Abstract
Exosomes, microvesicles of endocytic origin released by normal and tumor cells, play an important role in cell-to-cell communication. Angiogenesis has been shown to regulate progression of chronic myeloid leukemia (CML). The mechanism through which this happens has not been elucidated. We isolated and characterized exosomes from K562 CML cells and evaluated their effects on human umbilical endothelial cells (HUVECs). Fluorescent-labeled exosomes were internalized by HUVECs during tubular differentiation on Matrigel. Exosome localization was perinuclear early in differentiation, moving peripherally in cells undergoing elongation and connection. Exosomes move within and between nanotubular structures connecting the remodeling endothelial cells. They stimulated angiotube formation over a serum/growth factor-limited medium control, doubling total cumulative tube length (P = 0.003). Treatment of K562 cells with two clinically active tyrosine kinase inhibitors, imatinib and dasatinib, reduced their total exosome release (P <0.009); equivalent concentrations of drug-treated exosomes induced a similar extent of tubular differentiation. However, dasatinib treatment of HUVECs markedly inhibited HUVEC response to drug control CML exosomes (P <0.002). In an in vivo mouse Matrigel plug model angiogenesis was induced by K562 exosomes and abrogated by oral dasatinib treatment (P <0.01). K562 exosomes induced dasatinib-sensitive Src phosphorylation and activation of downstream Src pathway proteins in HUVECs. Imatinib was minimally active against exosome stimulation of HUVEC cell differentiation and signaling. Thus, CML cell-derived exosomes induce angiogenic activity in HUVEC cells. The inhibitory effect of dasatinib on exosome production and vascular differentiation and signaling reveals a key role for Src in both the leukemia and its microenvironment.
Keywords: Exosomes, Nanotubes, Chronic myeloid leukemia, Endothelial cells, Tyrosine kinase inhibitors
Introduction
Angiogenesis, the formation of new blood vessels from an existing vasculature, has been associated with growth and dissemination of solid tumors [1]. The role that angiogenesis plays in the pathogenesis and progression of hematologic neoplasias is now emerging [2]. There is a significant increase in the number of vessels in the bone marrow of patients with chronic myeloid leukemia (CML) [2, 3]. This is associated with an increased circulation of angiogenic factors such as vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF/ FGF2) and hepatocyte growth factor (HGF) [2, 4].
CML is an uncontrolled proliferation of hematopoietic progenitors containing a characteristic (9, 22) translocation resulting in the fusion of the Abelson (ABL) oncogene to the breakpoint cluster region (BCR) gene, known as the Philadelphia chromosome [5–7]. This oncogene encodes the constitutively activated bcr-abl tyrosine kinase that is involved in the pathogenesis of CML [8]. The introduction of the bcr-abl kinase inhibitor, imatinib mesylate, into the treatment of CML significantly improved patient survival [9]. However, some patients develop resistance to imatinib due to secondary point mutations in the bcr-abl tyrosine kinase domain or due to BCR-ABL gene amplification [10, 11]. This led to the development of new agents such as dasatinib, a bcr-abl and Src family kinase (SFK) inhibitor. Dasatinib is effective on all clinically demonstrated bcr-abl mutants identified to date, except T315I [12, 13]. A better understanding of the mechanisms involved in the disease progression may provide information to develop new treatment strategies. It has been proposed that the interaction between leukemic cells and the bone marrow microenvironment may play an important role in CML pathogenesis [14].
Exosomes are small vesicles, 40–100 nm in diameter, that participate in cell-to-cell communication [15]. They are released by cells in response to a broad spectrum of physiologic and pathologic conditions. Exosomes originate from the inward budding of the endosomal membrane, forming multivesicular bodies (MVBs). They are released into the extracellular space by fusion of the MVBs with the plasma membrane [15]. The biological role of exosomes depends on the cell of origin and recipient cell. Exosomes secreted by platelets contain tissue factor, involved in coagulation events [16]. Dendritic cells release exosomes containing major histocompatibility complex molecules able to activate T cells [17]. The influence of exosomes on cell polarity and developmental tissue patterning has been suggested to be related to their transport of morphogens and RNA [18]. Several roles of exosomes released by tumor cells to modulate the tumor microenvironment have been described. TGFβ expressed on the surface of exosomes released by different cancer cell lines is able to induce the differentiation of fibroblasts into myofibroblasts [19]. Ovarian cancer cell exosomes stimulate expansion of T regulatory cells that, in turn, suppress T cell activation [20]. Glioblastoma cell exosomes interact with endothelial cells stimulating their proliferation [21], and exosomes released by a pancreatic cell line transfected with D6.1A tetraspanin stimulate endothelial tubulogenesis [22]. The release of exosomes from K562 CML cells has been reported; however, the role of these exosomes in the modulation of the tumor bone marrow microenvironment has not been elucidated [23].
Here, we report that exosomes released by K562 CML cells are internalized by endothelial cells during tubular differentiation on Matrigel and are transferred to neighboring cells through the formation of nanotubular structures connecting the cells. Further, we show that these exosomes stimulate tube formation in endothelial cells through Src activation. While both imatinib and dasatinib reduced exosome release from K562 cells, only dasatinib blocked exosome effect on endothelial cells. These findings shed new light on the interaction of CML with and creation of a supportive bone marrow microenvironment.
Materials and methods
Cell cultures and reagents
The human K562 leukemia cell line was provided by Dr R. Childs (NHLBI, NIH, Bethesda, MD) and was cultured in DMEM with 10% FBS (Gemini Bio-Products, Sacramento, CA). Human umbilical vein endothelial cells (HUVEC) were purchased from Invitrogen (Carlsbad, CA) and were cultured in Medium 200 supplemented with 2% low serum growth supplement (LSGS), with penicillin and streptomycin. CCD-27Sk fibroblasts were purchased from American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% FBS. Imatinib mesylate (IM) was provided by Dr. R. Bertieri (Novartis, Milan, Italy). Dasatinib was purchased from Selleck Chemicals LLC (Houston, TX). Antibodies were obtained as follows: anti-CD63, Abcam (Cambridge, MA); anti-CD81 and anti-Tsg101, Santa Cruz (Santa Cruz, CA); monoclonal anti-α-tubulin, Sigma-Aldrich (Allentown, PA); anti-Src, -Erk, -Akt, -FAK, -phospho-416Y-Src, -phospho-202T/204Y-Erk, -phospho-308T-Akt, Cell Signaling Technology (Beverly, MA); anti-phospho-861Y-FAK, Millipore (Billerica, MA).
Isolation of exosomes
Exosome-free medium (EFM) was obtained by ultracentrifugation of DMEM containing 20% FBS at 100,000×g for 16 h at 4°C. Ultracentrifuged medium was then diluted 1:1 with serum-free DMEM for use. K562 cells were cultured for 24 h in EFM and the conditioned medium (CM) from 12 × 107 cells (160 ml) collected for exosome purification. CM was centrifuged progressively at 300×g for 10 min, 2000×g for 10 min, and then placed through a 0.22 lmfilter sterilization device. Effluent was ultracentrifuged at 100,000×g for 2 h in a fixed angle rotor. The exosome pellet was washed in PBS, ultracentrifuged, then exosome protein content measured. All centrifugation steps were performed at 4°C.
Electron microscopy
An aliquot of exosome suspension was loaded into a carbon-coated electron microscopy grid. The sample was fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. After two washes in distilled H2O, the sample was stained with 2% methylamine tungstate for 45 s and air-dried. EM samples were observed in a Zeiss transmission electron microscopy (TEM) 912.
