Abstract
Background
It is not known if the genes involved with endurance performance during young adulthood are also involved with changes in performance. We examined the associations of gene variants with symptom-limited exercise test duration at baseline and decrease in duration over 20 years.
Methods and Results
3,783 (1,835 Blacks 1,948 Whites) and 2,335 (1,035 Blacks 1,300 Whites) participants from CARDIA were included in the baseline and 20 year models, respectively. 217 SNPs in Blacks and 171 SNPs in Whites from 17 genes were genotyped. In Blacks, five SNPs in the ATP1A2, HIF1A, NOS3, and PPARGC1A loci tended to be associated (p<0.05) with baseline duration in a multivariate regression model. Blacks (n=99) with at least four of the most-favorable genotypes at these loci had approximately two minutes longer baseline duration than those with only two such genotypes (P<0.0001). In Whites, the HIF1A rs1957757 and PPARGC1A rs3774909 markers tended to be associated with baseline duration, but the association of a multimarker construct of the most-favorable genotypes at both SNPs with baseline duration was not statistically significant. In Whites, four SNPs in the AGT, AMPD1, ANG, and PPARGC1A loci tended to be associated with decrease in exercise duration over 20 years, and those (n=40) with all four favorable genotypes had 0.8 min less decline in duration compared to those with none or one (n=232) (P<0.0001).
Conclusion
In multimarker constructs, alleles at genes related to skeletal muscle Na+/K+ transport, hypoxia, and mitochondrial metabolism are associated with symptom-limited exercise test duration over time in adults.
Keywords: cardiorespiratory fitness, genotype, prospective study
INTRODUCTION
Low cardiorespiratory fitness (hereafter referred to as fitness) and decreases in fitness over time are associated with higher levels of risk factors for cardiovascular disease (CVD) and increased risk of mortality.1–4 Fitness is a multifactorial phenotype influenced by genetic and environmental factors. Heritability estimates for fitness-related phenotypes from twin and family studies range from 25–66%.5–7 The identification of genes and DNA sequence variants contributing to differences in fitness is a growing area of research. The seventh installment of the Human Gene Map for Performance and Health-Related Fitness Phenotypes contained 214 autosomal gene entries and quantitative trait loci, 7 loci on chromosome X, and 18 mitochondrial genes.8 However, no large longitudinal cohort study has examined the association of genetic variation with changes in fitness over time. Thus, it is not known if the genes involved with fitness during young adulthood are also involved with changes in fitness over time.
The Coronary Artery Risk Development in Young Adults (CARDIA) Study is a longitudinal study of Black and White young adults that includes measures of symptom-limited exercise test duration at three time points over 20 years of follow-up. Results from symptom-limited exercise testing are important and informative, as the protocol allows clinicians to test exercise tolerance in most of the general population. In CARDIA, subjects with low exercise test duration (<20th percentile) were 3- to 6-fold more likely to develop diabetes, hypertension, and the metabolic syndrome than participants with high duration (≥60th percentile),2 and the adjusted hazard of developing diabetes was 50% higher in women and double in men per 19% decline in duration over seven years.9 Thus, the purpose of the present study was to examine the association of single nucleotide polymorphisms (SNPs) in seventeen candidate genes with baseline symptom-limited exercise test duration and decrease in duration after 20 years of follow-up in the CARDIA Fitness Study.
METHODS
Study population
Details of recruitment, study design, and methods of the CARDIA study have been published elsewhere.10 The initial examination included 5115 Black and White men and women aged 18–30 years from four U.S. communities: Birmingham, AL; Chicago, IL; Minneapolis, MN; and Oakland, CA. All participants provided written informed consent, and institutional review boards from each field center approved the study annually.
Of the participants with genotype data (n=4244), 3960 had valid treadmill test data at baseline and 2618 had valid treadmill test data at both baseline and year 20 examinations. To minimize potential classification errors, parents’ ethnicity (reported by the participants) had to match that of the participant (n=177 excluded). We excluded participants who reported having or were unsure if they had cancer or HIV, pregnant women, and those missing data at either exam for age, BMI, weight, or hypertension and diabetes status. This resulted in 3783 participants (1835 Blacks, 1948 Whites) for baseline duration and 2335 participants (1035 Blacks, 1300 Whites) included in analyses involving decrease in duration over 20 years. Outliers, defined as phenotype value ≥4 standard deviations (SD) from the mean and at least 1 SD from the nearest value, were excluded from analyses (n=2 Whites).
Data Collection
Graded Exercise Testing
Symptom-limited graded exercise treadmill testing was performed at baseline and year 20 by eligible participants using a modified Balke protocol.11 The test consisted of up to nine 2-minute stages of progressively increasing difficulty. Stage 1 was at 3.0 mph and 2% grade, stages 2–6 were at 3.4 mph with grade beginning at 6% and increasing by 4% each stage, stages 7–8 were at 4.2 mph and 22% and 25% grade respectively, and stage 9 was at 5.6 mph and 25% grade. The exercise test consisted primarily of walking to facilitate performance by those unaccustomed to jogging and to allow for easier replication during future follow-up exams.11
For the present study, exercise test data were determined valid if participants achieved >2.0 min. on both the baseline and year 20 treadmill tests (99% met criteria). Baseline duration was defined as the duration of the symptom-limited exercise treadmill test in minutes at baseline. Decrease in duration over 20 years was calculated as the difference of year 20 exercise test duration minus baseline duration.
