an important resource to study Tetrahymena genes at the pathway level.
Construction
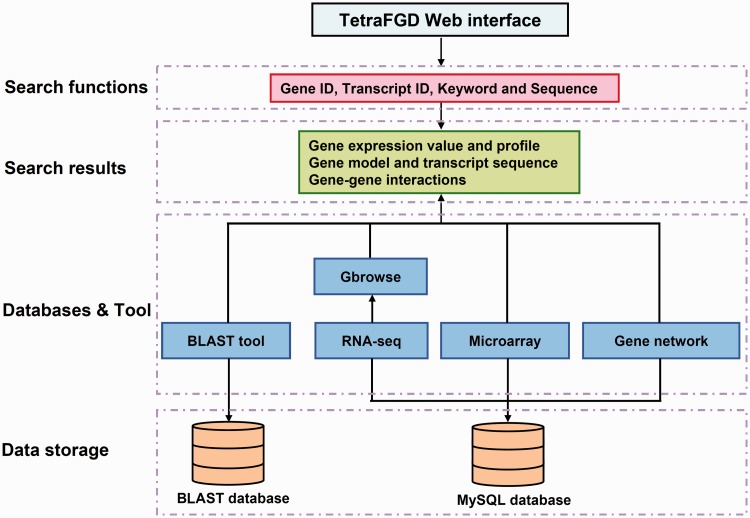
The schema of the TetraFGD is showed in Figure 1. Gene models, transcript sequences, microarray expression data and gene network data were stored in the MySQL database, and transcript sequences were formatted as a Basic Local Alignment Search Tool (BLAST) database. The web interface for searching these data was written by using Hypertext Preprocessor (PHP). Apache2 (http://httpd.apache.org/) in a CENTOS operating system was used as the web server. To provide a convenient way to check the gene models and the transcripts, Gbrowse2 (http://gmod.org/wiki/GBrowse) was set-up for graphically viewing the RNA-Seq data. A BLAST web server was also applied for the sequence-based searching against the formatted transcript database. Search functions make it easy to access to the three major resources in the TetraFGD.
Figure 1.
The schema of the TetraFGD.
Use and discussion
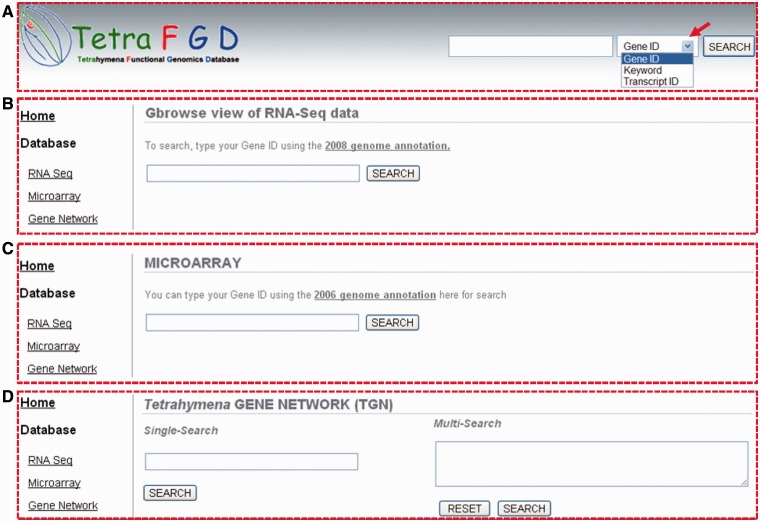
The TetraFGD can be accessed via the World Wide Web at http://tfgd.ihb.ac.cn/. An integrated searching box was designed to provide quick access to the data on the top of each page of the TetraFGD website. You can use ‘Gene ID’, ‘Keyword’ or ‘Transcript ID’ to search the database (Figure 2A). If you use the ‘Gene ID’ to search (Figure 2A), you should provide a ‘TTHERM_XXXXXXXX’ style ID, which was originally generated by gene prediction, and this style ID is also used in the Tetrahymena genome database (TGD, http://www.ciliate.org/). If you use a ‘Keyword’ to search (Figure 2A), you can type any word(s) in the search box, and the database will return the record(s) with a gene name (based on the gene annotation, but not the gene name from individual published studies) containing your keyword. If you use the ‘Transcript ID’ to search (Figure 2A), you should provide a transcript fragment ID from the RNA-Seq database. This ID can be found by either of two ways: (i) using the ‘TTHERM_XXXXXXXX’ to find its related transcript fragment ID(s) through searching the database or the look-up table (http://tfgd.ihb.ac.cn/index/version) and (ii) using your sequence to BLAST against the RNA-Seq assembled transcript database (http://tfgd.ihb.ac.cn/tool/blast).