Immunoblot analysis
Cells were pelleted after PBS washes; the pellet was lysed in modified RIPA buffer with phosphatase and protease inhibitors, as reported [24]. The lysate was clarified by centrifugation, and immunoblots were executed using standard techniques [24].
Endothelial tube formation assay
Subconfluent HUVECs were harvested and resuspended in limiting medium (Medium 200 with 0.2% LSGS) and treated with the indicated concentration of exosomes. This suspension was seeded (70,000 cells/well) in growth factor-reduced Matrigel-coated 24 well plate (BD Bioscience, San Jose, CA) and incubated up to 6 h at 37°C. Tube formation was examined under an inverted microscope and photographed at 40× magnification. Cumulative tube length was measured using the NIS elements software (Nikon Instruments Inc., Melville, NY). Results are the mean and SEM of triplicate experiments.
Labeling and internalization of exosomes
Exosomes fromK562 cells were labeled using PKH26 (red) or PKH67 (green) membrane-binding fluorescent labels according to manufacturer’s recommendations (Sigma-Aldrich, Allentown, PA). The exosome suspension was filtered with a 100 kDa MW cut-off Amicon Ultra Concentrator and the flow-through was used as the unbound dye control. HUVECs seeded on Matrigel-coated chamber slides (Thermo Scientific Inc., Rochester, NY) were incubated at 37°C with labeled exosomes at a concentration of 1 µg exosomes/10,000 cells or as described. Uptake was stopped by washing and fixation in 4% paraformaldehyde for 10 min. Where indicated, two labeled populations of HUVECs were generated after incubation of the cells in standard culture monolayer for 3 h with PKH26- or PKH67-labeled exosomes. Harvested labeled cells were mixed 1:1 and seeded on Matrigel-coated chamber slides for 4 h.
Immunofluorescence
Cells were prepared for fluorescence microscopy by permeabilization for 3 min with 0.1% Triton-X100, blocked with 5% BSA and incubated with phalloidin AlexaFluor 488 (Invitrogen, Carlsbad, CA) for 20 min. Some slides were incubated with mouse monoclonal anti-α-tubulin overnight after which they were exposed for 1 h at RT to secondary anti-mouse AlexaFluor 488 (Invitrogen, Carlsbad, CA) followed by incubation with phalloidin AlexaFluor 647 (Invitrogen, Carlsbad, CA) for 20 min. Chamber slides were mounted using DAPI Vectashield medium (Vector Labs, Burlingame, CA). Confocal images were sequentially acquired with Zeiss AIM software on a Zeiss LSM 510 Confocal system (Carl Zeiss Inc, Thornwood, NY) with a Zeiss Axiovert 100 M inverted microscope and UV laser tuned to 364 nm, a 25 mW Argon visible laser tuned to 488 nm, and a 1 mW HeNe laser tuned to 543 nm. A 40× Plan-Neofluar 1.3 NA oil immersion objective was used. Emission signals after sequential excitation of DAPI, AlexaFluor 488, and AlexaFluor 568 by the 364, 488 nm, or 543 nm laser lines were collected with a BP 385–470 filter, BP 505–530 or LP 560 filter respectively, using individual photomultipliers. Z-stacks consisted of 16–54 slices at 0.5 or 1 µm intervals and these stacks were examined with Bitplane’s Imaris software (v6.0; Zurich, Switzerland) for surface rendering. In some cases, a cutting plane was used to expose internal surface or the outer surface was made semi-transparent.
XTT cell viability assay
K562 cells and HUVECs were seeded into 96-well plates in full medium with increasing concentrations of imatinib or dasatinib, and incubated as shown. XTT assay was executed according to manufacturer’s instructions (Roche Applied Science, Indianapolis, IN). Absorbance was measured at 450 nm against a reference wavelength at 650 nm.
Matrigel plug assay
All animal experiments were conducted at University of Palermo in compliance with Italian Legislation for Animal Care. Four-week-old BALB/c nude mice (Charles River Laboratories International, Wilmington, MA) were injected subcutaneously with 400 µl Matrigel containing 100 µg/ plug K562-derived exosomes or PBS as control. IL-8 50 ng/plug was used as positive control of vascular induction. At the time of plug injection, mice were treated or not with imatinib 50 mg/kg or dasatinib 20 mg/kg by oral gavage daily for 14 days. The degree of vascularization was evaluated by determination of hemoglobin content (Drabkin’s reagent kit, Sigma, St Louis, MO).
Statistical analysis
Data are expressed as mean ± SEM of at least triplicate experiments. Statistical analyses were performed using the unpaired two-tailed Student’s t-test from GraphPad Prism software (San Diego, CA). Differences were considered statistically significant at P2 <0.05.
Results
Validation of K562 exosomes
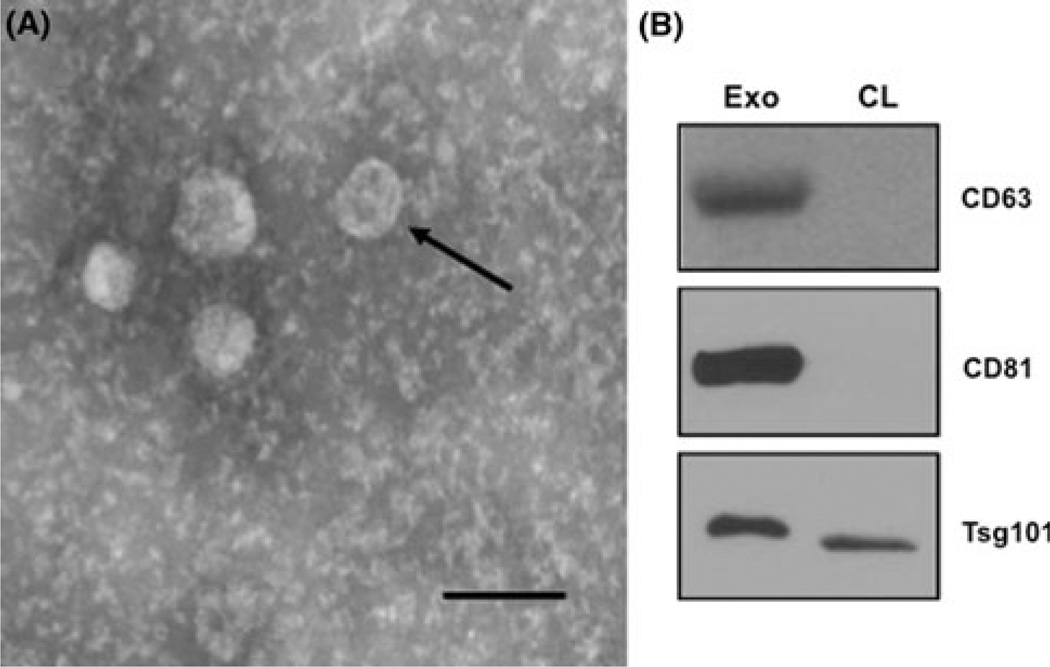
The K562 exosome purification procedure was validated in two ways: electron microscopy and selective protein expression. Figure 1a shows an example transmission electron micrograph of a representative exosome preparation. The arrow points to the characteristic cup-shaped morphology and the sizing bar indicates that the vesicles have the characteristic diameter range of 40–100 nm. CD63, CD81 and Tsg101 are known exosomal markers [15]. CD63 and CD81 were not detectable in cell lysate but were found in abundance in the exosomal preparation (Fig. 1b). Tsg101 was enriched in the exosomal compartment. This confirmed that K562 cells release true exosomal vesicles into their conditioned medium.
Fig. 1.