Gene and SNP selection
The following seventeen genes were selected from the 2005 update of the Human Gene Map for Performance and Health-Related Fitness Phenotypes8 based on their associations with fitness or interactions with physical activity in relation to outcomes such as CVD and obesity: angiotensin converting enzyme (ACE); actinin, alpha 3 (ACTN3); adiponectin receptor 1 (ADIPOR1); alpha-2A-adrenergic receptor (ADRA2A); beta-1-adrenergic receptor (ADRB1); angiotensinogen (AGT); adenosine monophosphate deaminase 1 (AMPD1); angiogenin (ANG); Na+/K+ ATPase alpha 2 subunit (ATP1A2); bradykinin receptor, beta 2 (BDKRB2); creatine kinase, muscle (CKM); endothelin 1 (EDN1); guanine nucleotide binding protein, beta polypeptide 3 (GNB3); hypoxia-inducible factor 1, alpha subunit (HIF1A); nitric oxide synthase 3 (NOS3); peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A); and titin (TTN) (Supplementary Table S1). TagSNPs within these genes with a minor allele frequency (MAF) of greater than 0.05 were identified using the Haploview Program12 from the Caucasian (CEU) and Yoruban (YRI) populations of the International HapMap database.13 The algorithm used for SNP selection was Haploview’s implementation of the Broad Institute’s Tagger software,14 with the r2 cut off for linkage disequilibrium (LD) clustering set to 0.8 and the logarithm (base 10) of odds threshold to 2. All tagSNPs selected by Tagger for the CEU population were included in the SNP panel. TagSNPs that were not in blocks, or only tagged themselves in the YRI population were not included. Nonsynonymous SNPs with a MAF >0.05 were also included. The final SNP set included 217 SNPs in Blacks and 171 SNPs in Whites. All included SNPs had a MAF ≥ 0.05 and Hardy-Weinberg equilibrium (HWE) p ≥ 0.0002 in the CARDIA cohort.
Genotyping
Genotyping of the SNPs was performed using the iPLEX MassARRAY genotyping system (Sequenom, Inc.; San Diego, CA). SNPs that did not perform well on this platform were genotyped using TaqMan Pre-Validated SNP assays (Applied Biosystems; Foster City, CA). Details for PCR conditions and primer sequences are available on request. Genotyping was successfully performed in 82% of the original SNP set (n=354 SNPs). Replicate samples (N=206) were randomly dispersed throughout the genotyping plate set. Only SNPs that had a minimum concordance of 99% were used for further analyses.
Statistical Analysis
All statistical analyses were performed with SAS version 9.1 (SAS Institute Inc, Cary, NC). Differences in continuous and categorical variables between ethnic groups were assessed using t-tests and chi-square tests, respectively. HWE was tested by comparing observed genotype frequencies to expected frequencies using the ALLELE procedure in SAS. The pair-wise LD among the SNPs was assessed using the ldmax program available in the GOLD software package.15
The association tests were conducted in three steps. First, general linear models were used for single SNP analyses of associations with baseline duration and decrease in duration over 20 years by ethnic group. In the baseline duration models, each SNP was tested individually with baseline values of age, sex, BMI, smoking, and hypertension and diabetes status (Y/N) included as covariates. In the decrease in duration over 20 years models, each SNP was tested individually with baseline values of age, sex, BMI, and exercise duration, smoking at year 20, Δweight over 20 years, and hypertension and diabetes status included as covariates. If a participant indicated they were hypertensive or diabetic at either exam or at both exams, then they were classified as having the condition in the decrease in duration models. For nominally significant SNPs (p<0.05) with a minor allele homozygote group size of N<7, genotypes were grouped by minor allele carrier status for analyses.
The second step involved stepwise multiple regression models (with backward elimination) including all nominally significant SNPs (p<0.05) from the single SNP analyses and all covariates. For each SNP nominally significant after stepwise selection, the genotype(s) associated with higher baseline duration or lesser decline in duration over 20 years were identified (i.e., high-treadmill duration genotypes and low-treadmill duration decrease genotypes) (Supplementary Table S2). To examine the combined effects of the favorable genotypes on baseline duration and decrease in duration, the number of genotypes associated with high-treadmill duration or low-treadmill duration decrease across all nominally significant SNPs was used as an index of genetic exercise test duration predisposition in a multiple regression model. Genotype effect size (R2) was defined as the proportion of total phenotypic variance explained by the genotype.
Since multiple SNPs were used in the multiple regression analyses, we applied a multiple testing correction as proposed by Nyholt.16 The effective number of independent SNPs (Meff) can be calculated based on the ratio of observed eigenvalue variance (λobs) and its maximum (M): Meff = 1 + (M-1) (1− (Var λobs/M)). The effective number of SNPs can then be used to adjust the standard α level (e.g., 5%). Since LD between the SNPs differed between Blacks and Whites, the effective number of SNPs was also different (effective number of SNPs in Blacks was 153 and 119 in Whites). Thus, in our study the corrected threshold for statistical significance was set to P<0.0003 in Blacks and P<0.0004 in Whites. Post-hoc power analyses were performed using QUANTO version 1.2.4 (http://hydra.usc.edu/gxe).17 We have 80% power to detect effect sizes of 1.0–1.1% in the baseline models and 1.5–1.9% in the decrease over 20 years models, under an additive genetic model using the multiple testing corrected alpha levels (Supplementary Figure S1).
RESULTS
Baseline and year 20 characteristics of the participants are summarized in Supplementary Table S3. Black participants were younger, heavier, had lower exercise test duration, and were more likely to smoke, be hypertensive, and have diabetes at baseline than Whites (Table S3a). Furthermore, Blacks experienced a significantly greater increase in weight and a greater decrease in exercise duration over 20 years compared to Whites (Table S3b). The MAF, HWE, and pairwise LD among all included SNPs (Tables S4 and S5) and the results for the associations of all SNPs with baseline duration and decrease in duration (Table S6) can be found in supplementary tables.
