Abstract
About 1 million people in the world die each year from diseases spread by mosquitoes, and understanding the mechanism of host identification by the mosquitoes through olfaction is at stake. The role of odorant binding proteins (OBPs) in the primary molecular events of olfaction in mosquitoes is becoming an important focus of biological research in this area. Here, we present a comprehensive comparative genomics study of OBPs in the three disease-transmitting mosquito species Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus starting with the identification of 110 new OBPs in these three genomes. We have characterized their genomic distribution and orthologous and phylogenetic relationships. The diversity and expansion observed with respect to the Aedes and Culex genomes suggests that the OBP gene family acquired functional diversity concurrently with functional constraints posed on these two species. Sequences with unique features have been characterized such as the “two-domain OBPs” (previously known as Atypical OBPs) and “MinusC OBPs” in mosquito genomes. The extensive comparative genomics featured in this work hence provides useful primary insights into the role of OBPs in the molecular adaptations of mosquito olfactory system and could provide more clues for the identification of potential targets for insect repellants and attractants.
Keywords: odorant binding proteins, OBP, mosquito, Culex quinquefasciatus, Aedes aegypti, Anopheles gambiae, olfaction, phylogeny
Introduction
The spread of infectious diseases among humans is mediated primarily by the world’s most dangerous animal, the mosquitoes among which the anthrophilic mosquitoes such as Anopheles gambiae, Anopheles funestus, Aedes albopictus, Aedes aegypti, and Culex quinquefasciatus are the most effective transmitters of viruses and parasites. They are responsible for the spread of a number of life-threatening diseases such as malaria, dengue, and West Nile encephalitis and recently Chikungunya with a lower mortality rate compared with the other diseases. According to the World Health Organization, global climate change is expanding mosquitoes range, heightening the risk of disease for millions of additional people. Primary prevention is one of the most important aspects to subside the spread of diseases either by controlling the population of these vectors or by preventing the interaction between the vector and the host.
Understanding the molecular mechanism for human host recognition mediated by olfaction would help in identifying new strategies for the prevention of the primary contact. Volatile products secreted by the human host in the process of metabolism are responsible for the attraction of these vectors to the host. The ability of recognizing and discriminating thousands of odorant molecules in insects as in mammals relies on specialized chemosensitive neural cells expressing olfactory receptor proteins (ORs) which reside within segregated compartments called sensilla. Each sensillum is a hair-like structure bathed in the sensillum lymph which contains a number of secreted proteins (McKenna et al. 1994; Pikielny et al. 1994; Wang et al. 1999). The odorant binding proteins (OBPs) are found to be important water-soluble components of this sensillum lymph. It was first identified in the moth as pheromone binding proteins (PBPs) (Vogt and Riddiford 1981). These globular proteins are believed to bind different odorant molecules (Plettner et al. 2000), owing to their high divergence within the family, and transport them to their respective olfactory receptors triggering the mechanism of olfaction (Pelosi and Maida 1995).
The arthropod OBPs form a large specific multi-gene family. They are 10–30 kDa globular and water-soluble proteins that are characterized by a specific six α-helical domain comprising of six highly conserved cysteines that have distinct disulphide connectivities. These structural features are now considered the hallmark of this protein family (Calvo et al. 2002; Valenzuela et al. 2002; Calvo et al. 2006). OBPs have been identified in a number of insect species, including four dipterian species Drosophila melanogaster (Galindo and Smith 2001; Graham and Davies 2002; Hekmat-Scafe et al. 2002; Valenzuela et al. 2002; Zhou et al. 2004; Vieira et al. 2007; Vieira and Rozas 2011), A. gambiae (Vogt 2002; Xu et al. 2003; Zhou et al. 2004; Li et al. 2005; Vieira and Rozas 2011), Aed. aegypti (Zhou et al. 2008), and C. quinquefasciatus (Pelletier and Leal 2009, 2011). These proteins are very divergent in terms of the sequences within the family, and sequence identities between the family members from the different species could drop as low as 8% (Vieira and Rozas 2011). In Drosophila, a subgroup of (i) OBPs lacking two of the six conserved cysteines, called MinusC OBPs and (ii) OBPs carrying additional conserved cysteines called PlusC OBPs have been identified (Hekmat-Scafe et al. 2002). The MinusC OBPs typically lack the second and fifth Cys residues. However, this definition appears to be somewhat ambiguous, since there are three Drosophila OBPs among this cluster which contain all the six hallmark cysteines (Pelosi and Maida 1995). MinusC OBPs have never been described to date in mosquito genomes.