K562 CML cells release exosomes in their CM. a Morphology of exosomes isolated from medium conditioned by K562 cells were analyzed by transmission electron microscopy. Arrow cup-shaped exosome. Scale bar 150 nm. b Exosome characterization by immunoblot. Equal amounts of total exosomal proteins (Exo) and K562 cell lysate (CL) were analyzed for the expression of CD63, CD81, and Tsg101
Exosomes induce vascular tube formation in vitro
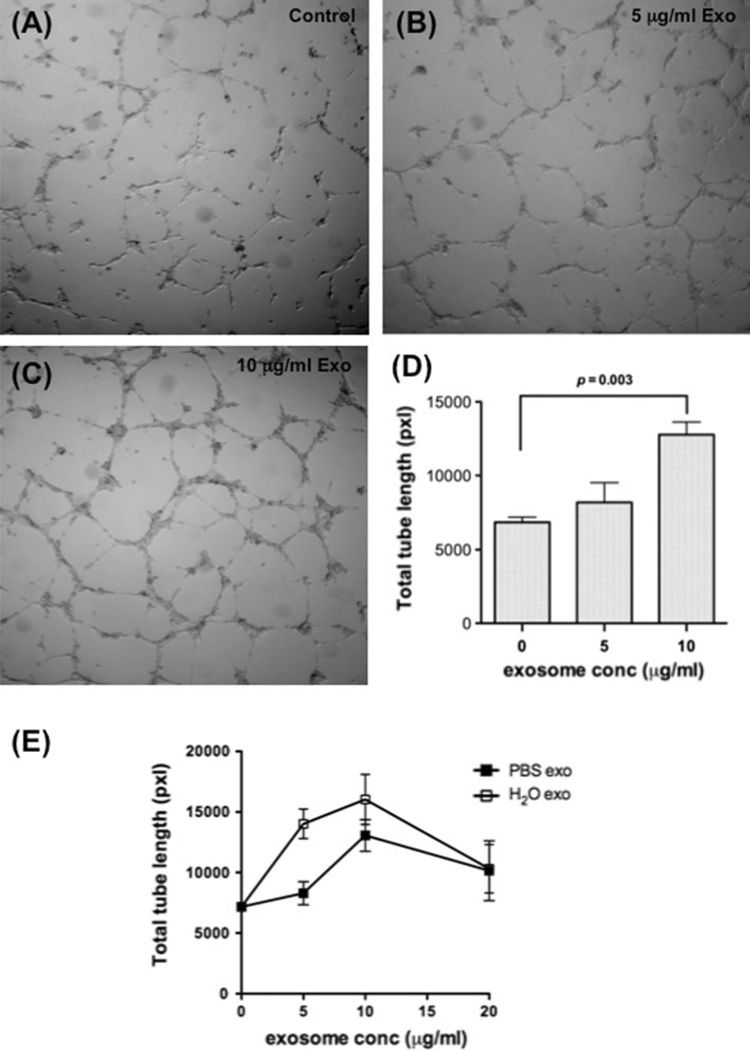
We hypothesized that exosomes from K562 CML cells would induce tubular differentiation of HUVECs in vitro. HUVECs plated on Matrigel in limiting medium with increasing concentrations of K562 exosomes formed more extensive tubes in a dose-dependent manner (Fig. 2a–c). Addition of 5 µg/ml exosomes stimulated a morphologic change in HUVECs without significantly increasing the tube network formation, where increasing the exosome dose to 10 µg/ml caused greater tube formation compared to controls (P = 0.003; Fig. 2d). Exosomes were then resuspended in water to induce hypotonic lysis and to release internal contents. HUVECs treated with lysed exosomes caused more tube formation at 5 µg/ml (P = 0.021) and 10 µg/ml compared to intact exosomes (Fig. 2e). Addition of exosomes 20 µg/ml resulted in reduced tubular network in both conditions.
Fig. 2.
Dose dependent stimulation of vascular tube formation by K562 exosomes. a–c Exosomes promote tube formation by HUVECs on Matrigel. a control; b 5 µg/ml exosomes; c 10 µg/ml exosome. d Quantitative analysis of the total tube length. Data are mean ± SEM of triplicate experiments. e Lysed K562 exosomes are equipotent or more active than intact exosomes. Data are mean ± SEM of triplicate experiments
Exosomes are internalized by endothelial cells during tube formation
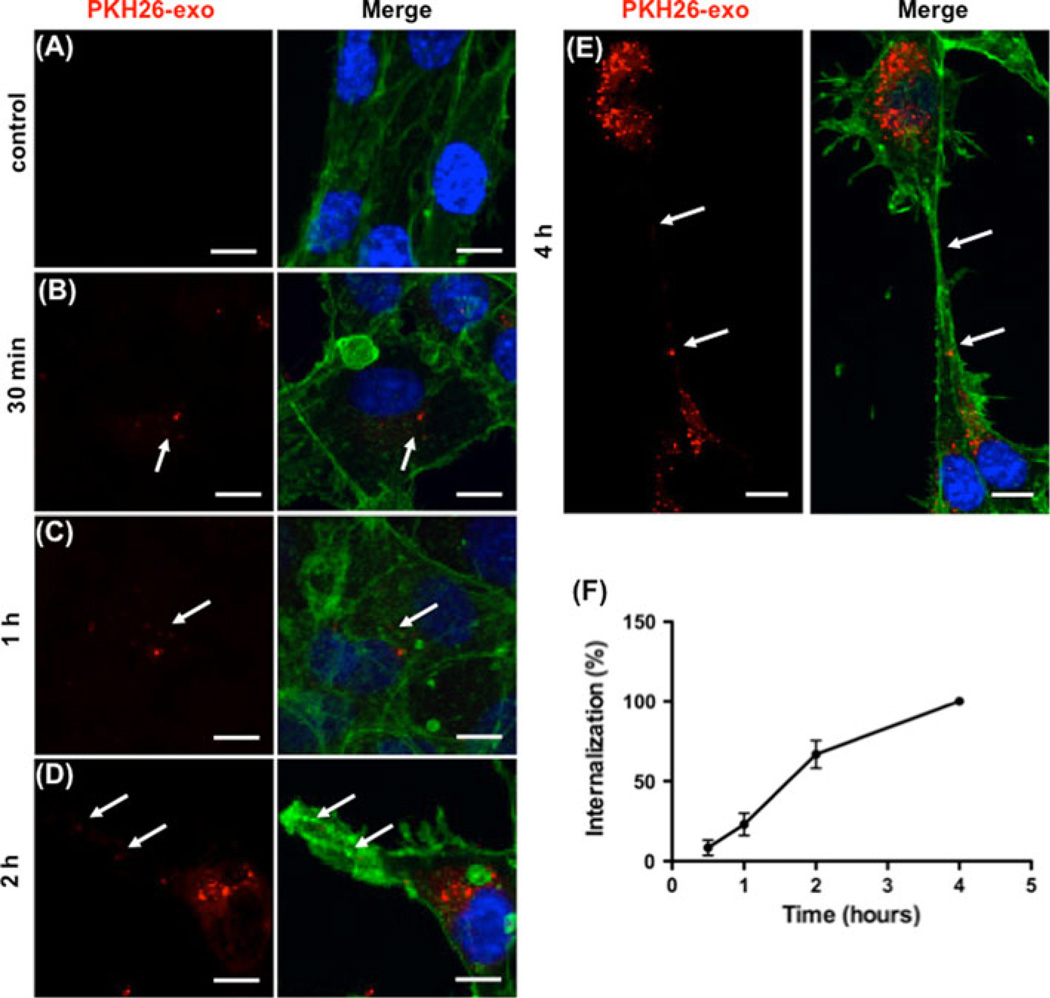
We posited that exosomes must be internalized in order to affect their function on endothelial cells. We first compared PKH26-labeled exosome internalization in HUVECs and normal human fibroblast CCD-27Sk cells. HUVECs internalized and accumulated exosomes in the perinuclear region (Online Resource 1a, b); whereas, few exosomes were taken up by CCD-27Sk cells with a more diffuse localization in the cytoplasm (Online Resource 1c, d). Thus, K562 exosomes were preferentially taken up by endothelial cells. HUVECs were next incubated on Matrigel with PKH26-labeled exosomes and uptake evaluated over time (Fig. 3). Early exosome localization was mainly perinuclear, remaining so during early tubular organization (up to 1 h, Fig. 3b, c, arrows). Localization remained perinuclear in cells that were not differentiating, with exosomes moving to the cell periphery and into extending pseudopods in those cells reorganizing and sprouting extensions. This was seen as early as 2 h (Fig. 3d, arrows). Tubes were nearly complete by 4 h at which time nanotubular structures connecting neighboring cells contained exosomes (Fig. 3e, arrows). Quantitation of fluorescence intensity was used to measure uptake of labeled exosomes and increased over time (Fig. 3f).