Figure 2.
Screenshots of searching function interfaces of the TetraFGD website. (A) Integrated searching box. The red arrow indicates the pull-down button to the searching menu for choosing the ‘Gene ID’, ‘Keyword’ or ‘Transcript ID’. (B–D) Individual searching boxes for RNA-Seq, microarray and gene network.
In addition to the integrated search box, we have also designed the individual searching function, such as RNA-Seq (Figure 2B), microarray (Figure 2C) and gene network (Figure 2D).
Searching RNA-Seq data
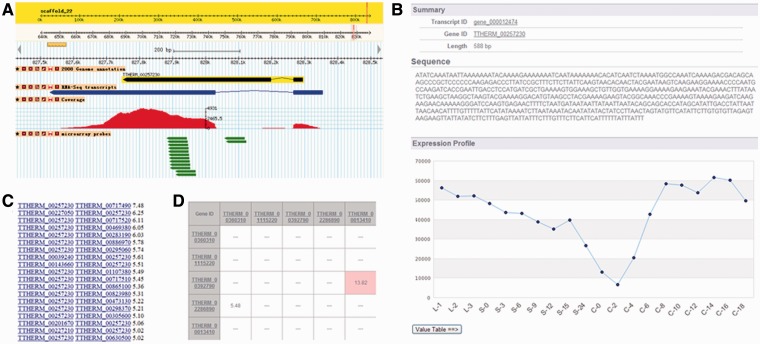
The TetraFGD displays the RNA-Seq data graphically and performs its search function through Gbrowse (http://gmod.org/wiki/Main_Page). Typically, Gene ID can be used to search the data (Figure 2B), and it also accepts a keyword (gene annotation), a transcript ID or a scaffold region. On the Gbrowse search result page, four tracks are shown, including a predicted gene model track, a RNA-Seq assembled transcript track (linked to the transcript sequence and its microarray expression information), a RNA-Seq coverage plot track and a microarray probe track (Figure 3A). Through these tracks, you can check whether there are any gene prediction mistakes, and retrieve the transcript sequence and the gene exon–intron structure information. These data are useful for studying downstream gene function. Through the Gbrowse, you can choose any specific interesting region and export the FASTA format sequence of any selected region by clicking the ‘Download Decorated FASTA File’ in the pull-down box. In addition, Gbrowse also allows the user to export the high-resolution image.
Figure 3.
Screenshots of search result interfaces of the TetraFGD website. (A) Gbrowse snapshot showing the RNA-Seq search result for the gene TTHERM_00257230. (B) Microarray result page for RNA-Seq assembled transcript, taking the gene_000012474 as an example (searching this ID in the top searching box by choosing the ‘Transcript ID’). (C and D) The screenshot montages of results for ‘Single-Search’ and ‘Multi-Search’ in TGN.
Searching microarray data
TetraFGD currently contains the microarray data of 20 time points during the three major physiological and developmental stages of the T. thermophila life cycle. Gene ID can be used to search the data either in the top search box or on the individual microarray search page (Figure 2C). You can search the microarray expression values either based on the predicted gene models or the RNA-Seq assembled transcripts. For the predicted gene models, the microarray result page follows the style of the TGED, and it also includes the Gene ID (hyperlink to the TGD), description, sequence (cDNA and protein) and shows the expression profile [see detail in (11)]. For the RNA-Seq assembled transcripts, re-normalized microarray expression data were added to the TetraFGD to help researchers get more accurate expression information, and the result gives the summary information, sequence, as well as the expression profile (Figure 3B). In addition, a new function was designed to retrieve the detailed gene expression values for each state by clicking the button ‘Value Table=>’ (Figure 3B).
Searching Tetrahymena gene network data
TGN was constructed with 15 049 genes (12) using the CLR algorithm (14), and it supplies a source to retrieve possible functionally related genes. On the gene network page, one can enter a gene ID for ‘Single-Search’ and multiple gene IDs for ‘Multi-Search’ (Figure 2D). These two types of search functions were designed for different needs. The ‘Single-Search’ was designed to find all the potential interacted candidates of the query gene, and the result will return a list of candidate genes with expression patterns similar to your query gene in the TGN (Figure 3C). The ‘Multi-Search’ was designed to find the potential interactions among a set of genes, and the result will return gene–gene interactions among your supplied genes in the TGN (Figure 3D). All the search results can be downloaded as a tab-delimited text file that can be visualized by using the Cytoscape software (http://www.cytoscape.org/) (16).