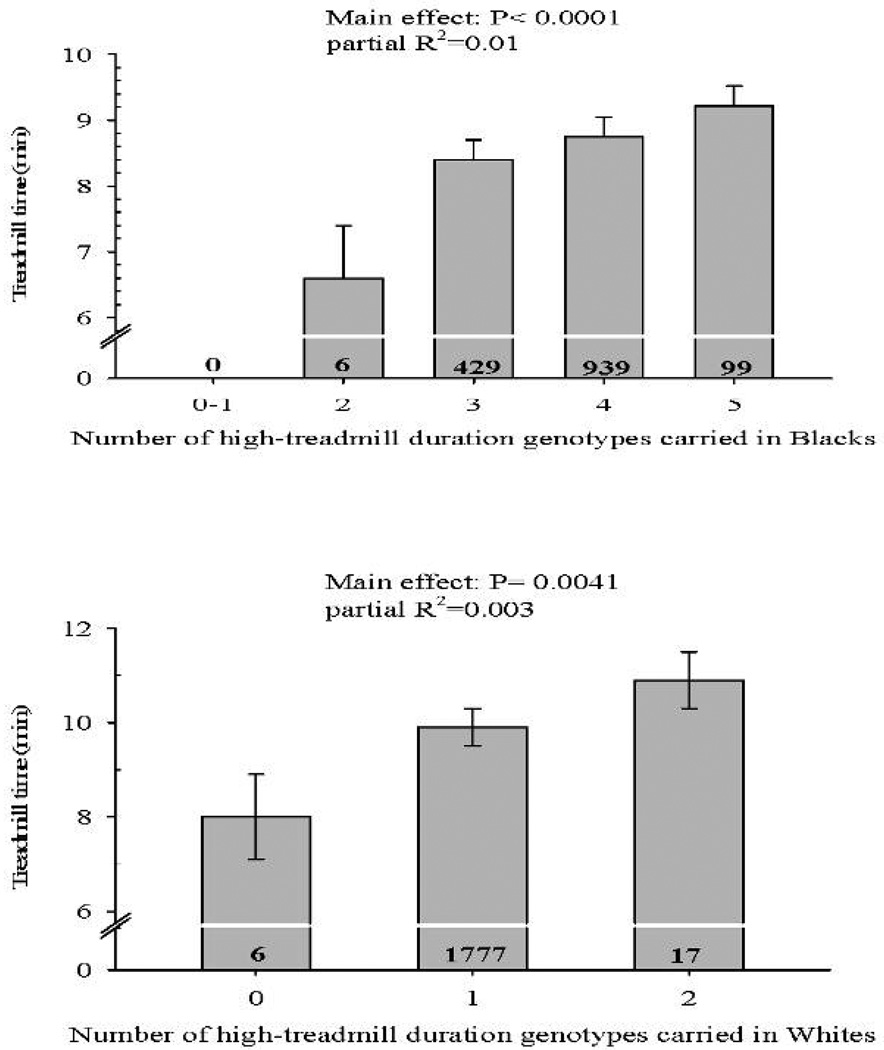
In Blacks, two SNPs in the PPARGC1A and NOS3 loci, three SNPs in the TTN gene locus, and one SNP each from the ACE, ACTN3, AGT, ATP1A2, and HIF1A loci were nominally (p<0.05) associated with baseline duration (Table 1). The stepwise regression model showed that sex, BMI, smoking, hypertension, ATP1A2 rs9660705, HIF1A rs1957755, NOS3 rs3918196, and PPARGC1A rs7657517 and rs2932971 were associated with baseline duration in Blacks (Table 2). None of the five SNPs reached the multiple testing corrected threshold for statistical significance. However, in Blacks, the number of high-treadmill duration genotypes carried at the five markers was significantly associated with baseline duration (P<0.0001), as those with five high-treadmill duration genotypes had a mean baseline duration of 9.2 min, compared to 6.6 min in those who only had two (Figure 1-top panel).
Table 1.
Nominally significant associations of SNPs with baseline exercise test duration in Blacks.
| Blacks | Mean (SE) Baseline duration (min) by genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GENE | SNP | p-value | R2 | Common allele homozygote |
N | Heterozygote/ or minor allele carrier |
N | Minor allele homozygote |
N |
| AGT | rs11568045 | 0.0454 | 0.001 | 8.7 (0.3) | 1240 | 8.4 (0.3) | 485 | 8.8 (0.4) | 49 |
| ATP1A2 | rs9660705 | 0.0148 | 0.002 | 8.6 (0.3) | 1391 | 8.6 (0.3) | 182 | 7.0 (0.7) | 9 |
| TTN | rs6732060 | 0.0146 | 0.002 | 8.7 (0.3) | 1053 | 8.5 (0.3) | 622 | 9.0 (0.4) | 65 |
| rs2366753 | 0.0147 | 0.002 | 8.7 (0.3) | 923 | 8.6 (0.3) | 723 | 9.0 (0.3) | 126 | |
| rs2627038 | 0.0222 | 0.002 | 8.7 (0.3) | 483 | 8.6 (0.3) | 859 | 8.9 (0.3) | 392 | |
| PPARGC1A | rs2932971 | 0.0198 | 0.002 | 8.6 (0.3) | 1064 | 8.7 (0.3) | 604 | 8.1 (0.3) | 88 |
| rs7657517 | 0.0363 | 0.001 | 8.5 (0.3) | 493 | 8.7 (0.3) | 846 | 8.8 (0.3) | 431 | |
| NOS3 | rs1799983 | 0.0015 | 0.003 | 8.5 (0.3) | 1424 | 8.9 (0.3) | 334 | 8.3 (0.5) | 23 |
| rs3918196* | 0.0039 | 0.002 | 8.5 (0.3) | 1576 | 8.9 (0.3) | 167 | --- | --- | |
| ACTN3 | rs2275998 | 0.0275 | 0.002 | 8.7 (0.3) | 871 | 8.4 (0.3) | 764 | 8.5 (0.3) | 141 |
| HIF1A | rs1957755 | 0.0228 | 0.002 | 8.6 (0.3) | 1518 | 8.8 (0.3) | 214 | 9.6 (0.6) | 12 |
| ACE | rs4316 | 0.0483 | 0.001 | 8.4 (0.3) | 656 | 8.6 (0.3) | 824 | 8.7 (0.3) | 289 |
genotypes grouped by minor allele carrier status
Table 2.