Fig. 3.
Time dependent uptake and localization of exosomes in HUVEC cells during differentiation. a–e Exosomes labeled with PKH26 (red) were added to HUVEC cells at time of plating and incubated as indicated. Effluent from a filtered suspension of PKH26- labeled exosomes was incubated with the HUVECs for 4 h as the control. Cells were fixed and stained for actin (green), and nuclei (DAPI, blue). Arrows indicate exosome localization. Scale bars 10 µm. f Quantitation of HUVEC uptake of PKH26-labeled K562 exosomes. The data represent the mean ± SEM of three independent experiments
Exosomes localize inside nanotubes
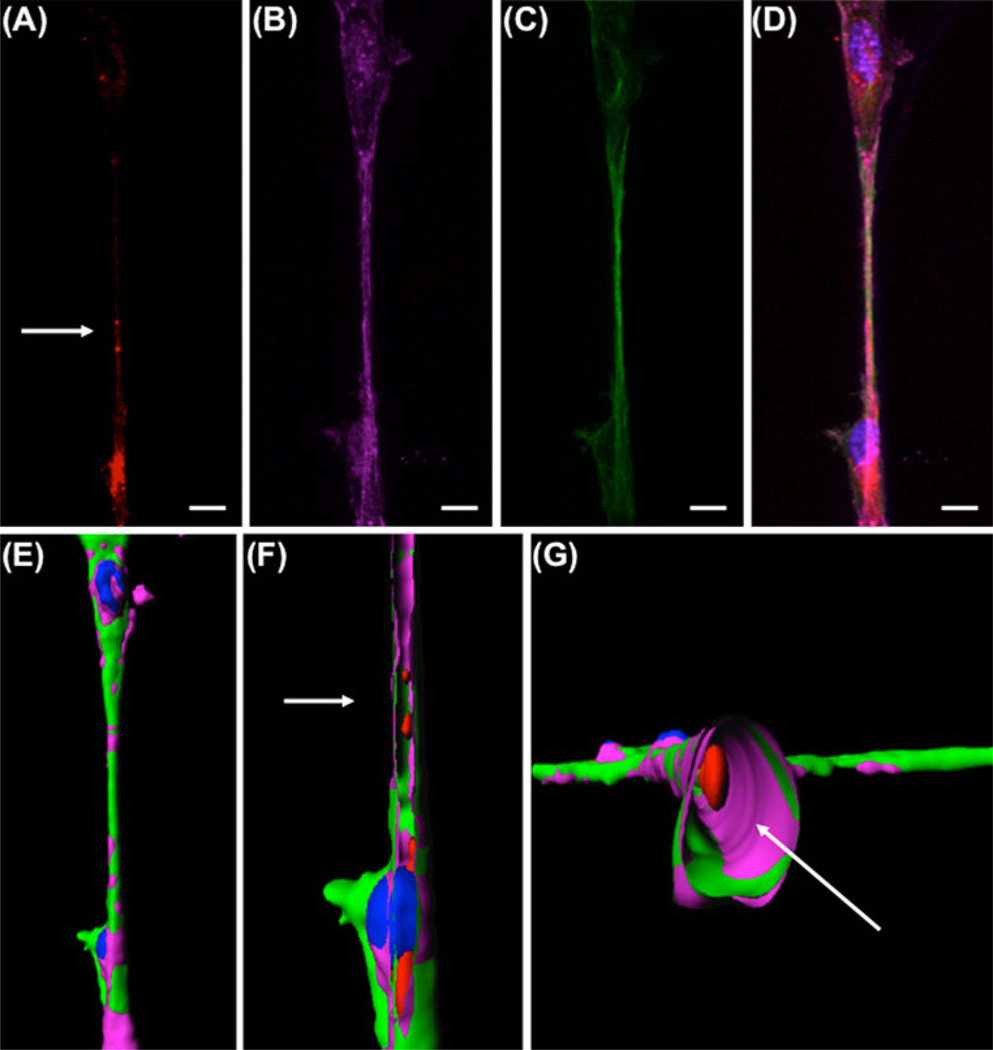
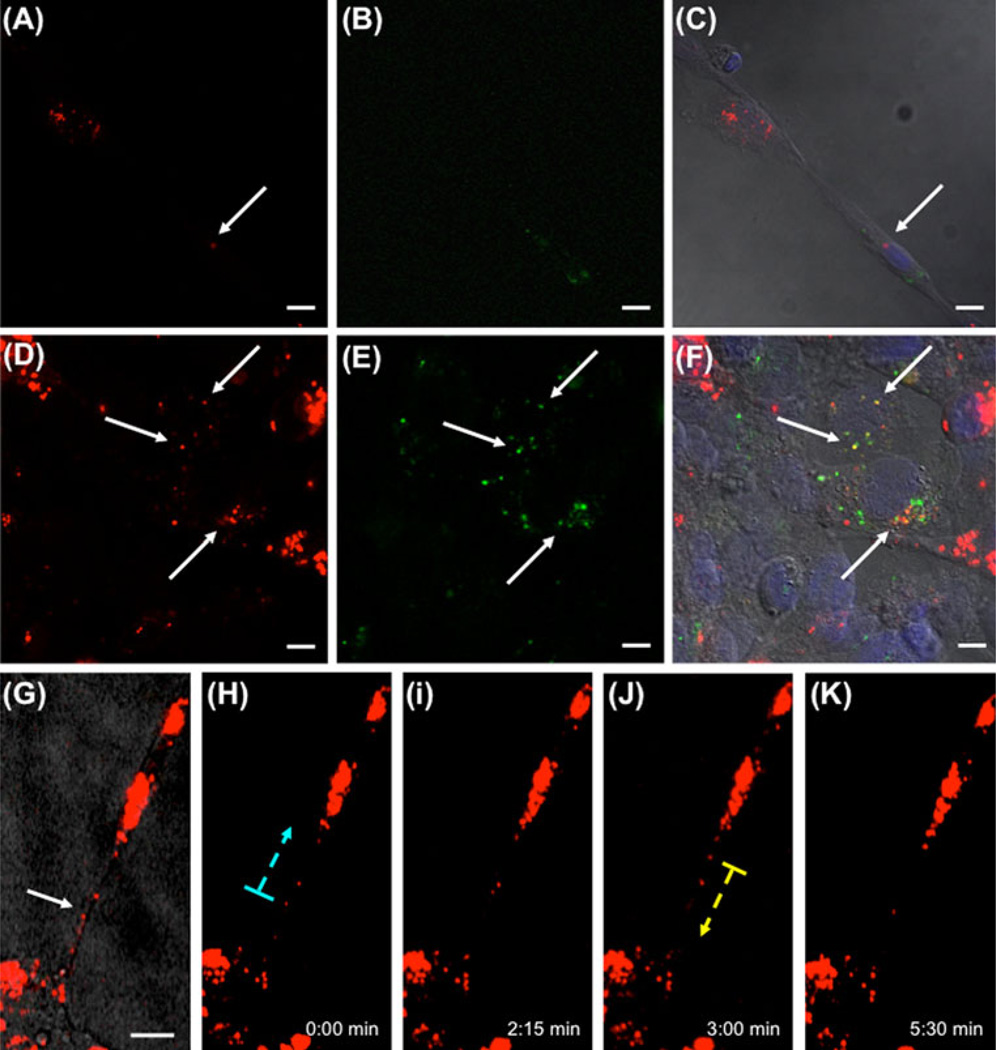
Large nanotubes are characterized as containing both actin and tubulin [25]. We demonstrated HUVECs formed large nanotubes during tubular differentiation, and that these nanotubes contained internalized exosomes (Fig. 4a–d, arrow). Three-dimensional reconstruction of confocal images using surface rendering allowed demonstration of exosomes within the nanotubes (Fig. 4e–g). Longitudinal sections (Fig. 4f) and cross sections (Fig. 4g) of the 3D image confirmed that exosomes were transported inside nanotubes. We next asked if exosomes were transferred between cells. This was accomplished by mixing HUVEC suspensions preincubated with either PKH26-stained red fluorescent or PKH67-stained green fluorescent exosomes. Four hours after plating on Matrigel, cells within the tubular network contained both red and green exosomes (arrows, Fig. 5a–f). This indicates a cell–cell exosome transfer occurred. This was further demonstrated using time-lapse video microscopy. Exosomes in the perinuclear area were mobile, but remained locally stationary within the cell; whereas, exosomes inside the nanotubes moved along the nanotube between cells (Fig. 5g–k, Online Resource 2/3). These experiments demonstrate that HUVECs internalize exosomes and transfer them to neighboring cells through extended large nanotube structures.