Other services in the TetraFGD
Besides the previously described functions, the TetraFGD now set-up a BLAST web server (17), allowing the RNA-Seq assembled transcripts to be found using the nucleotide sequence by BlastN or the protein sequence by TBlastN (http://tfgd.ihb.ac.cn/tool/blast). Moreover, detailed ‘Help’ information is provided to assist in using the database easily. On the TetraFGD ‘Search Help’ page (http://tfgd.ihb.ac.cn/index/schhelp), we have supplied detailed introductions and explanations of search functions to access to all the resources. The page ‘Sample Preparation’ (http://tfgd.ihb.ac.cn/index/smphelp) describes the standardization of culture conditions of growth, starvation and conjugation, used in preparing RNA for microarrays and RNA-Seq.
It is worth noting that T. thermophila gene annotations have been updated several times. The microarray platform was designed according to the 2006 version genome annotation, whereas the RNA-Seq data were analysed using the 2008 version genome annotation. There are some differences between these two versions of genome annotation. Therefore, we provide a look-up table (http://tfgd.ihb.ac.cn/index/version) for the conversions among 2006 version genome annotation IDs, 2008 version genome annotation IDs, transcript IDs and gene descriptions to conveniently use this database. The user will receive a warning when the search uses an improper gene ID.
Further development of the TetraFGD
Future development of the TetraFGD will include uploading and integrating additional Tetrahymena functional genomics data sets, such as the microarray gene expression, transcriptome, re-sequencing data of T. thermophila under stress from exposure to pollutants or displaying the effects of specific gene mutations and also the phosphorylation proteomics.
Conclusions
The TetraFGD website makes substantial improvement to the original TGED website through the addition of databases of RNA-Seq and gene network as well as BLAST searching. To facilitate access to these resources, a user-friendly web interface was developed. The TetraFGD website (also the earlier TGED website) has already attracted considerable interest from the worldwide scientists, and web traffic records indicated that they receive, on average, >150 unique visits from >40 countries per day during the past 2 years. In conclusion, the TetraFGD is an important integrated functional genomics resource, which is freely available to interested researchers.
Funding
Knowledge Innovation Program of CAS (KSCX2-EW-G-6-4); the Scientific Research Foundation for the Returned Overseas Chinese Scholars State Education Ministry; the open foundation of the State Key Laboratory of Genetics Resources and Evolution (GREKF10-09); National Scientific Data Sharing Platform for population and health, biologic medicine information center of China (2005DKA32402 to W.M.).
Conflict of interest. None declared.
Acknowledgements
The authors thank Prof. Martin Gorovsky (University of Rochester) for his critical review of the manuscript. They also thank the Gbrowse community members for their Email helps for installing the Gbrowse in the server and the Tetrahymena community for suggestions and for pointing out bugs in the database.
References
- 1.Asai DJ, Forney JD, editors. Tetrahymena thermophila. Methods in Cell Biology. Orlando, FL: Academic press; 2000. [Google Scholar]
- 2.Gibbons IR, Rowe AJ. Dynein—a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 3.Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 5.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- 7.Eisen JA, Coyne RS, Wu M, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stover NA, Krieger CJ, Binkley G, et al. Tetrahymena genome database (TGD): a new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res. 2006;34:D500–D5003. doi: 10.1093/nar/gkj054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stover NA, Punia RS, Bowen MS, et al. Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database (Oxford) 2012;2012:bas007. doi: 10.1093/database/bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao W, Xiong J, Bowen J, et al. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong J, Lu X, Lu Y, et al. Tetrahymena Gene Expression Database (TGED): a resource of microarray data and co-expression analyses for Tetrahymena. Sci. China Life Sci. 2011;54:65–67. doi: 10.1007/s11427-010-4114-1. [DOI] [PubMed] [Google Scholar]
- 12.Xiong J, Yuan DX, Fillingham JS, et al. Gene network landscape of the ciliate Tetrahymena thermophila. PLoS One. 2011;6:e20124. doi: 10.1371/journal.pone.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J, Lu XY, Zhou ZM, et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS One. 2012;7:e30630. doi: 10.1371/journal.pone.0030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith JJ, Hayete B, Thaden JT, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:54–66. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nina PB, Dudkina NV, Kane LA, et al. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8:e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]