Stepwise regression results for predictors of baseline exercise test duration in Blacks and Whites.
| Blacks | Whites | ||||||
|---|---|---|---|---|---|---|---|
| Variable | β | partial r2 | p-value | Variable | β | partial r2 | p-value |
| Sex (M/F) | −3.78 | 0.4855 | <0.0001 | Sex (M/F) | −3.29 | 0.3243 | <0.0001 |
| BMI, kg/m2 | −0.17 | 0.1197 | <0.0001 | BMI, kg/m2 | −0.23 | 0.1275 | <0.0001 |
| Smoking (Y/N) | 0.29 | 0.0097 | <0.0001 | Smoking (Y/N) | 0.34 | 0.0152 | <0.0001 |
| Hypertension (N/Y) | −0.48 | 0.0028 | 0.0012 | PPARGC1A rs3774909 | 2.17 | 0.0024 | 0.0044 |
| NOS3 rs3918196* | 0.44 | 0.0023 | 0.0032 | Hypertension (N/Y) | −0.36 | 0.0017 | 0.0176 |
| ATP1A2 rs9660705 | 1.41 | 0.0017 | 0.0096 | HIF1A rs1957757 | 0.95 | 0.0013 | 0.0349 |
| PPARGC1A rs2932971 | 0.61 | 0.0014 | 0.0192 | Diabetes (N/Y) | −1.36 | 0.0012 | 0.0445 |
| PPARGC1A rs7657517 | 0.16 | 0.0019 | 0.0071 | ||||
| HIF1A rs1957755 | 0.26 | 0.0012 | 0.0327 | ||||
| Model r2 0.6261 | Model r2 0.4735 | ||||||
β is the estimated regression coefficient for each variable in the model.
Figure 1.
Association of the number of high-treadmill duration genotypes with baseline exercise test duration at NOS3 rs3918196, ATP1A2 rs9660705, PPARGC1A rs7657517, PPARGC1A rs2932971, and HIF1A rs1957755 markers in Blacks (top panel) and HIF1A rs1957757 and PPARGC1A rs3774909 markers in Whites (bottom panel). P value for the main effect of high-treadmill duration genotypes along with variance in baseline duration explained by high-treadmill duration genotypes is shown at the top of each graph. Number of subjects with the selected number of genotypes is indicated inside each histogram bar.
In Whites, five SNPs from the PPARGC1A gene locus and one SNP each from the CKM, EDN1, and HIF1A loci were nominally associated with baseline duration (Table 3). Differences in baseline duration between genotypes were largest at HIF1A rs1957757, as minor allele homozygotes had about 1.0 min higher duration compared to the other genotypes (Table 3). The stepwise regression model showed that sex, BMI, smoking, hypertension, diabetes, PPARGC1A rs3774909, and HIF1A rs1957757 were associated with baseline duration (Table 2). However, the two SNPs were not statistically significant. Furthermore, the number of genotypes associated with higher baseline duration carried by Whites at the HIF1A rs1957757 and PPARGC1A rs3774909 markers was not significantly associated with baseline duration in a multivariate regression model (P=0.0041). In general, Whites with the most favorable genotypes at both SNPs had longer baseline duration (Figure 1-bottom panel), however since all white participants carried at least one of the nominally favorable genotypes, discriminatory ability was limited.
Table 3.
Nominally significant associations of SNPs with baseline exercise test duration in Whites.
| Whites | Mean (SE) Baseline duration (min) by genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GENE | SNP | p-value | R2 | Common allele homozygote |
N | Heterozygote/ or minor allele carrier |
N | Minor Allele homozygote |
N |
| PPARGC1A | rs2932977 | 0.0046 | 0.003 | 10.0 (0.4) | 1181 | 9.9 (0.4) | 550 | 9.3 (0.4) | 81 |
| rs3774909 | 0.0244 | 0.002 | 9.9 (0.4) | 1651 | 9.9 (0.4) | 169 | 7.9 (0.8) | 7 | |
| rs4697046 | 0.0106 | 0.003 | 9.9 (0.4) | 759 | 10.0 (0.4) | 801 | 9.6 (0.4) | 246 | |
| rs6448226 | 0.0211 | 0.002 | 9.9 (0.4) | 723 | 10.0 (0.4) | 850 | 9.6 (0.4) | 252 | |
| rs7665116 | 0.0233 | 0.002 | 9.9 (0.3) | 1381 | 10.1 (0.4) | 451 | 9.2 (0.5) | 38 | |
| EDN1 | rs1630736 | 0.0089 | 0.003 | 10.0 (0.4) | 559 | 9.9 (0.4) | 863 | 9.6 (0.4) | 416 |
| HIF1A | rs1957757 | 0.0065 | 0.003 | 9.9 (0.4) | 1560 | 10.0 (0.4) | 300 | 11.2 (0.6) | 21 |
| CKM | rs344816 | 0.0443 | 0.002 | 10.0 (0.4) | 501 | 9.8 (0.4) | 908 | 10.1 (0.4) | 410 |
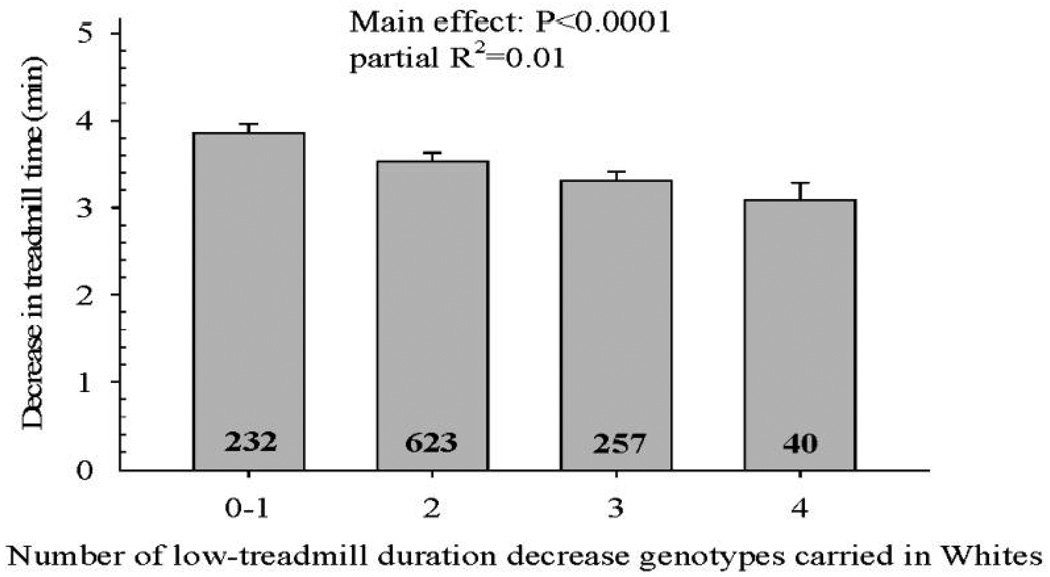