Fig. 4.
Exosomes localize inside nanotubes. a–d HUVEC cells incubated with PKH26-labeled exosomes (a, red) on Matrigel reorganized and sent out pseudopods that extended into nanotubes containing both F-actin (b, pink) and microtubules (c, green). Nuclei were stained with DAPI (blue). The merged image (d) shows the localization of exosomes at level of the nanotube. Scale bars 10 µm. e 3D reconstruction of the nanotube by surface rendering, showing outer view. f–g Sections of the nanotube showing exosome localization inside the nanotubes in longitudinal section (f) and cross section (g)
Fig. 5.
Exosomes are transferred cell-to-cell. a–f HUVECs carrying PKH26-stained exosomes (a/d, red) were mixed with HUVECs carrying PKH67-stained exosomes (b/e green) and plated on Matrigel for 4 h. After the incubation, cells were fixed and DAPI stained for nuclei (blue). Tube forming cells show the presence of both green and red exosomes inside the same cell (c/f, arrow). The arrows in figures d–f point to red exosomes co-localizing with green exosomes. g–k Exosomes move along nanotubes. HUVECs were incubated with PKH26-labelled exosomes and analyzed by fluorescent time-lapse video microscopy. g Brightfield view of HUVECs connected by nanotubes carrying exosomes (arrows). h–k Selected frames of a video sequence showing transfer of PKH26-labeled exosomes (dotted arrows indicate direction of movement). Scale bars: 10 µm
CML therapy modulates exosome secretion and function
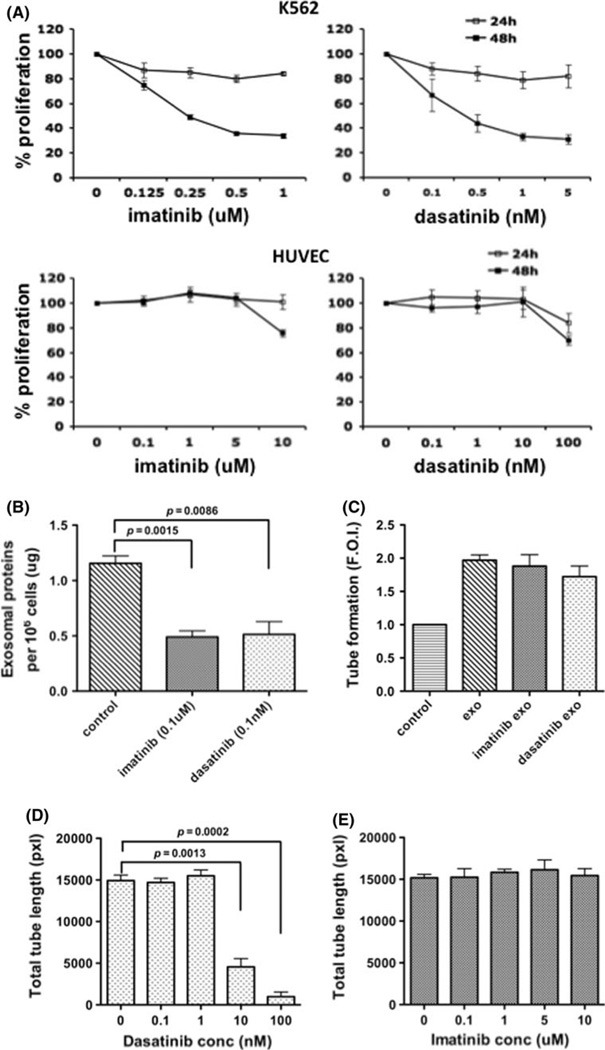
Non-toxic dose and time of exposure to imatinib and dasatinib were determined for K562 and HUVEC cells (Fig. 6a). Imatinib (0.1 µM) and dasatinib (0.1 nM) were selected for use to minimize the presence of apoptotic bodies in exosome-CM preparations. Drug treatment at these concentrations reduced released exosome protein content by 58 and 56% in the imatinib- and dasatinibtreated cells, respectively (Fig. 6b). Exosomes from both control cells and drug-treated cells worked equally well on HUVECs, yielding a 2-fold increase in total tube length (Fig. 6c). However, addition of dasatinib directly to the HUVECs inhibited tube formation in response to exosomes from untreated cells (Fig. 6d). In contrast, no reduction in exosome-stimulated tube formation was seen in presence of imatinib at all concentrations (Fig. 6e).
Fig. 6.
