Abstract
The type VI secretion system (T6SS) is a complex and widespread gram-negative export pathway with the capacity to translocate protein effectors into a diversity of target cell types. Current structural models of the T6SS indicate that the apparatus is composed of at least two complexes, a dynamic bacteriophage-like structure and a cell envelope-spanning membrane-associated assembly. How these complexes interact to promote effector secretion and cell targeting remains a major question in the field. As a contact-dependent pathway with specific cellular targets, the T6SS is subject to tight regulation. Thus, the identification of regulatory elements that control T6S expression continues to shape our understanding of the environmental circumstances relevant to its function. This review discusses recent progress toward characterizing T6S structure and regulation.
Keywords: apparatus, signals, bacteriophage-like, interbacterial interactions, host-cell interactions, expression
INTRODUCTION
The type VI secretion system (T6SS) is a recently discovered mechanism in gram-negative bacteria that targets proteins to both prokaryotic and eukaryotic cells (55, 97). Like the type III and IV secretion (T3S and T4S) pathways, T6S translocates substrates directly into recipient cells in a contact-dependent manner (3, 24). Also similar to these pathways, the T6 pathway accomplishes this feat using a complex machine–13 core subunits are essential for basic secretory functions of the apparatus, and additional components may be involved in translocation (14). Although clear functional parallels exist between T6 and other secretory pathways, with few exceptions the proteins that constitute the T6S apparatus are novel and cannot easily be assigned roles in the machine. Thus, determining the composition of the T6S apparatus, how its components interact, and how accessory factors modulate the function of the system, has been an active area of research over the past several years.
The components of a T6SS are generally encoded by a group of tightly clustered genes. Such genetic clusters average over 20 kb and are prevalent in proteobacterial genomes (14, 98). In many instances, multiple T6S gene clusters can be identified in a single organism. On the basis of sequence divergence, phenotypic profiles, and coordinate differential regulation, such systems do not appear redundant (39, 65, 98). Rather, the plasticity of T6S clusters among high taxonomic ranks suggests that evolutionarily divergent systems are frequently acquired horizontally. Recent analyses indicate that T6S gene clusters separate into at least five distinct phylogenetic groups (11, 14).
A T6SS can be recognized at a functional level by the robust transport of two proteins to the milieu, hemolysin coregulated protein (Hcp) and valine-glycine repeat protein G (VgrG). These proteins demonstrate codependency for export and together constitute part, or perhaps, the entire extracellular portion of the T6S apparatus (42, 49, 84, 116). Structurally, Hcp and VgrG resemble bacteriophage tail and tailspike proteins, respectively (57, 64, 83). The specific evolutionary mechanism driving this relatedness has not been investigated in detail; however, the proteins do not share significant primary sequence homology with their phage counterparts. Whether this structural relationship translates into functional similarity is also not known. Nonetheless, a popular model depicting the T6SS as an inverted bacteriophage-like structure on the bacterial cell surface has emerged from these observations (27, 47, 57).
Functionally, T6SSs separate into four categories: (a) bacterial cell targeting, (b) eukaryotic cell targeting, (d) bacterial and eukaryotic cell targeting, and (d) other. The last includes systems implicated in processes such as conjugation, gene regulation, and cellular adhesion (4, 28, 33, 107). Importantly, it remains to be determined whether these effects are direct or indirect consequences of T6S. Because the genetic factors underlying the functional diversity of T6S are not yet defined, and few systematic studies of its function have been reported, it remains plausible that the system is considerably more promiscuous than currently appreciated.
Although many T6SSs have been studied and ascribed functions, the effector proteins involved have been identified in only a small subset of T6SSs (49, 74, 84, 93, 101, 106). Yet even from this small sampling it is evident that the T6S apparatus can accommodate a structurally and functionally diverse array of substrates. Moreover, these substrates can be configured either as specialized domains fused to apparatus components or as more canonical, independently exported proteins. Genetic data suggest that the latter grouping of substrates transit the apparatus in a one-step mechanism that avoids periplasmic intermediates (93). In this review, we focus on two rapidly advancing topics in the field of T6S: the structure of the secretory apparatus and the conditions and signals that influence its expression and activation. We make no claims to have been exhaustive and we apologize in advance to our colleagues whose work we have omitted.
ARCHITECTURE OF THE TYPE VI SECRETION SYSTEM
The T6SS is composed of a minimal set of 13 subunits, which are thought to form the core apparatus (11, 21). Although there is not yet a high-resolution structure of the whole or subcomplexes of the T6SS, such as those available for the T3SS and T4SS (104, 109), a comprehensive model of T6SS assembly has recently emerged. Biochemical and structural studies support a general model in which T6SS components form two subassemblies. Aside from these core components, T6SS gene clusters usually encode accessory subunits. The function of most of these proteins is not yet known; however, several proteins are required for proper assembly or function of the apparatus, whereas others have been shown to trigger its timely assembly (6, 100). In this section, we summarize recent findings pertaining to the topology, structure, protein-protein interaction, and function of individual T6S machine components. Based on these data, we propose a structure-function model of the apparatus.
Core and Accessory Components
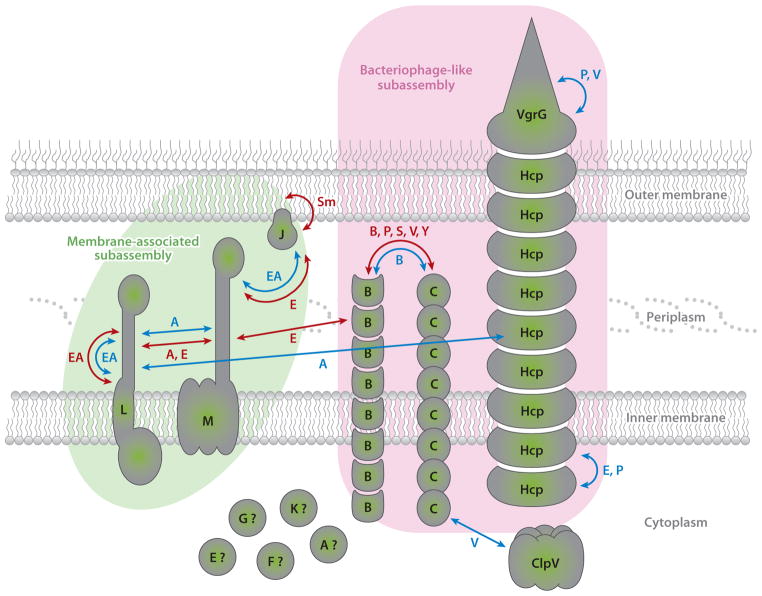
Systematic mutagenesis studies performed in Edwardsiella tarda and Vibrio cholerae have shown that each of the 13 conserved T6S genes is required for function (115, 116). On the basis of bioinformatic approaches, these genes can be classified into three categories. The first category includes genes encoding membrane-associated proteins, either integral membrane (TssL, TssM) or lipoproteins (TssJ) (Figure 1; outlined green). The second category of genes encodes proteins with relatedness to tailed bacteriophage components (Hcp, VgrG, TssB, TssC, TssE) (Figure 1; outlined pink). For the most part, this homology is apparent at the structural level, not at the sequence level. The last category contains proteins for which no function can be inferred from in silico analyses (TssA, TssF, TssG, TssK). Although we do not yet have a high-resolution structure of the T6SS, the machine can be viewed as an assemblage of two distinct substructures–a bacteriophage-like structure and a membrane complex–that interact to form an inverted bacteriophage-like structure anchored to the cell envelope (13, 57, 89).
