Abstract
Controlled vocabularies (CVs), i.e. a collection of predefined terms describing a modeling domain, used for the semantic annotation of data, and ontologies are used in structured data formats and databases to avoid inconsistencies in annotation, to have a unique (and preferably short) accession number and to give researchers and computer algorithms the possibility for more expressive semantic annotation of data. The Human Proteome Organization (HUPO)–Proteomics Standards Initiative (PSI) makes extensive use of ontologies/CVs in their data formats. The PSI-Mass Spectrometry (MS) CV contains all the terms used in the PSI MS–related data standards. The CV contains a logical hierarchical structure to ensure ease of maintenance and the development of software that makes use of complex semantics. The CV contains terms required for a complete description of an MS analysis pipeline used in proteomics, including sample labeling, digestion enzymes, instrumentation parts and parameters, software used for identification and quantification of peptides/proteins and the parameters and scores used to determine their significance. Owing to the range of topics covered by the CV, collaborative development across several PSI working groups, including proteomics research groups, instrument manufacturers and software vendors, was necessary. In this article, we describe the overall structure of the CV, the process by which it has been developed and is maintained and the dependencies on other ontologies.
Database URL: http://psidev.cvs.sourceforge.net/viewvc/psidev/psi/psi-ms/mzML/controlledVocabulary/psi-ms.obo
Introduction
Proteomics is the use of gel electrophoresis and/or chromatography combined with mass spectrometry (MS)-based methods to identify and quantify proteins from complex samples, e.g. blood or urine, with the aim of increasing our understanding of the proteins, their function, interactions, expression control and other properties under normal, diseased or other conditions. Information obtained in this manner can be useful for identifying new biomarkers and/or drug targets (1). Because of the establishment of high-throughput technologies for proteomics, the amount of data generated in MS-based proteomics experiments and stored in public repositories has grown rapidly (2). The HUPO-PSI (Human Proteome Organization–Proteomics Standards Initiative) is a proteomics community organization defining standard formats for the data representation in proteomics to facilitate the data comparison, exchange and verification. It developed a set of XML-based standard formats, including mzML (3) for raw and processed MS data, TraML (4) for input transitions to selected reaction monitoring (5) (SRM), i.e. targeted proteomics approaches, where only exactly determined m/z values are selected for detection by exactly specifying the precursor–product transitions (i.e. pairs of defined peptides and fragments to search for), mzIdentML (6) for peptide and protein identification data and mzQuantML (Walzer et al., in preparation) for proteomics quantification results.
The mentioned data formats are designed for representing proteomics data to support data sharing, re-analysis, database deposition and long-term storage of these data in public repositories like PRIDE (Protein IDEntifications database) (2) or PeptideAtlas (7). The formats make use of controlled vocabulary (CV) terms from different ontologies in a standardized manner (see Table 1) (Mayer et al., in preparation), to allow for future extensibility of standards (9) and to capture the true semantics of data, which is more difficult to achieve using purely XML-based techniques.
Table 1.
Proteomics standard formats making use of the PSI-MS ontology
| Standard format (reference) | Description |
|---|---|
| mzML (3) | Format for encoding of raw MS spectrometry output data. |
| mzIdentML (6) | Format for peptide and protein identification data. |
| mzQuantML (Walzer et al., in preparation) | Format for MS quantification information. |
| TraML (4) | Format for specifying SRM transitions. |
| PEFF (http://www.psidev. info/peff) | PSI Extended Fasta Format, a unified format for protein and nucleotide sequences. |
| imzML (8) | Format for MALDI imaging data. |
| mzTab (Griss et al., in preparation) | Tab-delimited format for MS identification and quantification information. |
In designing and development of such a CV, one should make sure that each modeled concept is represented by a unique preferred term and that synonyms are included as references to that term. In addition, one can define relations to express hierarchical or equivalence relationships or other associations between the CV terms. For the storage of the CV itself, several formats are possible (Mayer et al., in preparation). The PSI-MS CV is stored in the OBO (Open Biomedical Ontology) flat file text format described in more detail at http://www.geneontology.org/GO.format.obo-1_4.shtml.
The annotation of the data with CV terms is also the basis to ensure the compliance of the published data with the MIAPE (Minimum Information About a Proteomics Experiment) (10) and journal guidelines (11). The semantic validity of the usage of the CV terms inside an instance data file can be checked by semantic validators, which are based on the PSI validation framework (12) developed at the EBI (European Bioinformatics Institute), and that can be used to implement validators locally or in web environments.
In the following sections, we describe the PSI-MS CV as the central terminology reference used by the current proteomics standard formats defined by HUPO-PSI as well as by the upcoming mzQuantML format for MS quantification information; mzTab (Griss et al., in preparation), a tab-delimited file format used for MS identification and quantification information; PEFF (PSI Extended Fasta Format) (http://www.psidev.info/peff), a proposed unified format for protein and nucleotide sequence databases for a structured alternative to the generic Fasta (13) format; and some associated standards like imzML (8) for MS imaging data, and mz5 (14).
More general details about ontologies used in proteomics, the use of CV terms in connection with mapping files for semantic validation and MIAPE compliance checking and the OBO format and related tools for working with OBO files are presented in an overview article by Mayer et al. (in preparation).
The process of using CV values for semantic validation was first used by the PSI-MS group in the mzData (15) format, one of the two predecessors of the mzML (3) standard format, which was unified from mzData and the mzXML (16) format. Initially, there were two separate CVs in use: PSI:MS (corresponding to the current CV IDs between MS:1000000 and MS:1000934) and PSI:PI (corresponding to the current CV IDs greater than MS:1001000). Before the release of mzIdentML, they were merged to form the PSI-MS CV.