CML therapy regulates exosome release and exosomeinduced tube formation. a Effects of imatinib and dasatinib on K562 and HUVEC proliferation. Imatinib and dasatinib exposure continued for 24 or 48 h. b Both imatinib and dasatinib reduce total released exosomal protein. c Exosomes from imatinib- or dasatinib-treated K562 cells (0.1 µM, and 0.1 nM, respectively) are equipotent as those from vehicle-treated cells. d, e A dose-dependent inhibition by dasatinib (d), but not imatinib (e) on the HUVEC response to control exosomes. Data are mean ± SEM of triplicate experiments
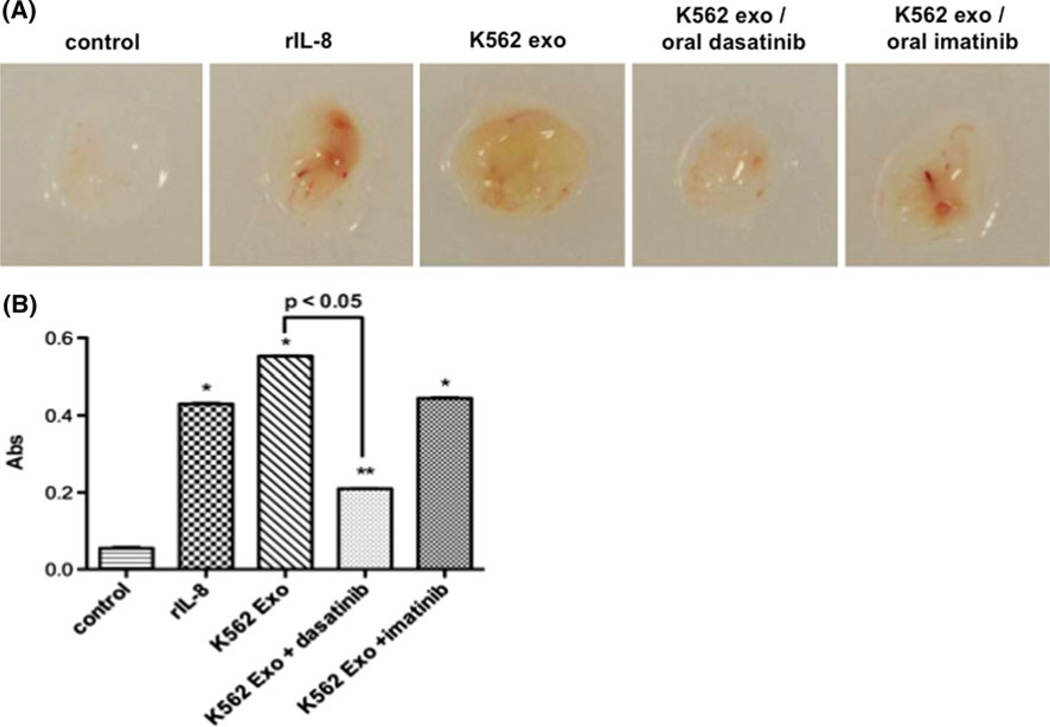
The in vivo Matrigel plug assay allowed demonstration that K562 exosomes stimulated angiogenesis that was at least equal to that caused by the positive IL-8 control (Fig. 7a, b). Daily oral dasatinib administration for 14 days starting at the time of Matrigel injection abrogated plug vascularization (P <0.01) consistent with our in vitro findings. As with the lack of imatinib inhibition of tubulogenesis in vitro or inhibition of key signaling events, oral imatinib treatment had no inhibitory effect on vascular ingrowth into the Matrigel plugs (Fig. 7).
Fig. 7.
K562 exosomes induce vascularization of Matrigel plugs in vivo. a Nude mice were injected with Matrigel plugs containing PBS or recombinant IL-8 (positive control), or 100 µg/plug K562 exosomes. Mice were treated with oral administration of dasatinib or imatinib as described in Materials and Methods, and analyzed for plug vascularization after 14 days of treatment. b Plug hemoglobin concentration as a measure of vascularity. Comparison of IL-8 or K562 exosome-containing plugs vs control: * P ≤ 0.01, ** P <0.05
Exosomes stimulate HUVECs through the activation of Src signaling
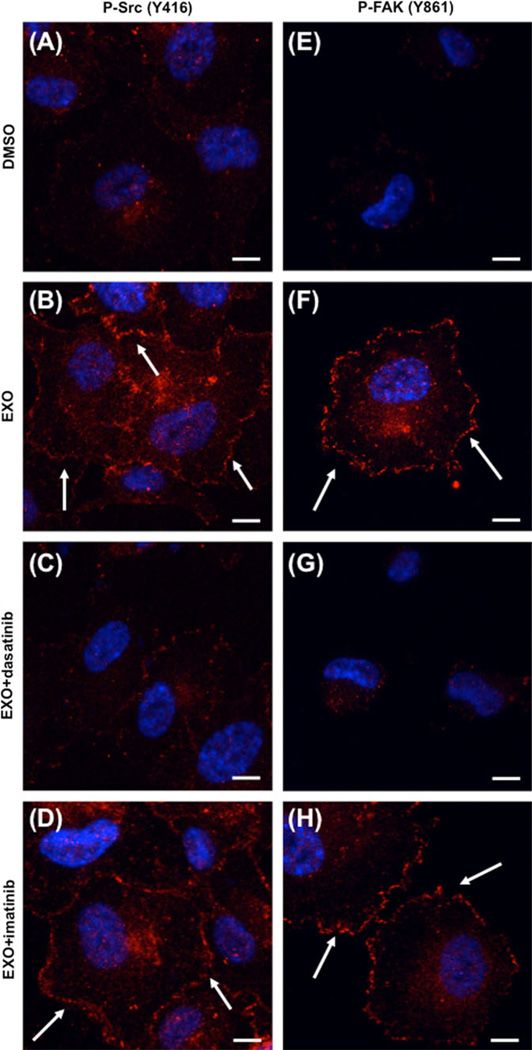
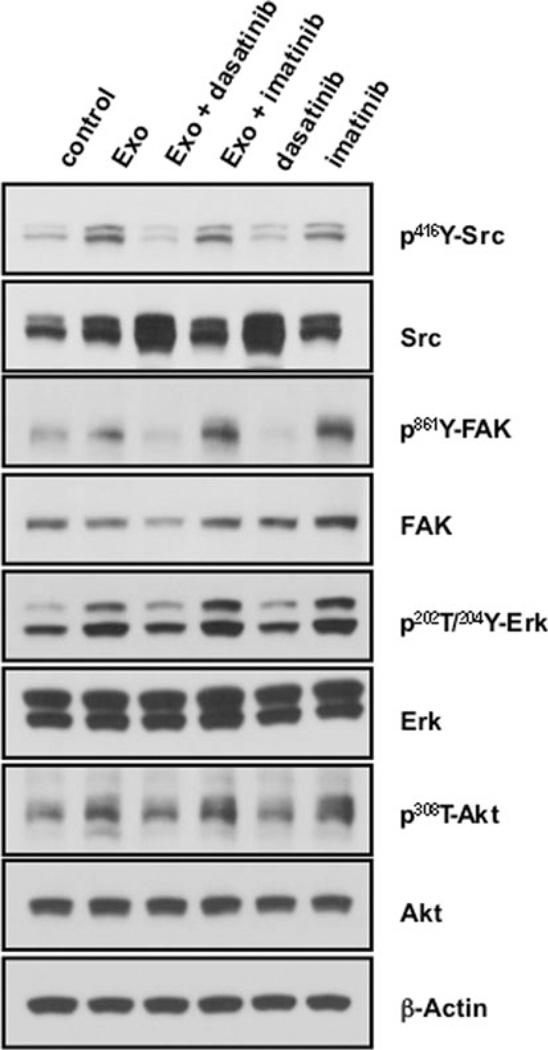
We evaluated potential signaling mechanisms through which dasatinib may differentially affect tube formation. Phosphorylation of 416Y-Src in its kinase domain is required for catalytic activity [26]. Figure 8 shows that HUVECs treated with 10 µg/ml control exosomes showed greater phosphorylated 416Y-Src compared to the untreated (DMSO) control, with increased p-Src localization at points of membrane contact (arrows). Dasatinib blocked Src phosphorylation in exosome-stimulated cells with loss of total and membrane p-Src staining. Imatinib did not alter exosome signaling. Focal adhesion kinase (FAK) is a known substrate of Src and is phosphorylated at Tyr861 [26]. Exosome exposure induced p-861Y-FAK and increased focal adhesions (arrows). FAK activation and focal adhesion staining was markedly attenuated by dasatinib treatment, while there was again little effect of imatinib. These data were confirmed by immunoblot analysis of monolayer HUVECs incubated with K562 exosomes (Fig. 9). The effects were broader, with dasatinib inhibition of exosome-stimulated downstream pathway activation. These findings place exosome effects and exosome trafficking upstream of Src signaling in exosome-induced HUVEC tubular differentiation.