Figure 1.
Protein interaction network between type VI secretion subunits. The localization and topologies of the core components of the T6SS are represented. Arrows indicate interactions detected among the subunits by biochemical/structural (blue) or two-hybrid approaches (red). The letter accompanying the arrow denotes the system where the interaction was detected. The membrane-associated subassembly and the bacteriophage-like subassembly are outlined in green and pink, respectively. The question mark represents subunits for which the localization has not been investigated. Relevant studies are discussed in the text. Abbreviations: T6SS, type VI secretion system; A, Agrobacterium tumefaciens; B, Burkholderia cenocepacia; E, Edwardsiella tarda; EA, enteroaggregative Escherichia coli; P, Pseudomonas aeruginosa; S, Salmonella enterica; Sm, Serratia marcescens; V, Vibrio cholerae; Y, Yersinia pseudotuberculosis Please add references (7, 12, 16a, 31a, 35, 42, 56, 64, 70, 70a, 76, 81, 84, 86 and 116)
In addition to these core components, accessory genes are usually associated with T6SS gene clusters. Because the bacteria that carry T6S gene clusters can be found in different environments and the function of T6S is highly versatile, these accessory proteins might be linked to regulation or provide supplemental functions to the apparatus. For example, accessory components critical for transcriptional regulation or posttranslational activation of the system have been characterized (see below). Thus, even though these proteins are referred to as accessory, their functions can be essential for the proper production, assembly, or activity of the apparatus.
Bacteriophage-Like Subunits
Rapidly after the identification of the T6SS, it appeared that at least two subunits, Hcp and VgrG, share secondary structure similarity with bacteriophage components. These similarities were confirmed soon after by structural data, which demonstrated that the fold of Hcp is related to that of the major tail protein of phage λ, gpV (76, 83), and that VgrG assumes a fold and quaternary arrangement similar to the gp27/gp5 complex, the spike of bacteriophage T4 (58, 64).
During bacteriophage tail assembly, gpV oligomerizes into a filamentous tubular structure of specific length (1, 25, 59). X-ray crystal packing of Hcp homologs from Pseudomonas aeruginosa and E. tarda revealed that under certain conditions Hcp hexamers assemble in a head-to-head or head-to-tail fashion (56, 76, 81). Using disulfide bond engineering, Ballister et al. (8) reported that Hcp nanotubes of defined length can be constructed in vitro. Although the current data suggest that Hcp might form a tubular structure, it remains to be determined whether such a structure exists in vivo and whether its length is regulated.
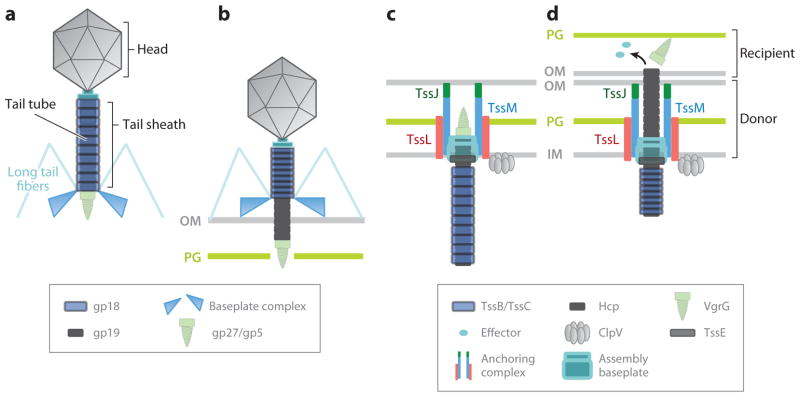
In the bacteriophage T4, a sheath surrounds the tail filament. The sheath is a contractile structure composed of >100 gp18 protomers helically assembled (Figure 2a,b) (2). In 2009, a study reported that two T6S subunits, TssB and TssC, form tubular structures observable by electron microscopy. Image reconstruction of the particles showed that TssB and TssC assemble into structures which exhibit cogwheel-like cross sections resembling the bacteriophage sheath (12, 22.). More recently, Basler et al. demonstrated that TssB/TssC tubule structures exist in two conformations, likely corresponding to extended and contracted sheath-like structures (8a). Time-lapse microscopy further showed that these tubules assemble dynamically into the cytoplasm and oscillate between the extended and contracted conformations through cycles of TssB/TssC assembly, disassembly and recycling. Interestingly, the internal diameter of the TssB/TssC complex (~10 nm) is sufficient to accommodate the Hcp tubule (external diameter of ~9 nm) (12, 22). On the basis of these observations, it has been proposed that TssB/TssC subunits would form a structure enclosing a hypothetical Hcp tube. Similar to the bacteriophage T4 sheath, contraction of the TssB/TssC tubules might propel the Hcp tube toward the cell exterior prior to secretion (Figure 2c,d). This is in agreement with the observation that secretion by the T6S apparatus is a one-step mechanism, in which substrates are directly transported from the cytoplasm of the donor to the recipient cell without transient accumulation in the donor periplasm (93). Whether substrates are preloaded in the Hcp tube or transit into the tube upon target cell perforation remains to be answered. A specific interaction between the ClpV AAA+-family ATPase and an N-terminal peptide of TssC has been defined, and ClpV disassembles TssB/TssC tubules in vitro (12). Given the findings that the purified tubules to which ClpV binds are in the contracted conformation and that TssB/TssC tubules are static in clpV mutant cells, it has been proposed that ClpV is not responsible for TssB/TssC tubule assembly or disassembly, but rather for recycling the TssB and TssC subunits upon contraction (8a).
Figure 2.
Schematic representation comparing proposed models of bacteriophage T4 and T6S. Homologous and analogous type VI secretion (T6S) and T4 proteins are colored the same as their T4 phage counterparts. (a) The bacteriophage T4 tail tube is surrounded by the tail sheath and terminated by the cell-puncturing device (gp27/gp5). (b) Upon host cell binding, the bacteriophage T4 baseplate undergoes a conformational change that triggers tail sheath contraction and results in puncturing of the outer membrane (OM) and DNA delivery. Models of (c) inactivated and (d) activated states of T6S based on protein localization and interactions between T6S subunits. The three membrane-associated proteins TssL, TssM, and TssJ form a complex bound to the peptidoglycan (PG) layer via TssL. The T6S appendix formed by an Hcp tube and a VgrG trimer is thought to be anchored at the cell envelope by the membrane-associated complex. It has been hypothesized that an assembly baseplate can participate in T6S appendix assembly. (c) TssB and TssC may form a sheath-like structure enclosing the Hcp tube within the periplasmic space. (d) Activation of the T6SS results in effector delivery to a target cell through the Hcp tube. By analogy with bacteriophage T4, the sheath-like structure could propel, through contraction, the Hcp tube toward the cell exterior or directly to the target cell. Abbreviation: IM, inner membrane.