Structure of the PSI-MS CV
The PSI-MS CV is a manually curated ontology stored in the OBO format, which is defined by the OBO-Edit working group (http://oboedit.org/?page=workinggroup) and is the format used by the open-source OBO-Edit (17) software. The details of the OBO format are described in detail at http://www.geneontology.org/GO.format.obo-1_2.shtml.
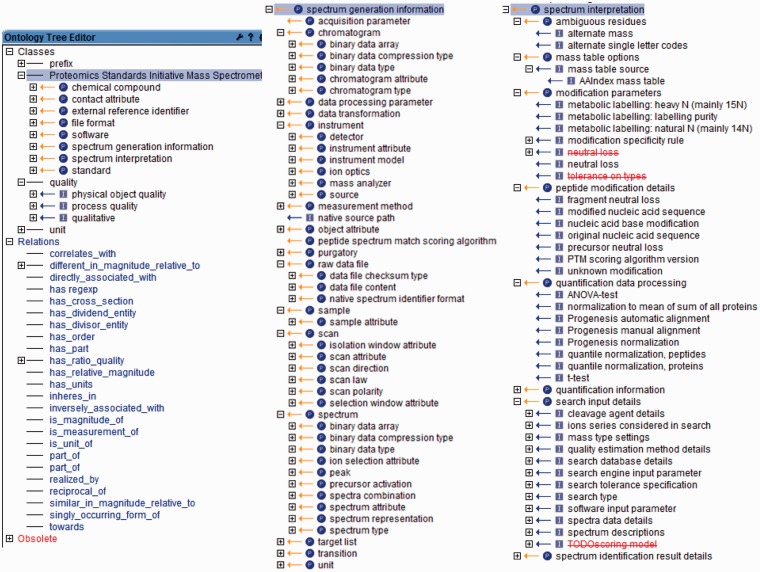
The PSI-MS CV is divided into the eight main branches, as shown on the left part of Figure 1 and shortly described in Table 2. In addition to the PSI-MS terms, it also contains different prefixes of SI (International System of Units, http://physics.nist.gov/cuu/Units/prefixes.html) units, and the definitions of used relations and terms included in the PATO (Phenotype Attribute Trait Ontology) (http://obofoundry.org/wiki/index.php/PATO:Main_Page) and Unit (18) ontologies (see ‘Dependencies on other ontologies’ section).
Figure 1.
The PSI-MS ontology, shown in screenshots from the OBO-Edit (17) software Left: the eight main branches of the PSI-MS ontology together with the terms and relations included from the PATO (quality) and ‘Unit’ ontologies. Middle: the ‘spectrum generation information’ branch of the PSI-MS ontology. Right: the ‘spectrum interpretation’ branch of the PSI-MS ontology. The striked out terms denote terms that are made obsolete.
Table 2.
Top branches of the PSI-MS ontology
| Top branch of the psi-ms.obo ontology | Description of the type of subordinate terms in the branches |
|---|---|
| Chemical compound | Terms about the chemical formula and attributes of chemical compounds, peptides and proteins. |
| Contact attribute | Terms about contact data (addresses, e-mail, fax, phone, URLs) of researchers, organizations and other roles and role types. |
| External reference identifier | Information about IDs, accession numbers, URIs (Uniform Resource Identifiers), hashes, DOIs (Digital Object Identifiers) or other identifiers referencing objects located in databases, repositories or in the web. |
| File format | Terms describing proprietary or standard formats used in proteomics. |
| Software | Terms about the different kinds of software (either specific for a vendor or an instrument or free software). It is divided into different groups like acquisition, analysis, data processing and quantitation software. |
| Spectrum generation information | Branch containing all terms describing the generation of a spectrum (see ‘Detailed Structure’ section). |
| Spectrum interpretation | Branch containing all the terms describing the interpretation of a spectrum (see ‘Detailed Structure’ section). |
| Standard | Terms about other standards, e.g. minimum information guidelines or retention time standards. |
At the heart of the PSI-MS CV are the two branches ‘spectrum generation information’ and ‘spectrum interpretation’, as shown in the middle and right parts of Figure 1 and described in the Tables 3 and 4. The file format mzML (3), representing raw or processed MS data, predominantly uses the first branch, whereas the mzIdentML (6) and mzQuantML file formats, which represent identification and quantitation results based on MS data, mostly use the second branch.
Table 3.
Sub-branches below ‘spectrum generation information’
| Branch below ‘spectrum generation information’ | Description of the type of subordinate terms in the branches |
|---|---|
| Chromatogram | Terms representing a detector response versus retention time. |
| Data processing parameter | Contains parameters and thresholds used in the data processing of data files. |
| Data transformation | Terms describing transforming data processing steps, e.g. file format conversion, baseline reduction, deconvolution, deisotoping, intensity normalization, peak picking, retention time alignment and smoothing operations. |
| Instrument | Branch containing instrument specific terms describing the different instrument models and their attributes, as well as general terms describing the source, ion optics, mass analyzer and detector of MS instruments. |
| Measurement method | Attribute of resolution terms, when recording the detector response in the absence of the analyte. |
| Object attribute | Branch containing attribute terms describing the sample preparation, the scans and runs, chromatogram, spectrum, inlet, instrument, isolation and window, etc. |
| Purgatory | A sort of predecessor of obsolete terms. |
| Raw data file | Branch for terms describing raw data files like e.g. checksums, data file content and the native spectrum identifier format, etc. |
| Sample | Branch for sample describing terms (sample number, sample concentration, sample volume, sample state, sample preparation, etc.) |
| Scan | Terms describing the recording of a spectrum, like scan polarity, isolation and selection window, etc. |
| Spectrum | Branch containing spectrum-related terms about spectrum type, spectrum representation (centroid or profile mode) and other spectrum and peak describing attributes, as well as terms for describing the binary representation of the spectra data. |
| Target list | CV terms for target lists (i.e. inclusion or exclusion terms) for specifying expected m/z coordinates, and for peptide or compound specific MS examinations. |
| Transition | Branch for terms describing SRM transition experiments. |
| Unit | Terms describing MS specific units, e.g. Th/s, etc. |
Table 4.