Fig. 8.
Dasatinib-sensitive exosome activation of Src signaling during HUVEC differentiation. HUVECs with 10 µg/ml control exosomes were plated on Matrigel in presence of DMSO vehicle control (a, e), dasatinib (10 nM), or imatinib (1 µM), and allowed to adhere for 30 min. Cells were fixed and red fluorescence labeling was performed to visualize p-Src (a–d) or p-FAK (e–h). Nuclei were stained with DAPI (blue). Scale bars 10 µm
Fig. 9.
Src-dependent exosome activation of signaling pathways in HUVECs. HUVECs growing in monolayer were incubated with or without exosomes and treated with DMSO (control), dasatnib (10 nM), or imatinib (1 µM) for 2 h. Data are representative of 2 or more blots
Discussion
Angiogenesis is now recognized to be a factor in CML progression [2]. The mechanisms involved in the stimulation of bone marrow angiogenesis are currently under investigation. We hypothesized that CML-released exosomes are taken up by and shared between endothelial cells during promotion of tubular differentiation. We demonstrated vesicle release from K562 CML cells and confirmed they were exosomes by morphological analysis, dimension, and expression of exosomal markers. These exosomes trafficked within the endothelial cells, moving between cells through large nanotubular structures involved in cellto- cell connections. Exosomes showed angiogenic activity in both in vitro and in vivo Matrigel assays. This was shown to be the result of phosphorylation and activation of Src and its downstream pathways. Dasatinib, a src family kinase inhibitor, but not imatinib, blocked endothelial cell response to CML exosomes. These data indicate that exosomes have a role in the physiologic organization of endothelial cells in a Src-dependent fashion.
Neoangiogenesis is an important step in development of a tumor [27]. In this process the release of angiogenic factors by tumor cells is required to activate a quiescent vasculature [1]. The role for exosomes in the cancer angiogenic process has been developing, with emergence of exosome support of the leukemic microenvironment a newer finding, presented by our collaborators [28] and now in this report. In their recent report, Taverna et al. [28] showed exosomes from LAMA84 CML cells affect vascular remodeling in vitro through an IL-8 mediated drive of VCAM-1. The mechanisms of interaction of CML exosomes with endothelial cells has not been elucidated. Exosomes may interact with their target cells in three ways: binding to cell surface receptors, fusion with the plasma membrane, or internalization [16]. We first confirmed CML exosomes stimulated angiogenesis using the K562 CML cell line and we showed that exosome content contributed to HUVEC stimulation. We next demonstrated fluorescent CML exosomes were taken up by HUVECs in a time-dependent fashion. Exosomes trafficked differentially over time and within quiet versus differentiating cells. Internalization starts immediately with HUVEC attachment onto Matrigel and increases over time with initial perinuclear localization. They moved to the cell periphery in cells involved in tube formation. We observed the presence of exosomes within the tunneling nanotubes (TNTs) connecting neighboring HUVEC cells, suggesting that these exosomes may be those involved in intercellular signaling. Transfer of angio-regulatory information between cells via TNTs, rather than secretion of growth factors or exosomes into the local milieu, would facilitate and optimize direct intercellular communication and could be more efficient for the cell.
TNTs are thin protrusions connecting adjacent cells and are involved in cell-to-cell communication [29]. They were described in cultured pheochromocytoma PC12 cells [30], and later in macrophages and T-cells [25, 31]. TNTs create membrane continuity between connected cells and are involved in the transfer of cytoplasmic organelles and proteins [29]. Mitochondria have been shown to move through myocyte nanotubes into endothelial progenitor cells, inducing the acquisition of a myocyte-like phenotype in these cells [32]. Heterogeneity in TNT dimension and cytoskeleton composition has been observed, with both thin (actin-only) and thick (actin and microtubules) forms [25]. We showed that HUVECs form thick TNTs during Matrigel differentiation, demonstrating both F-actin and microtubules in a non-structured organization. Live-cell imaging and three-dimensional reconstruction confirmed this and allowed visualization of the internal localization and movement of exosomes within the TNT cytoskeletal scaffolding.
The ability of exosomes to interact with and stimulate endothelial cells suggests exosomes as a new target for CML therapy. Imatinib is the initial treatment of choice in CML. It blocks CML proliferation and reduces VEGF expression through inhibition of the constitutively active bcr/abl kinase [33, 34]. However, in some patients imatinib alone is insufficient to prevent disease progression [35]. The use of dasatinib, the abl and Src family kinase inhibitor, as front-line therapy has been reported to be more effective than imatinib, with a lower rate of transformation to accelerated phase and blast crisis [35]. We show here that non-toxic concentrations of both imatinib and dasatinib effectively reduced K562 cell exosome release by more than 55%. However, those exosomes released from cells treated with imatinib or dasatinib maintained their ability to stimulate tubular differentiation by HUVECs on Matrigel at equivalent doses. Dasatinib, but not imatinib, blocked both HUVEC tubular differentiation in response to exosome treatment in vitro and vascularization of exosome- containing Matrigel plugs implanted in nude mice. This finding was supported by exosome activation of Src and its downstream signaling partners in a dasatinib-sensitive fashion. Src modulates angiogenesis in many different ways. It controls gene and protein expression of angiogenic growth factors and cytokines, including VEGF and IL-8 [36–39]. IL-8, induced by LAMA84 CML cell exosomes, was shown to be necessary for exosome-induced angiogenesis by Taverna et al. [28]. Src can also elicit downstream signaling in endothelial cells through activation of FAK, as we previously reported to be necessary for adhesion and spreading on matrix, and as components of HUVEC motility and differentiation [40]. Activation of FAK has been further shown to be involved in the activation of the phosphatidylinositol 3-kinase/Akt pathway inducing cell survival [38]. Src activates Erk leading to an increased cell proliferation [38]. K562 exosomes induced phosphorylation and activation of Src with concomitant phosphorylation of FAK, Erk and Akt. This and localization to focal adhesions was abrogated by dasatinib, but not imatinib. Surprisingly, our data showed an activation of all the pathways by non-toxic treatment with imatinib. This finding confirmed our paradoxical observation that imatinib- treated HUVECs responded more rapidly to K562 exosomes, although the final total tube length remained unchanged. Thus, dasatinib regulates exosome-induced FAK, Erk and Akt activity and subsequent HUVEC tubular organization by selective inhibition of Src.
We have advanced the recent findings of Taverna et al. demonstrating a pro-angiogenic effect of CML exosomes through demonstration of specific interactions of exosomes with and within endothelial cells. Our findings provide new information on how endothelial cells communicate with each other within the tube network, and show how endothelial cells share internalized exosomes with neighboring cells. Our finding that Src is central in the CML exosome pathway during endothelial cell stimulation may explain the greater activity of dasatinib in CML therapy. Dasatinib activity may be greater due to the involvement of Src in both the leukemic cell and the angiogenic microenvironment. This credentials exosome release and uptake as a potential new therapeutic target in CML.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Program of the Center for Cancer Research, NCI; Dr. Mineo was supported by a fellowship from Italian Association for Cancer Research (AIRC). The authors thank Drs. Virador and Muller for assistance with electron microscopy, and Mr. Lim and Ms. Mannan for confocal microscopy technical assistance.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10456-011-9241-1) contains supplementary material, which is available to authorized users.