The VgrG protein forms a trimer that structurally resembles the (gp27/gp5)3 complex of bacteriophage T4 (58, 64). The (gp27/gp5)3 complex assembles at the tip of the tail, and the β-helical needle-like structure formed by the C-terminal domain of gp5 is used to perforate the outer membrane of the target bacteria (58). By analogy, it is supposed that VgrG is displayed at the tip of the Hcp tube and that the Hcp tube propels VgrG toward the target cell. This model is supported by several observations. First, Hcp is required for VgrG release in culture supernatants (42, 84, 116). Second, specialized VgrG proteins carry a C-terminal extension that possesses specific properties. The C-terminal domain is located downstream of the β-prism and therefore is the first domain to enter into the recipient target cell (Figure 2). Bioinformatic analyses predicted several activities such as fibronectin or peptidoglycan binding, general adhesion, and actin modification. Whereas most of these predictions remain to be experimentally tested, the Vibrio cholerae VgrG1 protein carries a domain that catalyzes actin cross-linking (69, 84), and a VgrG of Aeromonas hydrophila acts as an actin ADP-ribosylating enzyme (101).
On the basis of the phage model of the T6SS, one would predict a physical interaction between Hcp and VgrG. Although a direct interaction between the proteins has not been detected, several groups have reported that Hcp and VgrG exhibit codependency for secretion (49, 84, 85, 116). Given the relative positions of VgrG and Hcp in the proposed phage-like structure, this suggests that Hcp polymerization requires, or is triggered by, VgrG recruitment to the apparatus. Interestingly, such a mechanism is consistent with what has been described for the corresponding phage proteins during particle morphogenesis (61, 73, 112).
Recent microscopy images obtained by Basler et al. showed that the base of the cytoplasmic T6SS sheath appears to be connected to the inner membrane by a large complex that may represent an assembly platform similar to the bacteriophage baseplate (8a). The baseplate is responsible for proper assembly of the hub and tail tube, as well as for contraction of the tail sheath (92). Among the T6S core components, the cytoplasmic submit, TssE, shares primary sequence homology with gp25, a structural component of the bacteriophage baseplate (11, 68). If the function of gp25 is conserved in the T6SS, TssE, in conjunction with a baseplate-like complex, might participate in assembly of the Hcp tube and the TssB/TssC sheath. In agreement with this hypothesis, the TssB/TssC tubules do not assemble in cells lacking tssE (8a).
The current model argues that canonical protein substrates are transported through the Hcp tube. Whereas the internal diameter of the Hcp hexamer (~4 nm) can accommodate a globular protein with a molecular mass less than approximately 50 kDa, the extremity of the VgrG β-prism is too narrow to allow substrate passage. One can speculate that the VgrG protein–as proposed for the bacteriophage T4–is released upon puncturing the target cell membrane, leaving the Hcp tube open and allowing delivery of substrates (Figure 2). Among the three characterized substrates of P. aeruginosa, Tse1 and Tse3 have peptidoglycan hydrolysis activities and are released in the periplasm of the bacterial target cell (93). By contrast, the biological target of Tse2, which is not yet identified, is located in the cytoplasm (49, 93).
Membrane-Associated Components
Recently, Aschtgen et al. (5) reported the isolation of a complex composed of four membrane proteins of the enteroaggregative E. coli (EAEC) Sci-1 T6SS: TssL, TssM, TssJ, and TagL. Further studies defined the localization and topology and delineated the interaction contacts of these different proteins. These and more recent biochemical and structural studies support a model in which these proteins form a complex that spans the cell envelope.
TagL is inserted into the inner membrane through three transmembrane segments (TMSs), and its large periplasmic domain carries a functional peptidoglycan-binding (PGB) motif. Mutagenesis studies have shown that TagL-mediated PGB is required for the activity of the T6SS; therefore, TagL may act as an anchor that tethers the apparatus to the cell wall (5). It is surprising that in the Sci-1 T6SS, the essential PGB activity is carried by an accessory protein. Indeed, this is often not always the case, and the PGB domain is instead fused to TssL or other subunits (6).
TagL interacts directly with TssL, an inner membrane protein anchored through a single C-terminal TMS. The bulk of TssL localizes in the cytoplasm and adopts a unique fold resembling a hook (6a, 31a). TssL has strong homology with IcmH (or DotU), a component of the type IVB secretion system (T4bSS). In the T4bSS, IcmH interacts with IcmF to form a complex that stabilizes core components of the apparatus (78). In the T6SS, the IcmF homolog is TssM. TssM is anchored to the inner membrane through three TMSs. A cytoplasmic domain of ~30 kDa, which usually contains functional ATP-binding and hydrolysis Walker motifs, is located within the cytoplasmic loop flanked by TMS2 and TMS3. Mutagenesis studies have been performed to investigate the importance of the Walker motif. Interestingly, the outcome of these studies depends on the T6SS model. In E. tarda, the Walker motif is dispensable for T6SS assembly, as the secretion of Hcp and VgrG proteins is not affected by its inactivation (116). By contrast, in Agrobacterium tumefaciens, the Walker motif is critical for Hcp secretion (70). TssM undergoes a conformational change dependent on ATP binding and hydrolysis that allows for the recruitment of Hcp to the TssM/TssL complex (70a). In several cases, Walker motifs cannot be identified in the cytoplasmic domain of TssM. It thus appears that TssM ATP binding and hydrolysis might be adapted to the specific needs of each T6SS.
The bulk of TssM (~80 kDa) is localized in the periplasm. It is composed of two subdomains: a large helical domain followed by a C-terminal β-domain (35). Yeast two-hybrid and coimmunoprecipitation experiments have shown that the periplasmic domain interacts with TssJ (35, 116). TssM associates with TssJ in a 1:1 stoichiometry and a Kd of 2–4 μM. Mapping studies showed that the TssM C-terminal β-domain is sufficient to interact with TssJ. TssJ is a lipoprotein that is anchored to the outer membrane through the acylation of its N-terminal cysteine residue (4). The crystal structures of the soluble domain of two TssJ proteins (from EAEC and Serratia marcescens) have been reported recently (35, 86). TssJ has a transthyretin fold (two parallel β-sheets) with additional elements, including a variable loop between β-strands 1 and 2 required for interaction with TssM. The conservation level of this loop is low, suggesting that it can act as a specificity determinant during assembly of the T6S apparatus (35). By virtue of interacting with TssL at the inner membrane and TssJ at the outer membrane, TssM spans the envelope. Rao et al. (86) reported that the S. marcescens TssJ lipoprotein is engaged in homomeric interactions. Interestingly, the TssL protein also oligomerizes (31a). It is therefore conceivable that the TssL-TssM-TssJ complex associates as a ring-like structure to shape a channel spanning the cell envelope, analogous to membrane spanning structures that have been observed for the T3SS and T4SS (37, 109).