In mosquitoes, three subfamilies of OBP genes have been characterized so far: (i) the Classic OBPs that carry the six conserved cysteines characteristic motif of the OBP family; (ii) the PlusC OBPs that have the same conserved cysteines and disulphide connectivity but which contain six additional cysteines with novel disulphide connectivities; (iii) the Atypical OBPs that are among the longest known OBPs and that have initially been described as containing a single Classic OBP domain in its N-terminal extended by a less characterized C-terminal extension. Very recently, it was shown that Atypical OBPs comprises two domains that are in fact homologous to the Classic OBP domain and were hence considered as “dimer OBPs” (Vieira and Rozas 2011).
In A. gambiae and Aed. aegypti, OBPs from the three different subfamilies have been reported to date while in C. quinquefasciatus, only the Classic and PlusC members of this family have been reported so far (Pelletier and Leal 2009, 2011). Atypical OBPs have not yet been reported in this genome.
An additional multi-gene family, known as D7 salivary proteins, is known to be distantly related to the arthropod OBP superfamily (Calvo et al. 2002, 2006, 2009). There are two types of D7 salivary proteins in the mosquito genome, the short and the long forms which contain one and two OBP-like domains, respectively (Valenzuela et al. 2002; Kalume et al. 2005; Choumet et al. 2007). The available structures of the D7 proteins indicate that the domains adopt a similar fold to the OBP domains but decorated with additional structural features and a seventh helix. In the two-domain D7 protein, the C-terminal OBP-like domain has been shown to bind to biogenic amines in A. gambiae and Aed. aegypti (Mans et al. 2008; Calvo et al. 2009), while the N-terminal domain in Aed. aegypti was shown to have a specific bioactive lipid-binding activity (Calvo et al. 2009). These members serve as important representatives for the construction of phylogenetic trees serving as outgroups for the OBP gene family in the current analysis.
This work describes the identification and extension of OBPs in the mosquito genomes of A. gambiae, Aed. aegypti, and C. quinquefasciatus. We provide a significant extension of the OBP gene family to a total of 110 new members in these three genomes and report the presence of all three classes of OBPs in the three mosquito genomes. In particular, we identified Atypical class of OBPs in C. quinquefasciatus. We further confirm that “Atypical OBPs” are composed of two domains that are homologous to Classic OBPs and provide in-depth characterization of their origin and structural features. This work also provides for a comprehensive and robust subclassification of the different OBP classes through structure-based alignments and phylogenetic analysis which could possibly reflect on the functional divergence of these proteins. We also provide a detailed primary structural and phylogenetic characterization of all these novel OBP subtypes. An extensive set of supplementary materials that detail our analyses and results are provided.
Results
Extension of OBPs Family in All Three Mosquito Genomes
In the already published works, 65 OBPs from A. gambiae (Vogt 2002; Xu et al. 2003; Zhou et al. 2004, 2008), 64 from Aedes aegyti (Zhou et al. 2008), and 53 OBPs from C. quinquefasciatus (Pelletier and Leal 2009) were previously identified. These OBPs have been characterized by these groups into three main subfamilies Classic, PlusC, and Atypical based on sequence features (fig. 1). Only very recently, Vieira and Rozas (2011) added four new putative genes to the A. gambiae OBP gene repertoire and 13 PlusC OBPs to the C. quinquefasciatus genome. These new genes were also identified by our sequence searches and bioinformatics analysis (see Materials and Methods) (table 1 and supplementary table S1a–e, Supplementary Material online). The fasta sequences of the identified genes are available for download as supplementary material.