In Blacks, two SNPs each from the ATP1A2, PPARGC1A, and TTN loci and one SNP each from the ADRB1, EDN1, and GNB3 loci were nominally associated with decrease in duration over 20 years (Table 4). None of the nine nominally significant markers remained in the model after backward elimination, as the stepwise model showed baseline values of age, BMI, and exercise duration, sex, Δweight, smoking at year 20, and diabetes were associated with decrease in duration (Supplementary Table S7). In Whites, one SNP at the AGT and AMPD1 loci, five PPARGC1A SNPs, and two ANG SNPs were nominally associated with decrease in duration over 20 years (Table 5). The stepwise regression model showed that baseline values of age, BMI, and exercise duration, Δweight, sex, smoking at year 20, diabetes, AGT rs5051, AMPD1 rs2010899, ANG rs1010458, and PPARGC1A rs4452416 were associated with decrease in duration (Table 6). Individually, none of the SNPs met the corrected threshold for statistical significance. However, carrying low-treadmill duration decrease genotypes at these four SNPs was significantly associated with preservation of exercise test duration over time (P<0.0001), as the decline in treadmill time over 20 years was graded across the number of favorable genotypes (Figure 2). Whites with four low-treadmill duration decrease genotypes had a mean change in duration of −3.1 min over 20 years, compared to −3.9 min in those with none or one (Figure 2).
Table 4.
Nominally significant associations of SNPs with decrease in exercise test duration over 20 years by genotype in Blacks.
| Blacks | Mean (SE) decrease in duration (min.) by genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GENE | SNP | p-value | R2 | Common allele homozygote |
N | Heterozygote/ or minor allele carrier |
N | Minor allele homozygote |
N |
| ATP1A2 | rs1016732 | 0.0003 | 0.009 | −3.3 (0.1) | 791 | −3.7 (0.1) | 186 | −4.0 (0.4) | 13 |
| rs2854248 | 0.0015 | 0.007 | −3.1 (0.1) | 271 | −3.3 (0.1) | 477 | −3.6 (0.1) | 218 | |
| TTN | rs12998857* | 0.0388 | 0.002 | −3.4 (0.09) | 869 | −3.1 (0.2) | 121 | --- | --- |
| rs3816781 | 0.045 | 0.003 | −3.3 (0.1) | 635 | −3.5 (0.1) | 318 | −3.0 (0.2) | 50 | |
| PPARGC1A | rs12374408 | 0.041 | 0.003 | −3.5 (0.1) | 485 | −3.3 (0.1) | 414 | −3.2 (0.2) | 78 |
| rs10002521* | 0.0271 | 0.003 | −3.4 (0.09) | 878 | −3.0 (0.2) | 106 | --- | --- | |
| EDN1 | rs6912834 | 0.047 | 0.003 | −3.3 (0.09) | 837 | −3.5 (0.1) | 164 | −4.3 (0.6) | 7 |
| ADRB1 | rs1801253 | 0.044 | 0.003 | −3.2 (0.1) | 383 | −3.4 (0.1) | 439 | −3.5 (0.1) | 168 |
| GNB3 | rs2301339 | 0.0026 | 0.006 | −3.5 (0.1) | 563 | −3.3 (0.1) | 353 | −2.9 (0.2) | 83 |
genotypes grouped by minor allele carrier status
Table 5.
Nominally significant associations of SNPs with decrease in exercise test duration over 20 years by genotype in Whites.
| Whites | Mean (SE) decrease in duration (min.) by genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GENE | SNP | p-value | R2 | Common allele homozygote |
N | Heterozygote/ or minor allele carrier |
N | Minor allele homozygote |
N |
| AGT | rs5051 | 0.0229 | 0.003 | −3.5 (0.1) | 444 | −3.5 (0.1) | 618 | −3.2 (0.1) | 238 |
| AMPD1 | rs2010899 | 0.0323 | 0.003 | −3.6 (0.1) | 396 | −3.5 (0.1) | 575 | −3.3 (0.1) | 259 |
| PPARGC1A | rs768695 | 0.0003 | 0.007 | −3.6 (0.1) | 335 | −3.3 (0.1) | 590 | −3.6 (0.1) | 321 |
| rs3774921 | 0.0442 | 0.003 | −3.6 (0.1) | 362 | −3.4 (0.1) | 560 | −3.7 (0.1) | 294 | |
| rs7657517 | 0.0071 | 0.004 | −3.6 (0.1) | 939 | −3.3 (0.1) | 276 | −4.0 (0.3) | 27 | |
| rs4452416 | 0.0134 | 0.003 | −3.4 (0.1) | 944 | −3.7 (0.1) | 316 | −3.7 (0.3) | 19 | |
| rs7677000 | 0.0067 | 0.004 | −3.6 (0.1) | 875 | −3.3 (0.1) | 314 | −3.9 (0.2) | 38 | |
| ANG | rs4470055 | 0.0037 | 0.005 | −3.5 (0.1) | 679 | −3.4 (0.1) | 483 | −3.9 (0.2) | 85 |
| rs1010458 | 0.0245 | 0.003 | −3.5 (0.1) | 812 | −3.5 (0.1) | 363 | −4.0 (0.2) | 52 | |
Table 6.