The observation that two distinct subassemblies exist in the T6SS raises the question of how these are connected. The periplasmic domain of the TssM protein interacts with TssB in a yeast two-hybrid assay, while Hcp was shown to co-immunoprecipitate with the periplasmic portion of the TssM-TssL complex (116, 70a). These results are compatible with the current model in which the sheath structure surrounding the Hcp tube is embedded in a channel-like structure formed by the assembly of the periplasmic domains of TssM.
INPUTS THAT MODULATE TYPE VI SECRETION EXPRESSION AND ACTIVITY
Even though our understanding of T6S function has grown considerably in recent years, its role in nature remains unclear. For instance, many bacteria of significant health concern possess one or more T6SSs; however, the pathogenic relevance of most of these systems is unknown. The majority of organisms with T6SSs are not pathogens and instead are found in marine environments, the rhizosphere, and in soil, or they are associated with higher organisms as symbionts or commensals (11, 14). Defining the signals and conditions that control the expression and activation of T6S under the highly varied environments such bacteria occupy will be critical for revealing its role in these contexts. Below we provide examples of regulatory pathways and signals that modulate T6S expression. We discuss how environmental cues that influence these pathways might provide insights into the physiological function of the system. Our review of T6S regulation is not comprehensive; therefore, we refer readers interested in further details to more exhaustive recent reviews (10, 66).
Environmental Signals
Recent findings have demonstrated various classes of regulators sensitive to environmental cues specifically modulate the activity of the T6SS. Further defining the signals that stimulate T6SSs is essential for understanding the physiological context in which these systems act.
Iron
The ferric-uptake regulator (Fur) protein is a key modulator of iron-dependent gene expression in bacteria (20). This regulator generally represses transcription through Fe(II)-dependent dimerization and subsequent DNA binding to a consensus sequence called the Fur box located within promoter regions. Transcriptional activation and iron-independent regulation by Fur also occur (19, 43). In addition to its important role in regulating iron acquisition and homeostasis, Fur regulates genes and processes that are not directly involved in iron metabolism. Some examples of these include toxins, adhesins, motility, and resistance to reactive oxygen species (18–20, 46, 88).
Expression of T6S in two opportunistic enteric pathogens, E. tarda and EAEC, is repressed directly at the transcriptional level by Fur (Figure 3) (17, 23). E. tarda is primarily a pathogen of fish; however, consumption of contaminated seafood can lead to gastroenteritis in humans (95). Infection models suggest that T6S plays an important role in the virulence of E. tarda and its close relative, Edwardsiella ictaluri, against fish (75, 91, 116). Deletion of genes encoding core components of the E. tarda Evp (E. tarda virulence proteins) T6SS, or insertional disruption of a gene encoding a putative effector of this system, evpP, attenuated the organism approximately 100-fold in blue gourami.
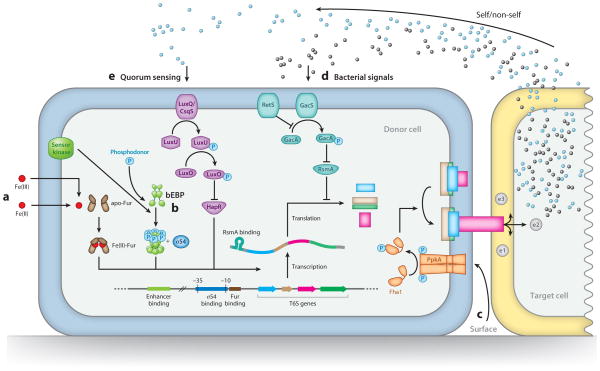
Figure 3.
Schematic representation of the diverse regulatory systems that modulate T6S expression and activation in assorted bacteria. Only those regulatory pathways emphasized in this review are depicted. Pathways are labeled A–E corresponding to the order of their presentation in the text. (A) Fur represses T6S transcription in the presence of iron. (B) bEBPs function in conjunction with σ54 to activate T6S transcription. (C) The TPP posttranslationally activates T6S in response to surface association. Self- and nonself-derived bacterial signals modulate T6S (D) posttranscriptionally through the Gac/Rsm pathway or (E) transcriptionally via quorum sensing. At right is a target bacterium undergoing intoxication by Tse1–3 effectors (e1–3). Abbreviations: TPP, threonine phosphorylation pathway; bEBP, bacterial enhancer binding protein; T6S, type VI secretion.
Fur-dependent regulation of the Evp T6SS was recently demonstrated by Mok and colleagues (23). Their work showed that Fur confers iron-dependent repression of production and export of an Hcp homolog (EvpC) and that the Fur protein binds directly to a Fur box sequence upstream of evpP, the first gene in the evp cluster. Fur-based repression of the evp genes is consistent with the contribution of the system to pathogenesis, as Fur repression would likely be alleviated inside iron-depleted host tissues.
EAEC is an emerging enteric pathogen characterized by its propensity to self-adhere and form biofilms on the intestinal mucosa (79). Infections with EAEC result in diarrhea and can be acute or chronic in nature; it is currently the second most common cause of diarrhea in persons traveling to developing countries (44, 102). The Sci-1 T6SS of EAEC is required for biofilm formation and is regulated by iron availability through a pathway involving DNA adenine methyltransferase (Dam)-catalyzed methylation and Fur repression (17). Two Fur boxes and three Dam methylation sites are present upstream of the Sci-1 gene cluster. Interestingly, one of the Fur-binding sites overlaps with a Dam methylation site, and Fur binding prohibits methylase access to the site. In the absence of iron, Fur dissociates and allows RNA polymerase to bind and initiate transcription. Similarly, the loss of Fur also permits methylation at the site, which inhibits the reassociation of Fur. Thus, low cytoplasmic Fe(II) yields stable on-state expression of the Sci-1 T6SS. It is not currently understood how sci-1 expression is returned to the off-state, as the binding of Fur to hemimethylated sequences that would be generated following DNA replication was not investigated (17).
The physiological consequences of sci-1 regulation by Fur are not yet known and depend on the abundance and form of iron present in a given environment. The anaerobic environment of the intestinal lumen favors the ferrous [Fe(II)] form of the ion. In support of this, studies of Salmonella have shown that genes under control of Fur remain repressed prior to tissue invasion (51, 54). Furthermore, Salmonella pathogenicity island I, which encodes a T3SS required for Salmonella invasion, is activated by iron-bound Fur (32). Extending these findings to EAEC Sci-1 suggests that the T6SS may remain repressed by Fur in vivo. This is congruent with studies demonstrating that T6S does not contribute to the virulence of EAEC in animal infection models (4, 31). However, an important consideration when interpreting these data is that the animal models employed are unlikely to accurately recapitulate chronic EAEC infection. It is conceivable that ferrous iron becomes depleted within stable intestinal biofilm communities of EAEC, thereby leading to Sci-1 T6SS activation. Sci-1 activation mediated by Fur depression is likely to occur in an environmental context, where its role in promoting adhesion could be exploited as an adaptation to oxidative stress or iron starvation (4, 29).