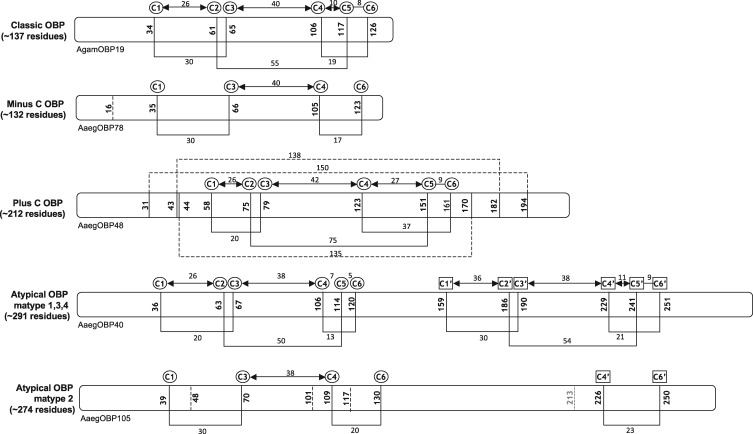
Fig. 1.—
Cysteine conservation patterns across the different subfamilies and subgroups of OBPs from Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus genomes. The six conserved cysteines in GOBP domain are denoted C1–C6. The six additional cysteines in the C-term of the Atypical OBPs are denoted C1′–C6′. The lines connecting cysteines represent the disulphide bonds and dotted lines represent the additional disulphide bonds in the PlusC OBPs.
Table 1.
Identification of OBPs in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus Genomes
| Subfamily |
|||||
|---|---|---|---|---|---|
| Classic | PlusC | Atypical | Not Determined | New Total | |
| A. gambiae | |||||
| Previously reporteda | 29 | 16 | 16 | 69 | |
| Newly identified | 4 | 4 | |||
| Aed. aegypti | |||||
| Previously reportedb | 33 | 17 | 14 | 111 | |
| Newly identified | 6 | 10 | 31 | ||
| C. quinquefasciatus | |||||
| Previously reportedc | 48 | 109 | |||
| Newly identified | 21 | 12 | 26 | 2 | |
Note.—The table shows statistics of previously and newly identified OBP members (AgamOBP65 to AgamOBP68, AaegOBP67 to AaegOBP114, CquiOBP54 to CquiOBP112) in all three mosquito genomes. Detailed results are provided in accompanying supplementary tables S1a–e, Supplementary Material online.
In this study, a major expansion is provided in the Atypical OBP subfamily of the mosquitoes where 31 new members (AaegOBP84 to AaegOBP114) are identified in Aed. aegypti which interestingly show high sequence similarities with the 26 (CquiOBP75–CquiOBP100) new Atypical members from the C. quinquefasciatus genome that are reported in this work (supplementary table S1c and d, Supplementary Material online).
In the Classic OBP subfamily, we have annotated six new members in the Aed. aegypti genome and 21 members in the C. quinquefasciatus genome. In addition to this, 10 new members have been added to the PlusC subfamily of the Aed. aegypti genome which sums up to the addition of 110 members to the OBP gene family of mosquitoes [which includes sequences identified by Vieira and Rozas (2011) and Pelletier and Leal (2011)].
Two-Domain OBPs and MinusC OBPs
Owing to the low sequence identity and length variations observed between the members of the OBP family, a structure-based alignment was used to align them (see Materials and Methods). It highly improved the quality of alignment compared with regular multiple sequence alignments namely for (i) the precise classification of the new OBPs into the three different subfamilies and (ii) the identification of residues in structurally conserved positions that would have been missed otherwise (supplementary fig. S3a–c, Supplementary Material online).
The conservation pattern of cysteines across the different classes were clearly highlighted in these structure-based alignments but could not be obtained otherwise with the ordinary sequence alignment methods. We further refer to the cysteine positions in this article by numbering them C1 to C6 with respect to the order of their positions in the Classic OBP proteins. A detailed schematic representation featuring the cysteine spacing and conservation together with their predicted disulphide patterns are given in figure 1. Overall, the six cysteine residues involved in disulphide bond formation, which are considered as the hallmark of this protein family (Calvo et al. 2002; Valenzuela et al. 2002; Calvo et al. 2006), are well conserved across the Classic, PlusC, and Atypical subclasses.