Sub-branches below ‘spectrum interpretation’
| Branches below ‘spectrum interpretation’ | Description of the type of subordinate terms in the branches |
|---|---|
| Ambiguous residues | Terms describing ambiguous amino acid residues and masses of non-standard amino acids. |
| Mass table options | Terms describing the sources of used mass tables. |
| Modification parameters | Terms for modification specificities, neutral losses or modifications used in metabolic labeling experiments. |
| Peptide modification details | Terms for describing the modification of peptides and proteins, e.g. PTMs (post-translational modifications). |
| Quantification data processing | Terms for the description of data processing steps in quantitative proteomics experiments, e.g. t-test, ANOVA, normalization and alignment steps. |
| Quantification information | Contains terms for quantification software, quantification data types and other quantification attributes; also, terms for use in the ‘AnalysisSummary’ element for supporting the validation of mzQuantML files. |
| Search input details | Contains terms about cleavage agents and their regular expressions, the considered ion series, about quality estimation, search database details, specification of search tolerances, the search type [PMF (peptide mass fingerprint), PFF (peptide fragment fingerprint), de novo, spectral library search, etc.), and terms for common and specific input parameters of software and search engines. |
| Spectrum identification result details | Terms for general and search engine–specific scores, false discovery rates and other peptide and protein results (e.g. protein ambiguity group assignments and taxonomy) details. |
The ‘spectrum generation information’ branch contains CV terms for the description of the sample, the chromatogram, the instrument used, the scan and the spectrum (Figure 1, middle part). It also contains parameters for the description of the acquisition parameters and the data processing, as well as CV terms describing the transitions in SRM (19, 20) experiments, the latter which is an integral part of the TraML standard (4) for the representation of SRM assays. For mzML, terms within this branch are required to make valid files, and features among other things a list of different identifier formats for spectra from different mass spectrometers in the ‘native spectrum identifier’ format node, which is a key for tracking spectra in mzML files back to the original raw data.
The branch ‘spectrum interpretation’ collects, for example, terms describing the use of an alternate mass table in isotope-labeled experiments (21). Additional terms are assembled here describing quantification information and quantification processing for annotations used in mzQuantML. Also, search input details containing CV terms defining the input parameters for software and database search engines as well as details about spectrum identification results [like scores, thresholds and false discovery rate values (22)] are grouped under the ‘spectrum interpretation’ branch (see Figure 1, right part).
An example excerpt using CV terms for reporting the scores of peptide identification results in an mzIdentML file is shown here, where the terms are included in cvParam XML elements:
<SpectrumIdentificationResult spectraData_ref="SID_1" spectrumID="index=137" id="SIR_1">
<SpectrumIdentificationItem passThreshold="false" rank="1" peptide_ref="RVDSGLHCPLLPDDR" calculatedMassToCharge="582.954" experimentalMassToCharge="582.931" chargeState="3" id="SII_1_1">
<PeptideEvidenceRef peptideEvidence_ref="PE1_2_0"/>
<cvParam accession="MS:1001328" cvRef="PSI-MS" value="0.0561" name="OMSSA:evalue"/>
<cvParam accession="MS:1001329" cvRef="PSI-MS" value="1.3475E-5" name="OMSSA:pvalue"/>
<cvParam accession="MS:1001171" name="mascot:score" cvRef="PSI-MS" value="56.16" />
<cvParam accession="MS:1001172" name="mascot:expectation value" cvRef="PSI-MS" value="2.4210e-006" />
</SpectrumIdentificationItem>
This example shows also how in a re-analysis, the scoring values of two or more different search machines (here OMSSA and Mascot) can, in principle, be reported. However, it must be stressed here that it is not possible to have a metric to compare the quality of the results of two different search machines. The CV allows one to document the used search machines, their versions, the used parameters for the searches and the resulting scores of them, so that they are easily reproducible by others.
The following example illustrates the usage of CV terms in an mzML file for the specification of a selection window (specifying the lower and upper limits of m/z values for detection):
<selectionWindowList count="1">
<selectionWindow>
<cvParam cvLabel="MS" accession="MS:1000501" name="scan m/z lower limit" value="110.000000"/>
<cvParam cvLabel="MS" accession="MS:1000500" name="scan m/z upper limit" value="905.000000"/>
</selectionWindow>
</selectionWindowList>
The next example shows the usage of CV terms in a TraML file for the specification of a transition by specifying the precursor and productions:
<TransitionList>
<Transition id="AAQVAQDEEIAR.2y8-1" peptideRef="AAQVAQDEEIAR.2">
<Precursor>
<cvParam unitCvRef="MS" unitName="m/z" unitAccession="MS:1000040" value="650.8288" accession="MS:1000827" name="isolation window target m/z" cvRef="MS"/>
</Precursor>
<Product>
<cvParam unitCvRef="MS" unitName="m/z" unitAccession="MS:1000040" value="931.4486" accession="MS:1000827" name="isolation window target m/z" cvRef="MS"/>
</Product>
Some special cases
A special case is the definition of terms for cleavage agents, as it requires two CV terms, one for the enzyme itself and one for the regular expression, which is referenced in the enzyme CV term by the ‘has_regexp’ relationship, as shown in the following example. In addition, the BRENDA (23) ontology is specified as the defining source for the enzyme by the database cross reference (‘dbxref’) (BRENDA:3.4.21.37). The regular expressions describing the restriction sites of the enzymes can be used for digesting a protein in silico, as used within search engines in proteomics.