σ54-dependent activators
Sigma factor 54 (σ54)-dependent activator proteins, also termed bacterial enhancer binding proteins (bEBPs), are a diverse group of proteins that mediate the translation of environmental signals to changes in gene expression in bacteria (87). bEBPs regulate gene expression by catalyzing the closed-to-open transition of σ54-RNAP holoenzyme transcription complexes. This activity is ATP-dependent and, as described below, can be regulated by signal binding and phosphorylation. bEBPs are generally composed of three domains: an N-terminal regulatory domain, an internal AAA+-family ATPase domain, and a C-terminal DNA-binding domain that typically mediates sequence-specific interactions with activator sequences 100 to 200 nucleotides upstream of the promoter (96). The N-terminal activation domain and the C-terminal DNA-binding domain are highly variable among bEBP homologs (99). This low conservation is partly responsible for the diversity of signals bEBP homologs detect and the distinct DNA sequences they bind.
A broad range of cellular processes, including nitrogen assimilation, motility, and virulence, are regulated by bEBPs (99). This class of proteins also serves a general role in the regulation of T6S (Figure 3). Bioinformatic analyses identified bEBPs encoded within phylogenetically diverse T6S gene clusters (9, 14). These analyses have further revealed probable σ54-binding sequences within the respective T6S promoter regions (9). A subset of T6S-associated bEBPs, including representatives from V. cholerae, A. hydrophila, Pectobacterium atrosepticum, and Marinomonas spp., were investigated using in vitro binding experiments and reconstituted transcriptional reporter assays. These studies confirmed the predicted role of the proteins in σ54-dependent transcriptional activation of T6S promoters.
The bEBP encoded within the virulence-associated secretion (vas) gene cluster, VasH, is a key regulator of V. cholerae T6S (62, 85). Reflecting its important regulatory role, T6S-dependent defense against Dictyostelium discoideum and killing of E. coli in this organism require VasH (71, 84). An analysis of vasH homologs in 26 V. cholerae strains identified an enrichment of nonsynonymous single nucleotide polymorphisms in the N-terminal regulatory domain (62). Amino acid changes within this domain could provide a mechanism for variable T6S expression response profiles within V. cholerae strains exposed to environmental signals. Interestingly, N-terminal domains of bEBPs can act as intramolecular activators or repressors (110). In V. cholerae V52, this domain appears to be a positive regulator, as vas T6S expression was decreased by an N-terminally truncated VasH allele (62). Notably, polymorphisms in vasH do not appear to underlie the significant differences in vas expression and activation observed between non-O1/non-O139 strains (e.g., V52) and pandemic disease strains (e.g., N16961, C6706, and A1552). Instead, this variability is explained likely by differences in the strength of repression through quorum-based signaling and by the novel regulator TsrA (type VI secretion system regulator A) (53, 117).
The activity of bEBPs can be regulated by specific phosphorylation events catalyzed by sensor histidine kinases and low-molecular-weight phosphodonors (72, 99). An example is the well-characterized bEBP NtrC (nitrogen regulatory protein C), which is activated through phosphorylation by NtrB (108). In addition, NtrC efficiently autophosphorylates in vitro in the presence of acetyl phosphate, and evidence suggests that acetyl phosphate is also relevant to its signaling properties in vivo (36). A sensor kinase that acts on T6S bEBPs has not been identified; however, these proteins autophosphorylate in vitro in the presence of acetyl phosphate (9). This finding suggests that upstream events such as environmental sampling by sensor kinases or changes in metabolism leading to the accumulation of low-molecular-weight phosphodonors could have a significant impact on σ54-dependent T6S expression.
Surface association
Surface association can promote dramatic changes to bacterial cell physiology. An analysis of global gene expression changes in Salmonella typhimurium demonstrated that one-third of its genes are altered during surface growth conditions (105). Such changes may be caused by signaling systems that directly detect surfaces or that respond to concomitant alterations in the local environment. Cells adhered to surfaces can develop into sessile communities, sometimes referred to as biofilms, in which long-term cell-cell contact is enhanced relative to planktonic cells (26). According to the current understanding of T6S-mediated effects, these conditions are favorable to its contact-dependent mechanism of effector delivery.
The Hcp secretion island-I encoded T6SS (H1-T6SS) of P. aeruginosa is posttranslationally activated by a threonine phosphorylation pathway (TPP) in response to surface growth of the organism (Figure 3) (100). Components of this pathway include PpkA, an inner-membrane-spanning serine/threonine kinase, and Fha1, a forkhead-associated domain-containing protein that is activated by PpkA via phosphorylation (50, 77). Activated Fha1 promotes H1-T6S-apparatus assembly and effector secretion. Unlike planktonically grown P. aeruginosa, strains placed on an agar surface for 4 h assemble an activated apparatus and contain elevated levels of phosphorylated Fha1 (100). Genetic analysis of the requirements for competitive fitness mediated by the H1-T6SS further established the role of the TPP in surface-dependent H1-T6SS activation and ruled out the involvement of a second phosphorylation-independent pathway. Surface activation of T6S by the TPP may be a general phenomenon, as components of the pathway are found in approximately 30% of identified T6S gene clusters (14).
Though the mechanism(s) is not yet clear, sessile growth may also serve as a cue for regulatory changes that elevate T6S expression. Recent findings demonstrated that cellular levels of TssC1, an H1-T6S component, are elevated in a biofilm compared with planktonically grown cells (113). Consistent with this observation, the protein Hcp is abundant in P. aeruginosa biofilms (94).
Bacteria-Derived Signals
Often functioning as a cell contact-dependent bacterial interaction pathway, it is not surpising that the sensing of other bacteria plays an important role in modulating T6S activity. Here we describe several examples of bacteria-derived signals that influence T6S expression.
The Gac/Rsm pathway
The Gac/Rsm signaling pathway couples extracellular bacteria-derived signals with marked changes in target mRNA translation (63). The pathway is initiated by the GacS/GacA two-component system, which upon stimulation leads to elevated expression of one or more small regulatory RNA (sRNA) molecules, variably termed rsmB, rsmX, rsmY, rsmZ, csrB, or csrC. These sRNA molecules interact with and sequester an mRNA-binding protein known as RsmA or CsrA. This protein generally acts as an inhibitor of translation by associating with sequences near or overlapping the ribosome-binding site. Thus, sRNA expression typically facilitates increased translation of specific mRNA targets. In some cases, mRNA binding by RsmA/CsrA can activate translation; however, the mechanism is less clear in these instances.