Interestingly, sequences that lack C2 and C5 cysteines were observed in the alignments. OBPs which lack these two particular cysteines, called the MinusC OBPs, have been characterized and expressed in other insect genomes such as Drosophila, Bombyx mori, Tribolium castaneum, and Apis mellifera (Vieira and Rosas 2011), but their presence in the mosquito genome has not been shown previously. AaegOBP78 from Aed. aegypti and 15 proteins from C. quinquefasciatus (CquiOBP59–CquiOBP62, CquiOBP64–CquiOBP74) were found to lack these two cysteines. As all these sequences retained the N-terminal signal peptide or the presence of the PBP/GOBP domain, they were retained in our analysis as MinusC OBPs (supplementary tables S3 and S4, Supplementary Material online).
We also observed interesting cysteine conservation patterns among the Atypical OBPs. The Atypical OBPs were previously described as proteins that hold a Classic OBP domain in the N-terminal end with an uncharacterized C-terminal domain. However, the close analysis of the extended C-terminal end of Atypical members highlighted the presence of six additional cysteines conserved within this subfamily, with a cysteine spacing pattern very similar to the conserved cysteines (C1–C6) at their N-terminal end. The observed cysteine conservation pattern in the case of the Atypcial OBPs is purely the reflection of the annotation of new members in this subfamily and has never been described before to our knowledge. We hence propose to annotate these cysteines as C1′–C6′. This remarkable conservation of cysteines is believed to hold important evolutionary information (Thangudu et al. 2005, 2008). Following this, we characterized the homologues of each of the two domains and identified their closest classic OBP homologue in their corresponding genomes and also the Drosophila genome which confirms that the Atypical OBPs are indeed “two-domain OBPs.” It is noteworthy that within the Atypical (two-domain) subfamily, a distinctive subtype called matype2 (see below and fig. 1) showed the presence of only six cysteines (C1, C3, C4, C6, C4′, and C6′), when compared with the other subtypes which carry the 12 cysteines. The Cys conservation pattern at the N-terminal domain of the OBP is similar to the MinusC OBPs; however, the C-terminal domain is found to have lost more cysteines comparatively.
Analysis of OBP Genes: Orthology across the Three Genomes and Their Corresponding Distribution
We investigated the orthology and gene distribution of OBPs in three genomes. Assembled genome is only available for A. gambiae at the date of this work in Ensembl Genomes and VectorBase 3.4 version. The chromosomal mapping for each of the OBP genes in Anopheles is hence known with precision (fig. 2). Their chromosomal distribution in the Anopheles genome is centrally featured in supplementary fig. S1a–e and further referenced in supplementary table S1a, Supplementary Material online. Though the syntenic relationship between the chromosome arms in A. gambiae and their corresponding orthologous chromosome arms in Culex and Aedes was established by Arensburger et al. (2010) with the help of genetic markers (supplementary table S2, Supplementary Material online), the genomic data of these two Culcinae species are only available in the form of supercontigs fragments (Nene et al. 2007; Arensburger et al. 2010) and are yet to be assembled. In these two genomes, a few supercontigs (about 10%) harbor markers that allow their chromosomal localization (Arensburger et al. 2010). Very few of these anchor supercontigs hosted OBP genes. Most supercontigs containing OBP genes did not harbor any genomic markers, hence cannot be assigned to a chromosome in Aedes and Culex. However, in many cases, direct orthologs in the Anopheles genome could be identified (fig. 2, supplementary fig. S1a–e and supplementary table S1a, c, and e, Supplementary Material online). OBP orthologs have been identified using the reciprocal BLAST hit approach (Moreno-Hagelsieb and Latimer 2008) which is widely used in the detection of orthologs. As illustrated in figures 2 and 3 and in supplementary figure S1a–e, Supplementary Material online, three-way orthology (1:1:1) between OBP genes in the three genomes were identified in 31 cases while two-way orthology (1:1) between OBP genes from only two genomes were identified in 5 cases between Anopheles and Culex, 6 between Anopheles and Aedes, and 19 between Aedes and Culex (fig. 3), thus confirming the genetic proximity between the Aed. aegypti and C. quinquefasciatus species. Our proposed analysis was found to be in complete agreement with the microsynteny analysis described very recently in Pelletier and Leal (2011), thus indicating that the orthology detected may serve as the basis of further syntenic analysis.