[Term]
id: MS:1001915
name: leukocyte elastase
def: "Enzyme leukocyte elastase (EC 3.4.21.37)." [BRENDA:3.4.21.37]
is_a: MS:1001045 ! cleavage agent name
relationship: has_regexp MS:1001957 ! (?<=[ALIV])(?!P)
[Term]
id: MS:1001957
name: (?<=[ALIV])(?!P)
is_a: MS:1001180 ! Cleavage agent regular expression
The list of allowed ‘dbxref’ terms is given at the GO (Gene Ontology) website at http://www.geneontology.org/cgi-bin/xrefs.cgi. Currently, the PSI-MS CV makes use of the following ‘dbxref’ terms: BRENDA, DOI, http://… resp. https://…, PubChem_Compound and PMID.
Dependencies on other ontologies
To avoid duplication of terms, the PSI-MS CV itself refers to terms defined in the PATO (http://obofoundry.org/wiki/index.php/PATO:Main_Page), and the Unit (18) ontologies. PATO (‘quality.obo’) describes phenotypic qualities, and ‘unit.obo’ contains general terms defining units of measurement. These two ontologies are imported into the PSI-MS CV in the document header by the following tags in the header part of PSI-MS:
It should be stressed here that by this reference mechanism, it is made sure that additions and updates of terms from the PATO and Unit ontologies are automatically available in the PSI-MS CV, so that the PSI-MS CV can easily stay in sync with new developments of the included PATO or Unit ontology.
An example of the use of PATO is the mapping rule in mzML for the validation of the allowed CV terms under a sample, where the term ‘quality of an object’ (PATO:0001241) can be used to describe the sample quality:
<CvMappingRule id="sample_may" cvElementPath="/mzML/sampleList/sample/cvParam/@accession" requirementLevel="MAY" scopePath="/mzML/sampleList/sample" cvTermsCombinationLogic="OR">
… … ..
<CvTerm termAccession="PATO:0001241" useTerm="false" termName="quality of an object" isRepeatable="true" allowChildren="true" cvIdentifierRef="PATO"></CvTerm>
… … ..
</CvMappingRule>
Such a mapping rule is a formal statement inside a mapping file, which exists for each of the HUPO-PSI standard formats and defines at which position and in which combination a certain CV term can occur inside the instance data file (Mayer et al., in preparation).
The units are used to specify the measurement unit for the CV terms that have a value; for example, the following example states that the value for the sample volume must be given in milliliters.
[Term]
id: MS:1000005
name: sample volume
def: "Total volume of solution used." [PSI:MS]
xref: value-type:xsd\:float "The allowed value-type for this CV term."
is_a: MS:1000548 ! sample attribute
relationship: has_units UO:0000098 ! milliliter
Measurement units that are specific to MS, e.g. ‘Thompson’, are defined in the PSI-MS CV itself under the ‘unit’ branch of ‘spectrum generation information’. Currently, there are also some general units, which are already defined in unit.obo and which are repeatedly redefined in PSI-MS. This is mainly due to historic reasons, and these terms are in the process of being removed or made obsolete.
Basic statistics for the PSI-MS CV
By November 2012, the ‘psi-ms.obo’ file (version 3.40.0) contained 2130 terms, from which 90 are obsolete and 20 were in the ‘purgatory’ branch. The ‘is_a’ relation, which is defined in the OBO relationship ontology (24), was used 2201 times. In addition, the ontology contains the definitions for other four types of relationships: ‘has_units’ (used by 166 terms), ‘part_of’ (131 usages), ‘has_regexp’ (19 usages) and ‘has_order’ (1 usage). Note that some ontology terms can contain more than one ‘is_a’ relationship, so that there are more usages of ‘is_a’ than the total number of terms (2062) in the PSI-MS ontology.
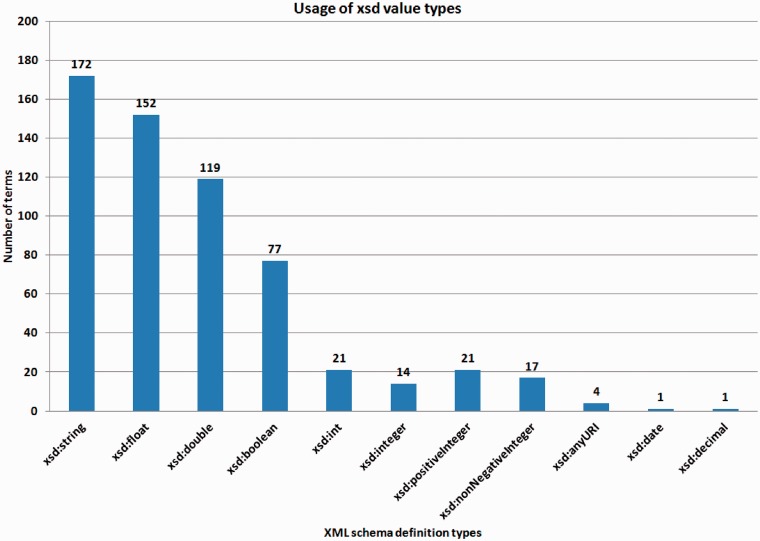
The majority of terms are only referenced inside HUPO-PSI standard proteomics data files in <cvParam> elements without specifying a value. However, 595 terms from psi-ms.obo are intended to be used with a value; most of them are of the type string (172 terms), float (152 terms), double (118 terms) or boolean (74 terms) (see Figure 2).