Stepwise regression results for predictors of decrease in exercise test duration over 20 years in Whites.
| Whites | |||
|---|---|---|---|
| Variable | β | partial r2 | p-value |
| Baseline duration, min | −0.52 | 0.1754 | <0.0001 |
| ΔWeight, kg | −0.04 | 0.2059 | <0.0001 |
| Baseline Age, yrs | −0.08 | 0.0196 | <0.0001 |
| Smoking at year 20 (N/Y) | −1.00 | 0.0209 | <0.0001 |
| Sex (M/F) | −1.01 | 0.0207 | <0.0001 |
| Baseline BMI, kg/m2 | −0.08 | 0.0221 | <0.0001 |
| Diabetes (N/Y) | −0.57 | 0.0052 | 0.0008 |
| Hypertension (N/Y) | −0.37 | 0.0044 | 0.0022 |
| PPARGC1A rs4452416 | 0.27 | 0.0035 | 0.0057 |
| ANG rs1010458 | 0.59 | 0.0035 | 0.0056 |
| AMPD1 rs2010899 | 0.26 | 0.0031 | 0.0088 |
| AGT rs5051 | 0.25 | 0.0025 | 0.0194 |
| Model r2 0.4868 | |||
Figure 2.
Association of the number of low-treadmill duration decrease genotypes with decrease in exercise duration over 20 years at AGT rs5051, ANG rs1010458, AMPD1 rs2010899, and PPARGC1A rs4452416 markers in CARDIA Whites. P value for the main effect of low-treadmill duration decrease alleles along with variance in decrease in duration explained by low-treadmill duration decrease alleles is shown at the top of each graph. Number of subjects with the selected number of genotypes is indicated inside each histogram bar.
DISCUSSION
We found multiple markers from several genes nominally (p<0.05) associated with symptom-limited baseline exercise test duration and decrease in duration over 20 years. Markers from three genes (ATP1A2, TTN, and PPARGC1A) in Blacks and PPARGC1A in Whites were nominally associated with both baseline duration and decrease in duration. Individually, none of the tested SNPs were significantly associated with the exercise duration phenotypes after correcting for multiple testing. However, multivariate regression models showed that Blacks with all five favorable fitness genotypes at five SNPs in the ATP1A2, HIF1A, NOS3, and PPARGC1A genes had the greatest baseline duration. Similarly, Whites with all four favorable genotypes at four SNPs in the AGT, AMPD1, ANG, and PPARGC1A genes had the lowest decline in exercise test duration over 20 years. All markers used in the present study reside in genes previously identified as candidates for fitness phenotypes. Nominally significant SNPs found in this study, except for ACE rs4316, ADRB1 rs1801253, NOS3 rs1799983, and TTN rs6732060, were located in non-coding regions (e.g., introns) and the mechanism of their association with symptom-limited exercise test duration is unknown. A description of the predictor SNPs in the regression models and the genes where they reside can be found in Supplementary Table S8.
The HIF1A rs1957577 marker in Whites and the rs1957755 marker in Blacks were associated with baseline duration in the stepwise regression models. In Whites, minor allele homozygotes (T/T) at rs1957577 had almost two minutes longer baseline duration than common allele carriers. HIF1A rs1957757 tags a cluster of 6 SNPs that cover the 5’-end of HIF1A (pairwise r2=1.0 between all SNPs in HapMap CEU). HIF1A is a transcription factor that regulates several genes in response to hypoxia, and these genes are involved in angiogenesis, erythropoiesis, and metabolism. Sequence variation in HIF1A was previously associated with baseline maximal oxygen consumption (VO2max) in older black men, while the Ser582Pro polymorphism was associated with ΔVO2max after 24 weeks of training in older white men.18 However, the Ser582Pro variant was not included in the present study and is not in strong LD (r2>0.8) with any of the included HIF1A SNPs.
The PPARGC1A rs7657517 and rs2932971 markers were significantly associated with baseline duration in Blacks, while the rs3774909 and rs4452416 markers were significantly associated with baseline duration and decrease in duration, respectively, in Whites. The PPARGC1A rs2932971 marker tags a cluster of four SNPs in blacks (pairwise r2=1.0 in HapMap YRI), while the rs3774909 and rs4452416 markers tag a cluster of two and three SNPs, respectively, in whites (pairwise r2=1.0 in HapMap CEU). PPARGC1A regulates the transcription of enzymes involved in oxidative phosphorylation through its role in mitochondrial biogenesis, skeletal muscle fiber-type formation, and glucose and lipid transport and oxidation. Previous studies reported that a common variant, Gly482Ser, was associated with endurance capacity in young adult European men19 and with changes in aerobic fitness after lifestyle intervention in adult European men and women.20 The Gly482Ser variant was not included in the present study and is not in strong LD (r2>0.8) with any of the included PPARGC1A SNPs.