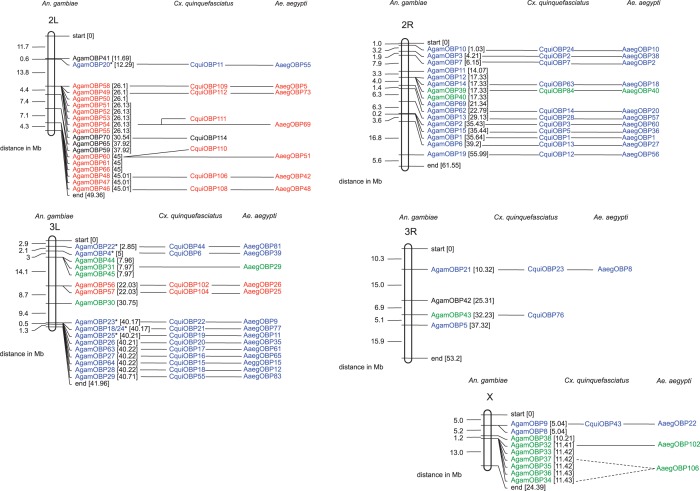
Fig. 2.—
Chromosomal localization of odorant binding proteins from Anopheles gambiae. Details about the gene names used here are shown in supplementary table S1a, Supplementary Material online. Shown to the right of the gene names are their direct three-way orthologs detected in the Aedes aegypti and Culex quinquefasciatus genomes by the reverse BLAST hit methodology. Classic OBPs are featured in blue, PlusC OBPs in red, and two-domain OBPs (or Atypical) in green. The chromosomes were drawn to scale in MapDraw software. The positions of and the distances between gene loci are indicated in megabases. When three-way orthology (1:1:1) was not detected but only two-way orthology (1:1), the two CquiOBP and AaegOBP orthologs are separately connected to the corresponding AgamOBP gene.
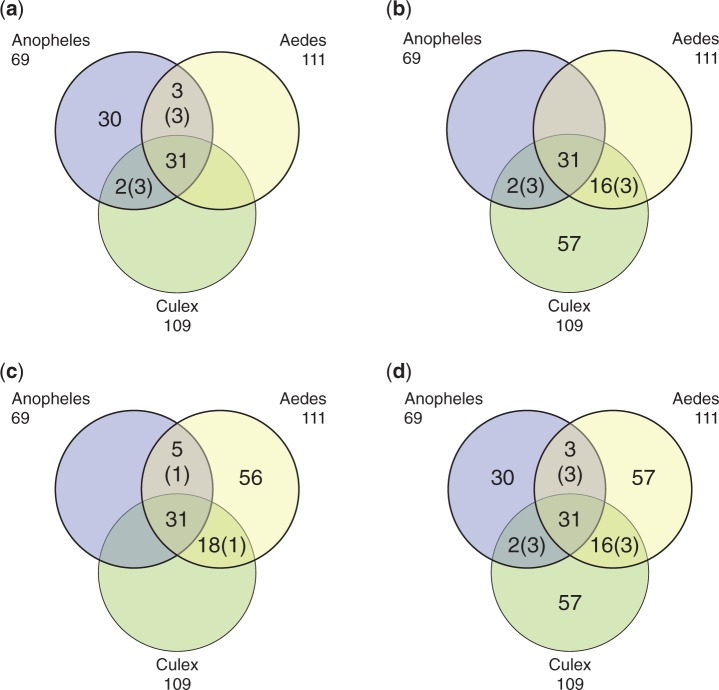
Fig. 3.—
Analysis of orthologous OBP genes shared across three mosquito species, Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. The Venn diagrams indicate the number of inferred orthologous genes shared among the mosquitoe species: (a) number of A. gambiae OBP genes orthologous to Aed. aegypti and C. quinquefasciatus; (b) number of Aed. aegypti OBP genes orthologous to A. gambiae and C. quinquefasciatus; (c) number of Culex OBP genes orthologous to A. gambiae and Aed. aegypti; (d) overall number of orthologous groups across the three mosquito species. The orthologs were identified using the reciprocal BLAST hit approach. The number of genes that share a three-way (1:1:1) orthology between the three species is 31. The number of genes in a species that have two-way orthology (1:1) with the two other species but not a three-way orthology is indicated between parenthesis and for a given species should be counted only once. For example, in (a), the total number of OBP genes in A. gambiae is 30 + 3 + 2 + 31 + (3) = 69, since three genes in A. gambiae have two-way orthology (1:1) with genes in both C. quinquefasciatus and Aed. aegypti but not a three-way orthology. Detailed listings of the orthology analysis are provided in supplementary table S1a, c, and e, Supplementary Material online.