Figure 2.
Number of used XML schema types as value types in the PSI-MS ontology (version 3.40.0). Note that xsd:integer and xsd:int are different in XML schema (http://www.w3.org/TR/xmlschema11-2/#built-in-datatypes), as the value space of the former is the infinite set.
In total, 202 terms with synonyms are contained in the PSI-MS CV, of which 179 were of type EXACT and 22 of the type RELATED.
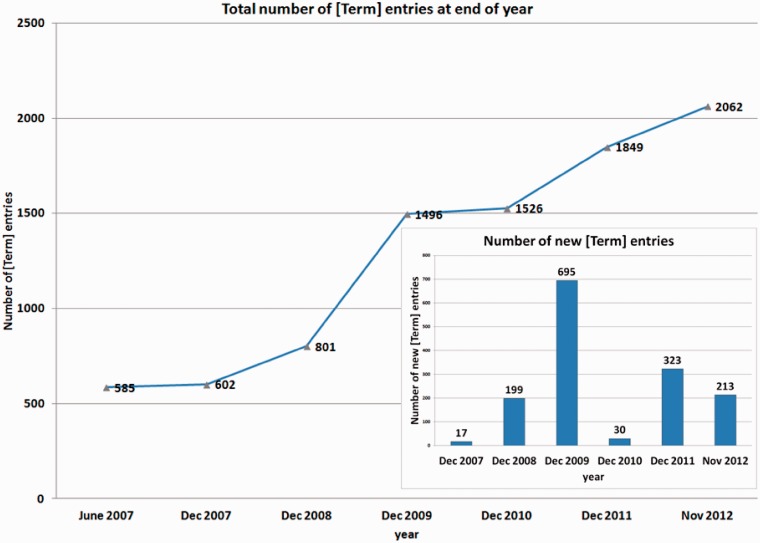
The growth of the PSI-MS ontology since June 2007 until November 2012 is depicted in detail in Figure 3. The high numbers of new terms in 2009 is probably due to the enacting of the mzML 1.1.0 specification by that year.
Figure 3.
Growth of the number of all terms (including obsolete terms and those included in the ‘purgatory’ branch) in the PSI-MS ontology since June 2007 (cf. http://psidev.cvs.sourceforge.net/viewvc/psidev/psi/psi-ms/mzML/controlledVocabulary/psi-ms.obo?view=log) and number of newly added terms per year to the PSI-MS ontology between June 2007 and November 2012 (inlet).
The statistical ontology metrics reported by BioPortal (25) are shown in Table 5.
Table 5.
Statistical ontology metrics for the PSI-MS ontology (version 3.40.0), according to the BioPortal (25) website (http://www.bioontology.org/wiki/index.php/Ontology_Metrics)
| Statistical metric according to BioPortal | Number |
|---|---|
| Number of classes | 4640 |
| Number of individuals | 0 |
| Number of properties | 10 |
| Maximum depth | 9 |
| Maximum number of siblings, i.e. terms on the same level | 157 |
| Average number of siblings | 1 |
| Classes with a single subclass | 151 |
| Classes with >25 subclasses | 23 |
| Classes with no definition | 991 |
Note that the numbers include the counts for the imported PATO and Unit (18) ontologies.
Maintenance of the PSI-MS CV
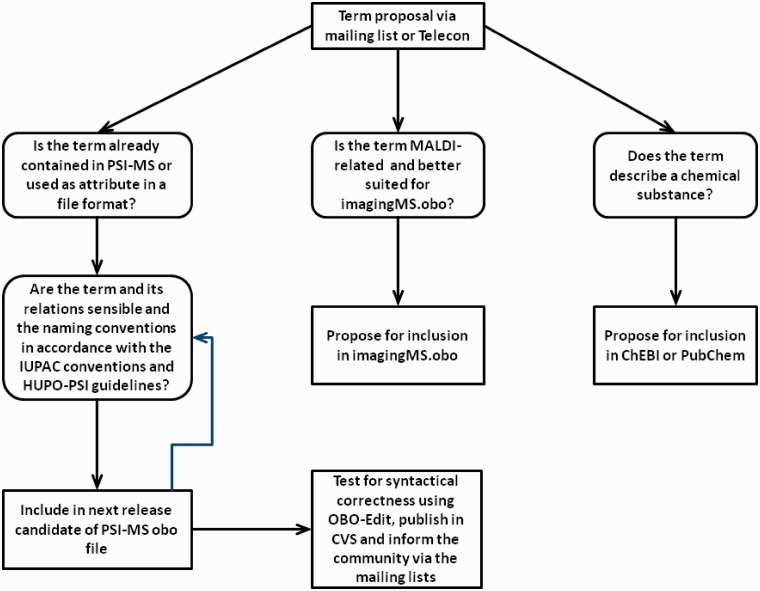
The PSI-MS CV evolved over time by important contributions of a wide community, including hardware and software vendors, which contributed much to the high-quality definition for many terms. The further development of the PSI-MS CV is an ongoing process. For this, the HUPO-PSI working groups defined some guidelines for the development of CVs (http://www.psidev.info/node/47). In addition, the detailed maintenance process advanced over the time, and some informal best practices evolved for it. Previously, requests for new terms were done by filling in a form on the PSI-PI website and by discussing the new-term proposals or terms in dispute via an issue tracker located at http://sourceforge.net/tracker/?group_id=65472&atid=848524. Now everyone in the proteomics community is free to subscribe to the ‘psidev-ms-vocab’ mailing list at https://lists.sourceforge.net/lists/listinfo/psidev-ms-vocab and to make proposals for new terms or improvements of the already existing ‘psi-ms.obo’ terms. Also, requests to restructure parts of the ontology are possible, for instance when it emerges that the current hierarchical structuring of terms is suboptimal or needs a reorganization because of new technological developments, but in all these cases, it is warranted that already existing terms are never deleted from the ontology because of the obsoletion mechnism. Often, there are also proposals discussed in the telephone conferences of the various PSI subgroups, so that the update can be done within ∼5 working days after such a request, provided that there are no objections and there is consensus about the requested terms. The current maintenance procedure is now described as it has been applied to the ‘psi-ms.obo’ ontology file since January 2012 (see Figure 4).