In the pseudomonads, the Gac/Rsm pathway is a key regulator of many important processes, including biocontrol and virulence factor production, cellular aggregation, and quorum sensing (40). Studies have revealed that P. aeruginosa, P. fluorescens, and P. syringae also use this pathway to regulate the expression of T6S (Figure 3) (45, 76, 90). This was first noted in P. aeruginosa, wherein microarray analyses of strains lacking retS or ladS, which encode a repressor and activator of GacA/S signaling, respectively, strongly implicated RsmA in the stability of HSI-I transcripts (41, 103). Later functional analyses demonstrated that assembly, activation, and effector secretion and targeting by the H1-T6SS are stimulated in the ΔretS background (49, 76, 77). The most definitive evidence for the involvement of the Gac/Rsm pathway in HSI-I regulation is found in recent work by Brencic & Lory (15), which demonstrates direct RsmA binding to the 5′ leader sequence of two HSI-I transcripts.
The incorporation of the H1-T6SS into the global regulon of the P. aeruginosa Gac/Rsm pathway has yielded valuable insights into the settings relevant to its function. In P. aeruginosa, the Gac/Rsm pathway directly or indirectly regulates the expression of approximately 500 genes (16, 41). Within this expansive set, researchers have noted reciprocal regulation of factors associated with planktonic and sessile growth–leading to the hypothesis that the Gac/Rsm pathway coordinates this physiological transition of the organism (111). In the Gac/Rsm regulon, H1-T6SS expression occurs coincident with factors having experimentally demonstrable roles in sessile community formation, such as two aggregation and adhesion-promoting exopolysaccharides, Pel and Psl. This early regulatory link implied that T6SS activity is relevant to closely interfacing bacteria; however, in what capacity remained unknown. This question has been answered–at least in part–by more recent studies demonstrating the role of the H1-T6SS in contact-dependent interbacterial interactions and the involvement of the system in biofilm-specific antibiotic resistance (49, 93, 113). Notably, the Gac/Rsm pathway of P. fluorescens, which is closely related to that of P. aeruginosa, responds to signals generated by other pseudomonads and certain Vibrio spp. (30).
Quorum Sensing
Quorum sensing is a bacterial regulatory mechanism that modulates gene expression based on cell population density (38, 80). A quorum is sensed by the accumulation of diffusible signaling molecules, which are themselves typically under quorum control. Regulation of quorum-controlled genes is achieved either by direct or indirect effects that signal molecules impart on the DNA-binding properties of dedicated regulatory proteins. A prevailing model is that quorum sensing regulates social behavior of bacteria, both within and between species (82). One piece of evidence in support of this model is that secreted products are disproportionally abundant in the quorum regulon of many species (48). Given this, it is not surprising that many instances of quorum-sensing-regulated T6SSs have arisen in the literature (39, 52, 60, 65, 67, 114, 117).
In V. cholerae, two chemically distinct quorum-sensing systems, autoinducer-2 (AI-2) and cholerae autoinducer-1 (CAI-1), collaborate to influence density-dependent gene expression (Figure 3) (80). Signal molecules from these pathways are detected by two sensor kinases, LuxQ and CqsS, respectively. The pathways converge on the phosphotransfer protein LuxU, which acts on LuxO, a DNA-binding response regulator protein. Phosphorylated LuxO activates the expression of sRNA molecules (qrr1–4) that in turn repress the production of HapR, a TetR-family global transcriptional regulator.
The majority of studies on V. cholerae T6S have been conducted in the serotype O37 strain V52. The V52 strain is a valuable model for studying T6S within the species, as Hcp and VgrG proteins are abundantly exported from the strain, its vas genes are highly expressed, and it exhibits strong Vas-dependent phenotypes against prokaryotic and eukaryotic cells (71, 84, 85). However, under similar in vitro conditions, most strains of V. cholerae, including O1 and O139 pandemic strains, display markedly lower vas expression and as a result do not exhibit Vas-dependent phenotypes (52, 53, 117).
Recent studies have suggested that differences in direct quorum-sensing-dependent expression of vas genes are partially responsible for T6S variability in V. cholerae. Ishikawa et al. (52) observed a striking correlation between HapR and Hcp expression among a panel of O1 isolates. Furthermore, a deletion of luxO was shown to strongly induce vas expression in two serotype O1 strains, A1552 and C6707 (52, 117). Consistent with current models of quorum-sensing circuitry in V. cholerae, activation of vas expression in ΔluxO required hapR and Δhfq recapitulated the effects of the luxO deletion.
Despite robust vas expression in O1 strains lacking luxO, the secretion system can remain functionally quiescent in this background (117). Thus, levels of HapR do not fully reconcile T6S-related phenotypic differences observed between V. cholerae strains. Briefly, complete activation of T6S in pandemic V. cholerae strains appears to involve additional factors such high osmolarity, low temperature, and relief of repression imposed on the system by the TsrA protein (53, 117).
The adaptive role of quorum control over T6S remains to be elucidated. In addition to its role in intraspecies sensing, quorum sensing might play a role in perceiving cells of other species (34). This seems particularly probable for the AI-2 pathway, as the dedicated signal synthase involved in AI-2 synthesis, LuxS, is widely conserved. Therefore, AI-2 signal levels could serve as a cue for V. cholerae to activate antibacterial defenses, such as its T6SS, in response to potential competitors. If the signal was self-derived, coregulated immunity proteins might ameliorate the detrimental consequences of self-targeting (93).
Not all T6SSs regulated by quorum sensing are induced at high cell densities. For example, in P. aeruginosa the H1-T6SS is repressed at high cell densities by a direct or indirect mechanism involving LasR, an acyl homoserine lactone-type quorum regulator (65). Interestingly, the two other T6SSs of P. aeruginosa are regulated reciprocally with the H1-T6SS by quorum sensing. The two T6SSs of Vibrio parahaemolyticus also display reciprocal regulation by quorum sensing (39). Differential regulation of T6S by quorum sensing, particularly those cases wherein this occurs within one bacterium, suggests that the system can act in a wide range of contexts and underscores its functional versatility.
SUMMARY POINTS.
The T6SS is a multicomponent secretory machine that delivers effector proteins to both prokaryotic and eukaryotic cells in a contact-dependent manner.
The T6SS is composed of five bacteriophage-like proteins that likely form a cell-puncturing device and participate in apparatus assembly.
Interactions between four essential membrane-associated proteins suggests a cell-envelope-spanning complex is central to T6S function.
Expression and activation of T6S are tightly controlled by diverse regulatory systems.
Fur is an iron-responsive regulator that directly represses T6S expression in EAEC and E. tarda.
σ54-dependent T6S expression is often controlled by bEBPs encoded within T6S gene clusters.
T6S accessory genes involved in signaling via threonine phosphorylation control posttranslational activation of T6S in response to surface association.
Self and nonself signals influence expression of T6S through quorum sensing and, in the case of pseudomonads, the Gac/Rsm pathway.
FUTURE ISSUES.
How similar is the mechanism of T6S effector secretion to bacteriophage DNA delivery to target cells?