Interestingly, the overwhelming majority of the OBP genes are organized in gene clusters in the three genomes (supplementary fig. S1a–e, Supplementary Material online). The clusters are mainly composed of gene duplicates. The genes in these genomic clusters hence share high sequence identity (data not shown) and are thereby phylogenetically very close (see below) as it is confirmed by inparalogy data from the inParanoid database (O’Brien et al. 2005). The extension of OBP gene repertoire in Aed. aegypti and C. quinquefasciatus with respect to A. gambiae was mainly driven by these gene duplication events which are more numerous in these two Culicinae species. There are a total of 12 OBP gene clusters in Aed. aegypti and 13 clusters in C. quinquefasciatus genomes when compared with 6 clusters in A. gambiae. The largest gene clusters are found in Aedes and Culex, and a few clusters contain as many as 12 genes. It is observed that 21 out of the 26 newly identified Atypical (two-domain) OBPs genes from C. quinquefasciatus are in fact distributed into three main gene clusters (fig. 2 and supplementary fig. S1a–e, Supplementary Material online). Similarly, 10 out of the 12 newly identified PlusC proteins are distributed into three gene clusters.
Phylogeny-Based OBP Clusters
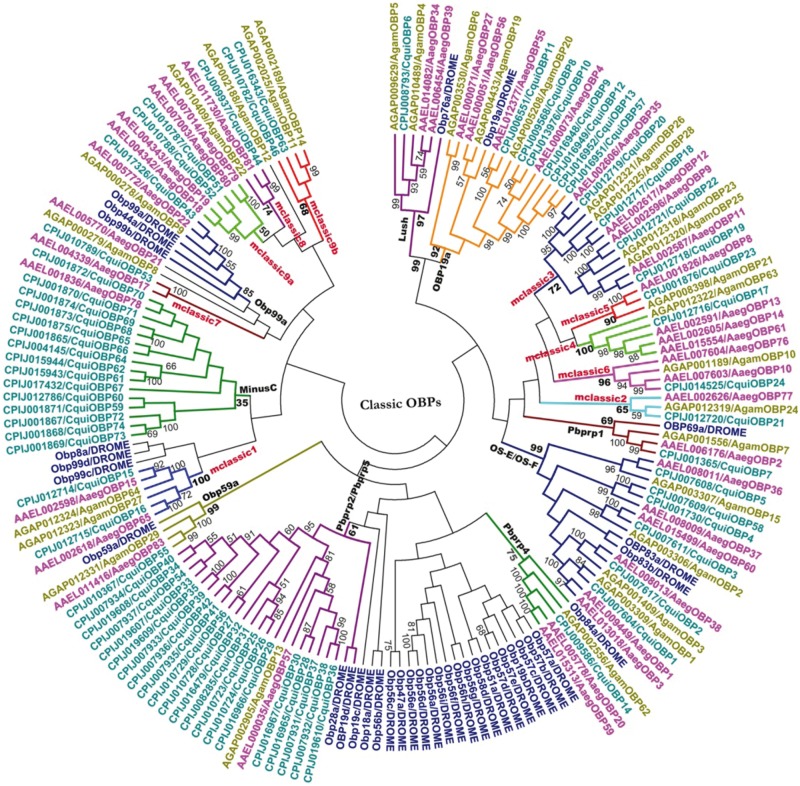
As expected and as already reported previously, OBP family members showed high divergence. The average sequence identity between OBP genes in A. gambiae, Aed. aegypti, and C. quinquefasciatus are 12.5%, 12.8%, and 13.1%, respectively, and their phylogenetic tree (see Material and Methods) also indicated a high sequence divergence (supplementary fig. S2a–c, Supplementary Material online). However, the comparative analysis of the different subfamilies of the OBPs in the mosquito genome provided more meaningful clustering patterns within each subfamily of the OBP members. The analysis was done based on the sequence alignment and phylogenetic trees constructed using sequences from individual subfamilies from all the three mosquito genomes used in this analysis and the Drosophila OBPs (Hekmat-Scafe et al. 2002) in the case of the Classic members. A bootstrap consensus tree was constructed using the neighbor joining method (Saitou and Nei 1987) with all the Classic OBPs from the three mosquito genomes and the D. melanogaster with 1000 bootstrap replicates (fig. 4). The clustering of the various Classic OBPs into clusters based on significant bootstrap values (50% cutoff) revealed the possibility of 18 different subtypes. These clusters carried orthologous and paralogous sequences from the three genomes. Few members of the mosquito genomes clustered with Drosophila OBPs (Hekmat-Scafe et al. 2002), and