Figure 4.
Simplified workflow of the PSI-MS maintenance procedure.
This maintenance work is coordinated by the PSI ontology coordinator. He/she is a member of the proteomics scientific community and is normally elected at the annual HUPO-PSI spring meeting or appointed by the steering committee in the case that an emerged vacancy for this position must be assigned between these meetings. After receiving a request for a new CV term, the PSI ontology coordinator checks whether the term and its description, data type, parent terms and relations are sensible. If necessary, any inconsistencies are clarified by consulting the proposer of the term. The ontology coordinator then checks whether a term with the same meaning is already present in the ontology or whether the term is necessary at all. The coordinator also checks whether the naming of the terms and synonyms are in accordance with the IUPAC (International Union of Pure and Applied Chemistry) nomenclature for MS terms (http://mass-spec.lsu.edu/msterms/index.php/Main_Page). If an attribute with the same meaning is already present in the schema of the corresponding data format, typically the CV term will not be added to avoid duplication of information.
An additional rule is used if a term is related to MALDI (Matrix Assisted Laser Desorption Ionization) checking: whether the term is already present in the MALDI imaging obo (http://www.maldi-msi.org/download/imzml/imagingMS.obo) and whether the term would be more suitable in that ontology. If there are proposals about chemical substances, e.g. used for matrix solutions, it is checked whether the substance is already defined in the Chemical Entities of Biological Interest (ChEBI) ontology (26). In that case, the request is denied, and the proposer is given notice that they should consider using a CV term referencing the corresponding ChEBI entry instead. If not, the CV coordinator can request the ChEBI team to incorporate the substance into their ontology if it fulfills the criteria for inclusion into ChEBI. If not, it is checked whether the substance is defined in the PubChem (27) database, and a new term in the PSI-MS CV is created, which references this PubChem entry by specifying the corresponding ‘dbxref’ term at the end of the def: tag line.
A term that passes all these checks then is included in the next release candidate of the obo file. This release candidate is then sent to the three mailing lists psidev-ms-vocab@lists.sourceforge.net, psidev-pi-dev@lists.sourceforge.net and psidev-ms-def@lists.sourceforge.net for public discussion. To allow a prompt update of the CV after requests for new or changed terms, there is no regular schedule, so that if there is no objection, the new terms of the release candidate become part of the next official release of the obo file, which is made public ∼5 working days after the release candidate. Otherwise, the term under question is further discussed by the subscribers of the mailing lists, either by email correspondence or, if necessary, in a telephone conference call, until everything gets clarified and the community comes to a consensus about the exact definition of the discussed term, whereat the consensus should be reached by the strength of the arguments. As far as possible, the term names should be general and non-proprietary. In case that vendor-specific terms are inevitable, for instance because they describe a proprietary software or product, the term name can be assembled by a leading identification for the proprietary product, followed by a colon and the actual CV term name. This naming mechanism can also help to prevent possible blockades resulting from conflicts of interest between rivaling companies. Then, the date and version are updated, and the new obo file is officially released by the ontology coordinator by first checking its syntactical correctness using the ‘Verification Manager’ of OBO-Edit and then transferring it to the CVS (Concurrent Versioning System) located on the SourceForge website (http://psidev.cvs.sourceforge.net/viewvc/psidev/psi/psi-ms/mzML/controlledVocabulary/psi-ms.obo?view=log). The release of the new obo version is then announced to the three mailing lists stated above together with a small summary of the new and/or changed terms. The version number of the PSI-MS CV has the format ‘x.y.z’. An increase in x means the release of a major build, i.e. that a change in a root level term took place, whereas an increase of y indicates the addition of new terms or the obsoletion of terms, and an increase of z means that only minor changes like the editing of names or definitions was done.
In cases where a merging, splitting, replacement or deprecation of an ontology term is necessary, e.g. owing to upcoming new technologies or instruments or changes in standard formats, the old terms must be set obsolete by assigning the ‘is_obsolete’ relation to them, but they must stay inside the ontology to ensure backward compatibility of instance data files already making use of these now obsoleted terms.
Future directions
Besides the usage by the proteomics standard formats of the HUPO-PSI group, the PSI-MS CV is used by six other projects (Table 6). With the further development of the standard formats and the appearance of new methods, software and instruments in proteomics, and—not to underestimate—the finalization of implementations of the PSI standards in conversion software, there is a steady growth of the PSI-MS CV over time (Figure 3). In addition, there is a demand for adjusting some aspects in the future, rooted in the history of PSI-MS. For instance, there are several units defined in PSI-MS, which are also defined in the ‘Unit’ ontology, as these terms pre-dated the existing of the unit ontology. Another example is the purgatory branch. It has its root also in the beginning of the PSI-MS development process, when there was no ‘is_obsolete’ relation for marking terms that should not be used any more. It can be expected that most of these terms will also be marked as obsolete in the future.
Table 6.