Contributor Information
Marco Mineo, Molecular Signaling Section, Medical Oncology Branch, Center for Cancer Research, National Cancer Institute, 10 Center Drive MSC 1906, Bethesda, MD 20892, USA.
Susan H. Garfield, CCR Confocal Microscopy Core Facility, Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA
Simona Taverna, Dipartimento di Biopatologia e Biotecnologie Mediche e Forensi, Sezione di Biologia e Genetica, Università di Palermo, Palermo, Italy.
Anna Flugy, Dipartimento di Biopatologia e Biotecnologie Mediche e Forensi, Sezione di Biologia e Genetica, Università di Palermo, Palermo, Italy.
Giacomo De Leo, Dipartimento di Biopatologia e Biotecnologie Mediche e Forensi, Sezione di Biologia e Genetica, Università di Palermo, Palermo, Italy.
Riccardo Alessandro, Dipartimento di Biopatologia e Biotecnologie Mediche e Forensi, Sezione di Biologia e Genetica, Università di Palermo, Palermo, Italy.
Elise C. Kohn, Molecular Signaling Section, Medical Oncology Branch, Center for Cancer Research, National Cancer Institute, 10 Center Drive MSC 1906, Bethesda, MD 20892, USA kohne@mail.nih.gov
References
- 1.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 2.Aguayo A, Kantarjian H, Manshouri T, Gidel C, Estey E, Thomas D, Koller C, Estrov Z, O’Brien S, Keating M, Freireich E, Albitar M. Angiogenesis in acute and chronic leukemias and myelodysplastic syndromes. Blood. 2000;96:2240–2245. [PubMed] [Google Scholar]
- 3.Lundberg LG, Lerner R, Sundelin P, Rogers R, Folkman J, Palmblad J. Bone marrow in polycythemia vera, chronic myelocytic leukemia, and myelofibrosis has an increased vascularity. Am J Pathol. 2000;157:15–19. doi: 10.1016/S0002-9440(10)64511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhelyazkova AG, Tonchev AB, Kolova P, Ivanova L, Gercheva L. Prognostic significance of hepatocyte growth factor and microvessel bone marrow density in patients with chronic myeloid leukaemia. Scand J Clin Lab Invest. 2008;68:492–500. doi: 10.1080/00365510701854991. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497–1499. [Google Scholar]
- 6.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 7.Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 1986;47:277–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- 8.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, Cervantes F, Hochhaus A, Powell BL, Gabrilove JL, Rousselot P, Reiffers J, Cornelissen JJ, Hughes T, Agis H, Fischer T, Verhoef G, Shepherd J, Saglio G, Gratwohl A, Nielsen JL, Radich JP, Simonsson B, Taylor K, Baccarani M, So C, Letvak L, Larson RA. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 10.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 11.O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- 12.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, Fairchild C, Hunt JT, Inigo I, Johnston K, Kamath A, Kan D, Klei H, Marathe P, Pang S, Peterson R, Pitt S, Schieven GL, Schmidt RJ, Tokarski J, Wen ML, Wityak J, Borzilleri RM. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 13.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O’Brien S, Nicaise C, Bleickardt E, Blackwood- Chirchir MA, Iyer V, Chen TT, Huang F, Decillis AP, Sawyers CL. Dasatinib in imatinib-resistant Philadelphia chromosome- positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 14.Gordon MY, Dowding CR, Riley GP, Goldman JM, Greaves MF. Altered adhesive interactions with marrow stroma of haematopoietic progenitor cells in chronic myeloid leukaemia. Nature. 1987;328:342–344. doi: 10.1038/328342a0. [DOI] [PubMed] [Google Scholar]
- 15.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 16.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 18.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 20.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5:e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gesierich S, Berezovskiy I, Ryschich E, Zoller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083–7094. doi: 10.1158/0008-5472.CAN-06-0391. [DOI] [PubMed] [Google Scholar]
- 23.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 24.Doong H, Rizzo K, Fang S, Kulpa V, Weissman AM, Kohn EC. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins. J Biol Chem. 2003;278:28490–28500. doi: 10.1074/jbc.M209682200. [DOI] [PubMed] [Google Scholar]
- 25.Onfelt B, Nedvetzki S, Benninger RK, Purbhoo MA, Sowinski S, Hume AN, Seabra MC, Neil MA, French PM, Davis DM. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol. 2006;177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 26.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 27.Folkman J, Shing Y. Angiogenesis. J Biol Chem. 1992;267:10931–10934. [PubMed] [Google Scholar]
- 28.Taverna S, Flugy A, Saieva L, Kohn EC, Santoro A, Meraviglia S, De Leo G, Alessandro R. Role of exosomes released by chronic myelogenous leukemia cells in angiogenesis. Int J Cancer. 2011 doi: 10.1002/ijc.26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerdes HH, Carvalho RN. Intercellular transfer mediated by tunneling nanotubes. Curr Opin Cell Biol. 2008;20:470–475. doi: 10.1016/j.ceb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 31.Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Kohler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 32.Koyanagi M, Brandes RP, Haendeler J, Zeiher AM, Dimmeler S. Cell-to-cell connection of endothelial progenitor cells with cardiac myocytes by nanotubes: a novel mechanism for cell fate changes? Circ Res. 2005;96:1039–1041. doi: 10.1161/01.RES.0000168650.23479.0c. [DOI] [PubMed] [Google Scholar]
- 33.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 34.Legros L, Bourcier C, Jacquel A, Mahon FX, Cassuto JP, Auberger P, Pages G. Imatinib mesylate (STI571) decreases the vascular endothelial growth factor plasma concentration in patients with chronic myeloid leukemia. Blood. 2004;104:495–501. doi: 10.1182/blood-2003-08-2695. [DOI] [PubMed] [Google Scholar]
- 35.Cortes J, Hochhaus A, Hughes T, Kantarjian H. Front-line and salvage therapies with tyrosine kinase inhibitors and other treatments in chronic myeloid leukemia. J Clin Oncol. 2011;29:524–531. doi: 10.1200/JCO.2010.31.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanda S, Miyata Y, Kanetake H, Smithgall TE. Nonreceptor protein-tyrosine kinases as molecular targets for antiangiogenic therapy (Review) Int J Mol Med. 2007;20:113–121. [PubMed] [Google Scholar]
- 37.Mukhopadhyay D, Tsiokas L, Sukhatme VP. Wild-type p53 and v-Src exert opposing influences on human vascular endothelial growth factor gene expression. Cancer Res. 1995;55:6161–6165. [PubMed] [Google Scholar]
- 38.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 39.Summy JM, Trevino JG, Lesslie DP, Baker CH, Shakespeare WC, Wang Y, Sundaramoorthi R, Metcalf CA, 3rd, Keats JA, Sawyer TK, Gallick GE. AP23846, a novel and highly potent Src family kinase inhibitor, reduces vascular endothelial growth factor and interleukin-8 expression in human solid tumor cell lines and abrogates downstream angiogenic processes. Mol Cancer Ther. 2005;4:1900–1911. doi: 10.1158/1535-7163.MCT-05-0171. [DOI] [PubMed] [Google Scholar]
- 40.Masiero L, Lapidos KA, Ambudkar I, Kohn EC. Regulation of the RhoA pathway in human endothelial cell spreading on type IV collagen: role of calcium influx. J Cell Sci. 1999;112(Pt 19):3205–3213. doi: 10.1242/jcs.112.19.3205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.