these clusters were named after their closest Drosophila OBPs. Among these OS-E/OS-F, Pbprp1, LUSH, OBP19a, and Pbprp4 have already been described previously (Xu et al. 2003; Zhou et al. 2008; Pelletier and Leal 2009). However, one member from C. quinquefasciatus in each of the two subtypes OS-E/OS-F (CquiOBP58) and OBP19a (CquiOBP57) have been annotated. The huge expansion of sequences (CquiOBP25–CquiOBP42) observed by Pelletier and Leal (2009) were found to be homologous to AaegOBP57 and AgamOBP13 and were indeed closely related to the Pbprp2/Pbprp5 of Drosophila. CquiOBP55 and AaegOBP83 identified in this analysis are orthologs of AgamOBP29 and homologous to OBP59a of Drosophila and have an unusually long sequence as recently mentioned by Vieira and Rozas (2011). Clustering of three orthologous OBP sequences AgamOBP9, AaegOBP22, and CquiOBP43 with the Drosophila MinusC members OBP99a, OBP44a, and OBP99b was observed with a considerable bootstrap support, among which OBP99a alone retains all the six cysteines, while the two others lack the C2 and C5 cysteines (see below). Among the Drosophila MinusC OBPs, three members of the MinusC subfamily (Obp83f, Obp99a, and Obp99d) retain all six conserved cysteines, whereas four members of the subfamily (Obp8a, Obp44a, Obp99b, and Obp99c) have C2 and C5 cysteines lacking. Therefore, the mosquito OBPs, which cluster with these Drosophila OBPs, do not represent true MinusC OBPs. The other clusters which do not have a close Drosophila homologue are named as mclassic1–9 (fig. 4 and supplementary fig. S3a, Supplementary Material online). In addition to these subtypes, one group, displaying outstanding sequence features (supplementary fig. S3a, Supplementary Material online) with 16 members lacking C2 and C5 cysteines, has been named as “Bombyx mori MinusC” due to their homology with the B. mori MinusC sequences though its branch holds a bootstrap value of only 35%. This homology was determined using BLAST analysis and confirmed with the inParanoid eukaryotic ortholog database (O’Brien et al. 2005). Other subtype classifications of the Classic members were also similar to the clustering seen in the inParanoid database.
Fig. 4.—
Unrooted phylogenetic tree of Classic OBPs in the three mosquito genomes and in Drosophila melanogaster. The Anopheles gambiae, Aedes Aegypti, and Culex quinquefasciatus members are colored in mustard, pink, and turquoise, respectively. The bootstrap values of the branches are indicated on the nodes in percentage values. The names of identified clusters inside the Classic OBPs subfamily are indicated on the branches. Detailed alignments of the members inside each cluster are provided in supplementary figure S3a, Supplementary Material online.