Other projects using the PSI-MS ontology [adapted from the BioPortal (25) website at http://bioportal.bioontology.org/ontologies/1105]
| Project (Reference) | Description |
|---|---|
| ISA software suite (28) (http://isa-tools.org) | Open source software suite for assisting in the annotation and local management of experimental metadata from high-throughput studies. |
| NCBO (National Center for Biomedical Ontology) Annotator (29) (http://www.bioontology.org/annotator-service) | Web service that annotates textual metadata (e.g. journal abstracts). |
| NCBO Resource Index (30) (http://www.bioontology.org/resources-index) | The NCBO Resource Index is a system for ontology-based annotation and indexing of biomedical data; the key functionality of this system is to enable users to locate biomedical data resources related to a particular concept. |
| OntoCAT (31) (http://www.ontocat.org) | Provides high-level abstraction for interacting with ontology resources including local ontology files in standard OWL and OBO formats and public ontology repositories. |
| MeRy-B (Metabolomic Repository Bordeaux) (32) (http://bioportal.bioontology.org/ontologies/1105) | A plant metabolomics knowledge base allowing the storage and visualization of metabolic profiles from plants. |
| OntoMaton (https://github.com/ISA-tools/OntoMaton) | Facilitates ontology search and tagging functionalities within Google Spreadsheets. Now part of isa-tools. |
It was demonstrated here that the use of CVs in proteomics made the proteomics standard formats more independent of changes of names or definitions of terms. The obo file also helps to keep pace with technological advancements by allowing the addition of new terms for upcoming technologies. This helps to keep the proteomics formats stable and independent of the set of used vocabulary terms. This approach can also be used in other -omics disciplines (genomics, transcriptomics, proteomics, interactomics, metabolomics, fluxomics, …), so that the use of CVs by these formats can, for example, help to integrate data set in so-called multi-omics studies, or to match terms in meta-analyses in case that the single analyses are using synonyms in their naming schemes for the same concepts, even if the synchronization of terms from ontologies from different -omics areas still remains a challenge for the future.
Of course, it can also be expected that new technological developments, like MSE (33); ion mobility (34) (a method where the ionized molecules are separated according to their mobility in a carrier gas instead of the separation according to their mass–charge ratio) and hybrid multi-dimensional ion-separation approaches combining both ion separation techiques; SWATH [a DIA (Data Independent Acquisition) method, where one has to specify a series of isolation windows called ‘swaths’); QITL (Quantitative Isobaric Terminal Labeling) (35), where the C termini of the peptides are labeled with 16O or 18O and the N-termini with normal or d(2)formaldehyde to allow the quantitation of the peptides; GeLC-MS, a combination of Gel-based and liquid chromatography-MS–based proteomics (36); or other upcoming methods will require the addition of new terms to the PSI-MS CV.
Another future direction will be the integration of vocabulary terms representing metabolic information, e.g. terms related to the standard gas chromatography–MS method of metabolomics (37) (in metabolomics, mostly gas chromatography–MS is used instead of liquid chromatography–MS, as it is relatively easy to convert the low-molecular-weight metabolites into gaseous form by derivatization, i.e. chemical modifications), for usage in mzML files or other standard formats. Also, the planned novel NMR Markup Language (NMR-ML) under development by the COSMOS (Coordination of Standards in Metabolomics, http://www.cosmos-fp7.eu/wp2) project of the Metabolomics Standards Initiative (MSI) (38) for storing NMR spectroscopy data (39) can be imagined to make use of the PSI-MS CV and to contribute new terms to it, e.g. to describe the chemical shifts, which are dependent of the shielding of the external magnetic field by the local chemical environment of the hydrogen nuclei and can be used for detecting and elucidating the structure of the molecules.
Although it is expected that by all the mentioned and other new technologies, the PSI-MS CV will grow over time, we do not await that there will be an exponential growth like, for instance, in the sequencing domain. Instead, we rather expect that the number of terms will only grow moderately in the future. This is because the growth of the CV (Figure 3) in the past was mainly driven by the definition of the various proteomics data formats of the HUPO-PSI. These formats are already defined now, and because of the usage of the PSI-MS CV, they are relatively independent of changes in the used terms, and therefore relatively stable. So it is not to expect that a complete redesign will be necessary—this would also contradict the idea of the CV that obsolete terms must stay inside the CV forever, so that all already existing data files still stay reproducible. Of course, it may be necessary that owing to technological developments, a splitting of branches or terms will become necessary, as it happened, for instance, in the medical field, where the term non-A non-B hepatitis became obsolete and must be replaced by the hepatitis forms caused by the C, D and E viruses. In such a case, this of course means that it is up to the software programs respective curators of the public repositories to interpret and handle resp. update such obsolete terms properly in already existing data files or databases, because this cannot be done automatically.
Funding
G.M., J.A.V., A.R.J. and P.A.B. are funded by the European Union project ProteomeXchange (http://www.proteomexchange.org, EU FP7 grant number 260558). J.A.V. is also supported by the Wellcome Trust (grant number WT085949MA). P.A.B. is also funded by the Swiss Federal Government through the Federal Office of Education and Science. M.E. is funded by P.U.R.E. (http://www.pure.rub.de, Protein Unit for Research in Europe), a project of Nordrhein-Westfalen, a federal state of Germany. F.L. is supported by the Swedish Research Council through the BILS infrastructure. A.R.J. also acknowledges funding from the UK BBSRC (BB/I000909/1 and BB/H024654/1). E.W.D. is funded in part by NIGMS grants R01 GM087221 and P50 GM076547/Center for Systems Biology, and from the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg.
Conflict of interest. None declared.
Acknowledgements
In memoriam Andreas Bertsch, who was a former ontology coordinator of the PSI-PI group and lost his life far too early. We want to acknowledge also all the former coordinators and contributors to the PSI-MS CV throughout the years.