What is the overall architecture of the T6SS? Can the entire apparatus be isolated in vitro or visualized in vivo?
How did two phylogenetically unrelated complexes, bacteriophage-like and bacterial membrane-associated, evolve to form the T6SS? How do the two subassemblies of the T6SS interact with each other?
What are the physiological relevant targets of T6SSs? Defining the regulatory networks that influence T6SS expression and the environmental context in which these systems are functional will advance our understanding of the role of T6SSs.
What are the upstream regulators and corresponding signals that activate dedicated T6S-associated regulators and regulatory systems such as bEBPs and the TPP?
What role does competitor sensing play in bacterial cell-targeting T6SSs? Is specificity determined by regulatory factors, effectors, or surface receptors?
Acknowledgments
J.M.S. was supported in part by Public Health Service, National Research Service Awart, T32 GM07270, from the National Institute of General Medical Services. J.D.M was funded by grants from the NIH (AI080609 and AI057141). J.D.M holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Work in the E.C. laboratory is supported by the CNRS and funded by the Agence Nationale de la Recherche (ANR-10-JCJC-1303-03). Y.R.B is supported by a doctoral fellowship from the French ministry of Research. We thank members of the Mougous and Cascales laboratories and Pacome-Aude Lavieille for helpful discussions.]
Key Terms and Definitions
- bEBP
bacterial enhancer-binding protein
- EAEC
enteroaggregative Escherichia coli
- Effector proteins
proteins secreted by bacteria that influence target cell physiology
- Ferric uptake regulator (Fur)
a conserved Fe(II)-dependent regulator of gene expression
- Hcp
hemolysin coregulated protein
- PGB
peptidoglycan binding
- Secretion systems
mechanistically distinct pathways for passaging proteins through membranes
- T6SS
type VI secretion system
- T6S
type VI secretion
- Tag
type VI associated gene
- TMS
transmembrane segment
- TPP
threonine phosphorylation pathway
- VgrG
valine-glycine repeat protein C
- σ54
a sigma factor that promotes a stable, closed RNAP complex requiring activators to open; also known as RpoN
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Eric Cascales, Email: cascales@imm.cnrs.fr.
Joseph D. Mougous, Email: mougous@u.washington.edu.
LITERATURE CITED
- 1.Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199:301–10. doi: 10.1006/viro.1994.1128. [DOI] [PubMed] [Google Scholar]
- 2.Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, et al. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–29. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–31. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol Microbiol. 2010;75:886–99. doi: 10.1111/j.1365-2958.2009.07028.x. Provides evidence that four T6S proteins assemble a membrane-associated complex. [DOI] [PubMed] [Google Scholar]
- 6.Aschtgen MS, Thomas MS, Cascales E. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence. 2010;1:535–40. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 6a.Aschtgen MS, Zoued A, Lloubes R, Journet L, Cascales E. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 type VI secretion system, is inserted by YidC. Microbiology Open. 2012;1:71–82. doi: 10.1002/mbo3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aubert D, MacDonald DK, Valvano MA. BcsKC is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsLB. J Biol Chem. 2010;285:35988–98. doi: 10.1074/jbc.M110.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA. 2008;105:3733–38. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–6. doi: 10.1038/nature10846. Describes a dynamic tubular structure associated with the T6S apparatus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. Regulation of type VI secretion gene clusters by σ54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–67. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–60. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–25. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76:815–21. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 14.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–32. doi: 10.1111/j.1365-2958.2009.06670.x. Shows that T6S in P. aeruginosa is directly regulated by the Gac/Rsm pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–45. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Bröms JE, Lavander M, Sjöstedt A. A conserved α-helix essential for type VI secretion-like system on Francisella tularensis. J Bact. 2009;8:2431–2446. doi: 10.1128/JB.01759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. An epigenetic switch involving overlapping Fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 2011;7:e1002205. doi: 10.1371/journal.pgen.1002205. Demonstrates direct regulation of T6S by Fur and DNA methylation in EAEC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calderwood SB, Mekalanos JJ. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–64. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campoy S, Jara M, Busquets N, de Rozas AM, Badiola I, Barbe J. Intracellular cyclic AMP concentration is decreased in Salmonella typhimurium fur mutants. Microbiology. 2002;148:1039–48. doi: 10.1099/00221287-148-4-1039. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77:2590–601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9:735–41. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:1102–11. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty S, Sivaraman J, Leung KY, Mok YK. Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem. 2011;286:39417–30. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–25. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 25.Cornelis GR, Agrain C, Sorg I. Length control of extended protein structures in bacteria and bacteriophages. Curr Opin Microbiol. 2006;9:201–6. doi: 10.1016/j.mib.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 27.Cotter P. Microbiology: Molecular syringes scratch the surface. Nature. 2011;475:301–3. doi: 10.1038/475301a. [DOI] [PubMed] [Google Scholar]
- 28.Das S, Chakrabortty A, Banerjee R, Chaudhuri K. Involvement of in vivo induced icmF gene of Vibrio cholerae in motility, adherence to epithelial cells, and conjugation frequency. Biochem Biophys Res Commun. 2002;295:922–28. doi: 10.1016/s0006-291x(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 29.de Pace F, Boldrin de Paiva J, Nakazato G, Lancellotti M, Sircili MP, et al. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology. 2011;157:2954–62. doi: 10.1099/mic.0.050005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubuis C, Haas D. Cross-species GacA-controlled induction of antibiosis in pseudomonads. Appl Environ Microbiol. 2007;73:650–54. doi: 10.1128/AEM.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 31a.Durand E, Zoued A, Spinelli S, Watson PJ, Aschtgen MS, Journet L, Cambillau C, Cascales E. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J Biol Chem. 287:14157–68. doi: 10.1074/jbc.M111.338731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellermeier JR, Slauch JM. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol. 2008;190:476–86. doi: 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol. 2005;55:1160–82. doi: 10.1111/j.1365-2958.2004.04453.x. [DOI] [PubMed] [Google Scholar]
- 34.Federle MJ, Bassler BL. Interspecies communication in bacteria. J Clin Invest. 2003;112:1291–99. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, et al. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng J, Atkinson MR, McCleary W, Stock JB, Wanner BL, Ninfa AJ. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J Bacteriol. 1992;174:6061–70. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–14. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–68. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 39.Gode-Potratz CJ, McCarter LL. Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol. 2011;193:4224–37. doi: 10.1128/JB.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33:279–94. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 41.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–54. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, et al. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286:12317–27. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall HK, Foster JW. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol. 1996;178:5683–91. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrington SM, Dudley EG, Nataro JP. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett. 2006;254:12–18. doi: 10.1111/j.1574-6968.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 45.Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- 46.Hassett DJ, Sokol PA, Howell ML, Ma JF, Schweizer HT, et al. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 48.Hense BA, Kuttler C, Muller J, Rothballer M, Hartmann A, Kreft JU. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5:230–39. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 49.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. Reports the finding that the T6SS can target bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu F, Schwarz S, Mougous JD. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:1111–25. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikeda JS, Janakiraman A, Kehres DG, Maguire ME, Slauch JM. Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J Bacteriol. 