In Blacks, markers from ATP1A2 were nominally associated with both baseline duration and decrease in duration, and ATP1A2 rs9660705 was significantly associated with baseline duration in the multiple regression model. In skeletal muscle, the Na+/K+ ATPase regulates transsarcolemmal [Na+] and [K+] gradients and is critical for the maintenance of membrane excitability and contractility. Elevated muscle Na+-K+ ATPase content and maximal activity have been shown in trained individuals and after endurance training in untrained subjects.21, 22 The Na+/K+ ATPase is constituted by a catalytic subunit (α) with three isoforms (α1, α2, and α3), with the α2-gene (ATP1A2) expressed mainly in skeletal muscle. Genetic variation at the ATP1A2 gene locus was found to be associated with the trainability of VO2 max in sedentary adults.23
Although aerobic endurance typically declines with age,24 it is influenced by multiple factors such as initial level, habitual physical activity, body composition, and diseases.25,26 In the present study, we controlled for many of these factors including baseline BMI, baseline duration, Δweight, as well as the presence of diabetes and hypertension. However, there was still marked variation in the decrease in symptom-limited exercise test duration over 20 years in our cohort, as evidenced by the large SD. It should be noted that there may not have been equal effort during the exercise tests over time. Because we had a symptom-limited test and no objective marker of a true maximum, it is likely that individuals had varying levels of motivation during the two exercise tests and this differential effort was probably not random. However, the Balke protocol has been shown to correlate with VO2max in adults. The correlation between Balke treadmill time and VO2max was 0.92 in 51 healthy men (57% sedentary) between 35 to 55 years of age27, and 0.94 in 49 healthy women (59% sedentary) between 20 and 42 years of age.28
Few studies have examined the effects of multiple markers from multiple genes on fitness. We found that carrying several favorable genotypes, at loci shown to be suggestively associated with baseline exercise test duration or decrease in duration, resulted in two and a half minutes longer baseline treadmill time in Blacks and a one minute lesser decrease in treadmill time over 20 years in Whites. These data provide evidence of both the independent and combined effects of multiple markers on exercise test duration in young adults and with decrease in duration over 20 years. Symptom-limited exercise tests have important clinical relevance as an indication of exercise tolerance, as many individuals may not be physically able to perform a true maximal exercise test to exhaustion.
Recently, a quantitative molecular classifier (predictor) relating ΔVO2max to baseline muscle gene RNA expression identified 29 predictor genes loci.29 Regression analysis of SNPs from 25 of these loci combined with SNPs from 10 other candidate genes yielded a model where 11 SNPs explained 23% of the variance in exercise training induced gains in VO2max. Comparison of these results to the current study suggests that the genes associated with responsiveness to exercise training differ from those associated with fitness level or its decrease over time.
The present study was limited to selected candidate genes. We were powered to detect effect sizes of ~1.0% at baseline and between 1.5–1.9% in the decrease over 20 years models. The SNPs we tested did not contribute large effect sizes, as nominally significant SNPs showed only small individual effects on the overall variance in each exercise test duration phenotype. However, pooling the markers associated with each phenotype after stepwise regression analyses explained more of the variance in each exercise test duration phenotype. Overall, these pooled markers still only explained a relatively small proportion of the variance in baseline duration (≤1.0%) or decrease in duration (1.0%), which provides support for the polygenic and environmentally influenceable nature of fitness phenotypes.
In summary, multi marker constructs consisting of alleles at genes related to skeletal muscle function and Na+/K+ transport, hypoxia, and mitochondrial metabolism are associated with baseline symptom-limited exercise test duration and decrease in duration over 20 years in adults.
Cardiorespiratory fitness-related phenotypes have been shown to have a substantial genetic component in numerous epidemiological studies. However, identifying common genetic variants associated with changes in health-related fitness phenotypes over time in large population-based studies has proven difficult due to the lack of appropriate datasets and the feasibility of measuring fitness in a large group of people. We observed in the CARDIA fitness Study that carrying several favorable alleles, at loci shown to be independently associated with baseline exercise test duration or with change in exercise duration, resulted in an approximately two and a half minutes longer baseline treadmill time in Blacks and a one minute lesser decrease in treadmill time over 20 years in Whites. Thus, these data provide evidence of both the independent and conjoint effects of multiple markers from several candidate genes on symptom-limited exercise test duration in young adults and with change in exercise test duration over 20 years. Symptom-limited exercise tests have important clinical relevance for evaluating exercise tolerance, as many individuals may not be able to perform a true maximal exercise test to exhaustion. These results are of interest to clinicians, as exercise duration in CARDIA participants was shown to be predictive of the development of cardiovascular disease risk factors such as diabetes, hypertension, and metabolic syndrome. Understanding the underlying genetic basis of the ability to benefit from regular exercise can likely facilitate custom-tailored programs and therapies for individuals with less favorable genotypes.