References
- 1.Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov. Today. 2012;17(Suppl.):S16–S23. doi: 10.1016/j.drudis.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Vizcaíno JA, Coté R, Reisinger F, et al. The proteomics identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens L, Chambers M, Sturm M, et al. mzML—a community standard for mass spectrometry data. Mol. Cell Proteomics. 2011;10:R110000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutsch EW, Chambers M, Neumann S, et al. TraML—a standard format for exchange of selected reaction monitoring transition lists. Mol. Cell Proteomics. 2012;11:R111.015040. doi: 10.1074/mcp.R111.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holman SW, Sims PF, Eyers CE. The use of selected reaction monitoring in quantitative proteomics. Bioanalysis. 2012;4:1763–1786. doi: 10.4155/bio.12.126. [DOI] [PubMed] [Google Scholar]
- 6.Jones AR, Eisenacher M, Mayer G, et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell Proteomics. 2012;11:M111.014381. doi: 10.1074/mcp.M111.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutsch EW. The PeptideAtlas project. Methods Mol. Biol. 2010;604:285–296. doi: 10.1007/978-1-60761-444-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schramm T, Hester A, Klinkert I, et al. imzML—a common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteomics. 2012;75:5106–5110. doi: 10.1016/j.jprot.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Jones AR, Paton NW. An analysis of extensible modelling for functional genomics data. BMC Bioinformatics. 2005;6:235. doi: 10.1186/1471-2105-6-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor CF, Paton NW, Lilley KS, et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez H, Snyder M, Uhlen M, et al. Recommendations from the 2008 International Summit on Proteomics Data Release and Sharing Policy: the Amsterdam principles. J. Proteome Res. 2009;8:3689–3692. doi: 10.1021/pr900023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montecchi-Palazzi L, Kerrien S, Reisinger F, et al. The PSI semantic validator: a framework to check MIAPE compliance of proteomics data. Proteomics. 2009;9:5112–5119. doi: 10.1002/pmic.200900189. [DOI] [PubMed] [Google Scholar]
- 13.Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 14.Wilhelm M, Kirchner M, Steen JA, Steen H. mz5: space- and time-efficient storage of mass spectrometry data sets. Mol. Cell Proteomics. 2012;11:O111.011379. doi: 10.1074/mcp.O111.011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orchard S, Jones P, Taylor C, et al. Proteomic data exchange and storage: the need for common standards and public repositories. Methods Mol. Biol. 2007;367:261–270. doi: 10.1385/1-59745-275-0:261. [DOI] [PubMed] [Google Scholar]
- 16.Pedrioli PG, Eng JK, Hubley R, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 17.Day-Richter J, Harris MA, Haendel M, Lewis S. OBO-Edit—an ontology editor for biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 18.Gkoutos GV, Schofield PN, Hoehndorf R. The Units Ontology: a tool for integrating units of measurement in science. Database (Oxford) 2012;2012:bas033. doi: 10.1093/database/bas033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J. Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 20.Kiyonami R, Domon B. Selected reaction monitoring applied to quantitative proteomics. Methods Mol. Biol. 2010;658:155–166. doi: 10.1007/978-1-60761-780-8_9. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, Wisniewski JR, Cox J, et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protocols. 2011;6:147–157. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 22.Dudoit S, Shaffer JP, Boldrick JC. Multiple hypothesis testing in microarray experiments. Stat. Sci. 2003;18:71–103. [Google Scholar]
- 23.Scheer M, Grote A, Chang A, et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degtyarenko K, de Matos P, Ennis M, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Xiao J, Suzek TO, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W33. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocca-Serra P, Brandizi M, Maguire E, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit on Translat. Bioinforma. 2009;2009:56–60. [PMC free article] [PubMed] [Google Scholar]
- 30.Jonquet C, Lependu P, Falconer S, et al. NCBO resource index: ontology-based search and mining of biomedical resources. Web Semant. 2011;9:316–324. doi: 10.1016/j.websem.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamusiak T, Burdett T, Kurbatova N, et al. OntoCAT - simple ontology search and integration in Java, R and REST/JavaScript. BMC Bioinformatics. 2011:12. doi: 10.1186/1471-2105-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferry-Dumazet H, Gil L, Deborde C, et al. MeRy-B: a web knowledgebase for the storage, visualization, analysis and annotation of plant NMR metabolomic profiles. BMC Plant Biol. 2011;11:104. doi: 10.1186/1471-2229-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plumb RS, Johnson KA, Rainville P, et al. UPLC/MSE; a new approach for generating molecular fragment information for biomarker structure elucidation (vol 20, pg 1989, 2006) Rapid Commun. Mass Spectrom. 2006;20:2234–2234. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 34.Holcapek M, Jirasko R, Lisa M. Recent developments in liquid chromatography-mass spectrometry and related techniques. J. Chromatogr. A. 2012;1259:3–15. doi: 10.1016/j.chroma.2012.08.072. [DOI] [PubMed] [Google Scholar]
- 35.Yang SJ, Nie AY, Zhang L, et al. A novel quantitative proteomics workflow by isobaric terminal labeling. J. Proteomics. 2012;75:5797–5806. doi: 10.1016/j.jprot.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Roepstorff P. Mass spectrometry based proteomics, background, status and future needs. Protein Cell. 2012;3:641–647. doi: 10.1007/s13238-012-2079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koek MM, Jellema RH, van der Greef J, et al. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics. 2011;7:307–328. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sansone SA, Fan T, Goodacre R, et al. The metabolomics standards initiative. Nat. Biotechnol. 2007;25:846–848. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- 39.Zhang A, Sun H, Wang P, et al. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]