2005;187:912–22. doi: 10.1128/JB.187.3.912-922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. Demonstrates V. cholerae O1 T6S is negatively regulated by quorum sensing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–84. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janakiraman A, Slauch JM. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol Microbiol. 2000;35:1146–55. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 55.Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jobichen C, Chakraborty S, Li M, Zheng J, Joseph L, et al. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One. 2010;5:e12910. doi: 10.1371/journal.pone.0012910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanamaru S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc Natl Acad Sci USA. 2009;106:4067–68. doi: 10.1073/pnas.0901205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–57. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 59.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–98. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 60.Khajanchi BK, Sha J, Kozlova EV, Erova TE, Suarez G, et al. N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology. 2009;155:3518–31. doi: 10.1099/mic.0.031575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971;58:693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- 62.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–82. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–53. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 64.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–59. doi: 10.1073/pnas.0813360106. Together with Reference 83, this study illustrates the structural conservation between T6SS and phage-tail components. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009;155:2845–55. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leung KY, Siame BA, Snowball H, Mok YK. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol. 2011;14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 67.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008;4:e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lossi NS, Dajani R, Freemont P, Filloux A. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology. 2011;157:3292–305. doi: 10.1099/mic.0.051987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–43. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma LS, Lin JS, Lai EM. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker A motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 2009;191:4316–29. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70a.Ma LS, Narberhaus F, Lai EM. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 2012;287:15610–21. doi: 10.1074/jbc.M111.301630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107:19520–24. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCleary WR, Stock JB, Ninfa AJ. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–98. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meezan E, Wood WB. The sequence of gene product interaction in bacteriophage T4 tail core assembly. J Mol Biol. 1971;58:685–92. doi: 10.1016/0022-2836(71)90033-7. [DOI] [PubMed] [Google Scholar]
- 74.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79:2941–49. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore MM, Fernandez DL, Thune RL. Cloning and characterization of Edwardsiella ictaluri proteins expressed and recognized by the channel catfish Ictalurus punctatus immune response during infection. Dis Aquat Organ. 2002;52:93–107. doi: 10.3354/dao052093. [DOI] [PubMed] [Google Scholar]
- 76.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. Identifies T6S proteins that posttranslationally regulate the T6SS. [DOI] [PubMed] [Google Scholar]
- 78.Nagai H, Kubori T. Type IVB secretion systems of Legionella and other gram-negative bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Osipiuk J, Xu X, Cui H, Savchenko A, Edwards A, Joachimiak A. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J Struct Funct Genomics. 2011;12:21–26. doi: 10.1007/s10969-011-9107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–65. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–13. doi: 10.1073/pnas.0706532104. Describes the activity of a eukaryotic cell-targeting T6S effector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rao VA, Shepherd SM, English G, Coulthurst SJ, Hunter WN. The structure of Serratia marcescens Lip, a membrane-bound component of the type VI secretion system. Acta Crystallogr D. 2011;67:1065–72. doi: 10.1107/S0907444911046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rappas M, Bose D, Zhang X. Bacterial enhancer-binding proteins: unlocking σ54-dependent gene transcription. Curr Opin Struct Biol. 2007;17:110–16. doi: 10.1016/j.sbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 88.Rashid RA, Tarr PI, Moseley SL. Expression of the Escherichia coli IrgA homolog adhesin is regulated by the ferric uptake regulation protein. Microb Pathog. 2006;41:207–17. doi: 10.1016/j.micpath.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 2011;24:751–57. doi: 10.1094/MPMI-11-10-0262. [DOI] [PubMed] [Google Scholar]
- 90.Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol. 2010;192:3584–96. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rogge ML, Thune RL. Regulation of the Edwardsiella ictaluri type III secretion system by pH and phosphate concentration through EsrA, EsrB, and EsrC. Appl Environ Microbiol. 2011;77:4293–302. doi: 10.1128/AEM.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–80. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 93.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–47. doi: 10.1038/nature10244. Describes the activity of cell wall–targeting antibacterial T6S effectors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–54. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schlenker C, Surawicz CM. Emerging infections of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2009;23:89–99. doi: 10.1016/j.bpg.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 96.Schumacher J, Joly N, Rappas M, Zhang X, Buck M. Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation. J Struct Biol. 2006;156:190–99. doi: 10.1016/j.jsb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 97.Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–37. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6(8):e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shingler V. Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–16. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 100.Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–90. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein VgrG1 from Aeromonas hydrophila that induces host cell toxicity by ADP-ribosylation of actin. J Bacteriol. 2010;192:155–68. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor DN, Bourgeois AL, Ericsson CD, Steffen R, Jiang ZD, et al. A randomized, double-blind, multicenter study of rifaximin compared with placebo and with ciprofloxacin in the treatment of travelers’ diarrhea. Am J Trop Med Hyg. 2006;74:1060–66. [PubMed] [Google Scholar]
- 103.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA. 2006;103:171–76. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waksman G, Fronzes R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem Sci. 2010;35:691–98. doi: 10.1016/j.tibs.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 105.Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–87. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 106.Wang X, Wang Q, Xiao J, Liu Q, Wu H, et al. Edwardsiella tarda T6SS component evpP is regulated by esrB and iron, and plays essential roles in the invasion of fish. Fish Shellfish Immunol. 2009;27:469–77. doi: 10.1016/j.fsi.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 107.Weber B, Hasic M, Chen C, Wai SN, Milton DL. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. 2009;11:3018–28. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 108.Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:8919–23. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Worrall LJ, Lameignere E, Strynadka NC. Structural overview of the bacterial injectisome. Curr Opin Microbiol. 2011;14:3–8. doi: 10.1016/j.mib.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 110.Xu H, Hoover TR. Transcriptional regulation at a distance in bacteria. Curr Opin Microbiol. 2001;4:138–44. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- 111.Yahr TL, Greenberg EP. The genetic basis for the commitment to chronic versus acute infection in Pseudomonas aeruginosa. Mol Cell. 2004;16:497–98. doi: 10.1016/j.molcel.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 112.Yap ML, Mio K, Ali S, Minton A, Kanamaru S, Arisaka F. Sequential assembly of the wedge of the baseplate of phage T4 in the presence and absence of gp11 as monitored by analytical ultracentrifugation. Macromol Biosci. 2010;10:808–13. doi: 10.1002/mabi.201000042. [DOI] [PubMed] [Google Scholar]
- 113.Zhang L, Hinz AJ, Nadeau JP, Mah TF. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J Bacteriol. 2011;193:5510–13. doi: 10.1128/JB.00268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang W, Xu S, Li J, Shen X, Wang Y, Yuan Z. Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch Microbiol. 2011;193:351–63. doi: 10.1007/s00203-011-0680-2. [DOI] [PubMed] [Google Scholar]
- 115.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and antibacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 117.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 2010;107:21128–33. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]