Supplementary Material
Acknowledgments
Funding Sources: The Coronary Artery Risk Development in Young Adults study is supported by National Heart, Lung, and Blood Institute (grants N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, and N01-HC-95095). The CARDIA Fitness study is supported by R01 HL 078972.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures: None
REFERENCES
- 1.Blair SN, Kohl HW, 3rd, Paffenbarger RS, Jr, Clark DG, Cooper KH, Gibbons LW. Physical fitness and all-cause mortality. A prospective study of healthy men and women. JAMA. 1989;262:2395–2401. doi: 10.1001/jama.262.17.2395. [DOI] [PubMed] [Google Scholar]
- 2.Carnethon MR, Gidding SS, Nehgme R, Sidney S, Jacobs DR, Jr, Liu K. Cardiorespiratory fitness in young adulthood and the development of cardiovascular disease risk factors. JAMA. 2003;290:3092–3100. doi: 10.1001/jama.290.23.3092. [DOI] [PubMed] [Google Scholar]
- 3.Blair SN, Kohl HW, 3rd, Barlow CE, Paffenbarger RS, Jr, Gibbons LW, Macera CA. Changes in physical fitness and all-cause mortality. A prospective study of healthy and unhealthy men. JAMA. 1995;273:1093–1098. [PubMed] [Google Scholar]
- 4.Sternfeld B, Sidney S, Jacobs DR, Jr, Sadler MC, Haskell WL, Schreiner PJ. Seven-year changes in physical fitness, physical activity, and lipid profile in the CARDIA study. Coronary Artery Risk Development in Young Adults. Ann Epidemiol. 1999;9:25–33. doi: 10.1016/s1047-2797(98)00030-1. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, Province MA, Leon AS, Rao DC, Skinner JS, Wilmore JH. Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Med Sci Sports Exerc. 1998;30:252–258. doi: 10.1097/00005768-199802000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Fagard R, Bielen E, Amery A. Heritability of aerobic power and anaerobic energy generation during exercise. J Appl Physiol. 1991;70:357–362. doi: 10.1152/jappl.1991.70.1.357. [DOI] [PubMed] [Google Scholar]
- 7.Lesage R, Simoneau JA, Jobin J, Leblanc J, Bouchard C. Familial resemblance in maximal heart rate, blood lactate and aerobic power. Hum Hered. 1985;35:182–189. doi: 10.1159/000153540. [DOI] [PubMed] [Google Scholar]
- 8.Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41:35–73. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- 9.Carnethon MR, Sternfeld B, Schreiner PJ, Jacobs DR, Jr, Lewis CE, Liu K, Sidney S. Association of 20 year Changes in Cardirespiratory Fitness with Incident Type 2 Diabetes: The CARDIA Fitness Study. Diabetes Care. 2009;32:1284–1288. doi: 10.2337/dc08-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, Liu K, Savage PJ. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 11.Sidney S, Haskell WL, Crow R, Sternfeld B, Oberman A, Armstrong MA, Cutter GR, Jacobs DR, Savage PJ, Van Horn L. Symptom-limited graded treadmill exercise testing in young adults in the CARDIA study. Med Sci Sports Exerc. 1992;24:177–183. [PubMed] [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 13.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 14.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 15.Abecasis GR, Cookson WOC. GOLD--Graphical Overview of Linkage Disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 16.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 2006; Available from: http://hydra.usc.edu/gxe. [Google Scholar]
- 18.Prior SJ, Hagberg JM, Phares DA, Brown MD, Fairfull L, Ferrell RE, Roth SM. Sequence variation in hypoxia-inducible factor 1alpha (HIF1A): association with maximal oxygen consumption. Physiol Genomics. 2003;15:20–26. doi: 10.1152/physiolgenomics.00061.2003. [DOI] [PubMed] [Google Scholar]
- 19.Lucia A, Gomez-Gallego F, Barroso I, Rabadan M, Bandres F, San Juan AF, Chicharro JL, Ekelund U, Brage S, Earnest CP, Wareham NJ, Franks PW. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol. 2005;99:344–348. doi: 10.1152/japplphysiol.00037.2005. [DOI] [PubMed] [Google Scholar]
- 20.Stefan N, Thamer C, Staiger H, Machicao F, Machann J, Schick F, Venter C, Niess A, Laakso M, Fritsche A, Haring HU. Genetic variations in PPARD and PPARGC1A determine mitochondrial function and change in aerobic physical fitness and insulin sensitivity during lifestyle intervention. J Clin Endocrinol Metab. 2007;92:1827–1833. doi: 10.1210/jc.2006-1785. [DOI] [PubMed] [Google Scholar]
- 21.Green HJ, Chin ER, Ball-Burnett M, Ranney D. Increases in human skeletal muscle Na(+)-K(+)-ATPase concentration with short-term training. Am J Physiol Cell Physiol. 1993;264:C1538–C1541. doi: 10.1152/ajpcell.1993.264.6.C1538. [DOI] [PubMed] [Google Scholar]
- 22.Madsen K, Franch J, Clausen T. Effects of intensified endurance training on the concentration of Na,K-ATPase and Ca-ATPase in human skeletal muscle. Acta Physiol Scand. 1994;150:251–258. doi: 10.1111/j.1748-1716.1994.tb09684.x. [DOI] [PubMed] [Google Scholar]
- 23.Rankinen T, Perusse L, Borecki I, Chagnon YC, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. The Na(+)-K(+)-ATPase alpha2 gene and trainability of cardiorespiratory endurance: the HERITAGE family study. J Appl Physiol. 2000;88:346–351. doi: 10.1152/jappl.2000.88.1.346. [DOI] [PubMed] [Google Scholar]
- 24.Fleg JL, Morrell CH, Bos AG, Brant LJ, Talbot LA, Wright JG, Lakatta EG. Accelerated longitudinal decline of aerobic capacity in healthy older adults. Circulation. 2005;112:674–682. doi: 10.1161/CIRCULATIONAHA.105.545459. [DOI] [PubMed] [Google Scholar]
- 25.Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc. 2001;33:S446–S451. doi: 10.1097/00005768-200106001-00013. [DOI] [PubMed] [Google Scholar]
- 26.Kriegsman DM, Deeg DJ, Stalman WA. Comorbidity of somatic chronic diseases and decline in physical functioning: the Longitudinal Aging Study Amsterdam. J Clin Epidemiol. 2004;57:55–65. doi: 10.1016/S0895-4356(03)00258-0. [DOI] [PubMed] [Google Scholar]
- 27.Pollock ML, Bohannon RL, Cooper KH, Ayres JJ, Ward A, White SR, Linnerud AC. A comparative analysis of four protocols for maximal treadmill stress testing. Am Heart J. 1976;92:39–46. doi: 10.1016/s0002-8703(76)80401-2. [DOI] [PubMed] [Google Scholar]
- 28.Pollock ML, Foster C, Schmidt D, Hellman C, Linnerud AC, Ward A. Comparative analysis of physiologic responses to three different maximal graded exercise test protocols in healthy women. Am Heart J. 1982;103:363–373. doi: 10.1016/0002-8703(82)90275-7. [DOI] [PubMed] [Google Scholar]
- 29.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NBJ, Nielsen S, Akerstrom T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJC, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–1496. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.