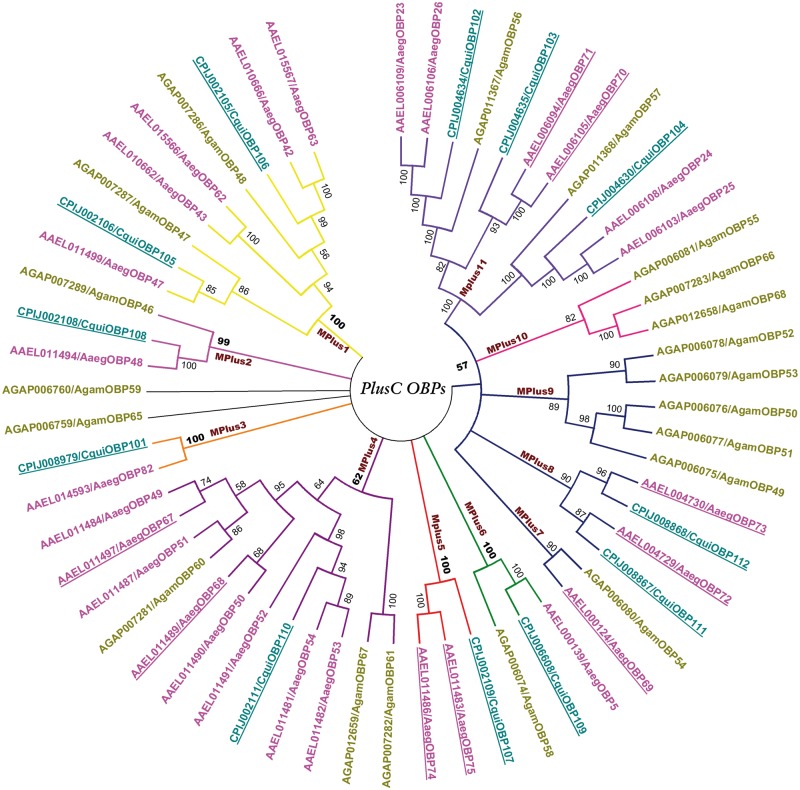
As shown in figure 5, the PlusC OBPs clustered as seven major phylogenetic clusters based on bootstrap cutoff value of 50%, but we further subdivided them into 11 subtypes (mplus1–mplus11). Indeed, though the interior node of mplus7–11 cluster hold a bootstrap value of 57%, we separated them as different subtypes because they clearly hold distinct sequence features (supplementary fig. S3b, Supplementary Material online). Furthermore, analysis of chromosomal localization of PlusC members from A. gambiae shows that mplus11 subtype members are specific to chromosome 3L while all other PlusC OBPs were specifically distributed on chromosome 2L. At this stage, it is difficult to interpret the molecular background behind this clustering.
Fig. 5.—
Unrooted phylogenetic tree of PlusC OBPs in the three mosquito genomes. The Anopheles gambiae, Aedes Aegypti, and Culex quinquefasciatus members are colored in mustard, pink, and turquoise, respectively. The bootstrap values of the branches are indicated on the nodes in percentage values. The names of identified clusters inside the PlusC OBPs subfamily are indicated on the branches. Detailed alignments of the members inside each cluster are provided in supplementary figure S3b, Supplementary Material online.
The Classic subfamily members from the three genomes share an average sequence identity of 15.5%, while the PlusC OBPs share 17.3% average sequence identity. No distinct sequence features could be observed at the subfamily level (Classic, PlusC, and Atypical) because of high sequence divergence. Nevertheless, a close examination of the alignments for the different clusters which contain orthologous sequences from the three genomes within each subfamily indicates that the phylogenetic clusters established in this study tend to have specific sequence patterns (supplementary fig. S3a and b, Supplementary Material online). Some subgroups are characterized by a very low average sequence identity like the B. mori MinusC subgroup within the Classic OBPs (21.5%), the mclassic9 (23.3%), or the mplus9 (24.3%), while other subgroups share significantly higher sequence identities like OS-E/OS-F (55.2%), Pbprp4 (60.2%), or mclassic4 (77.3%).
Sequence Specific Clustering of Two-Domain OBPs
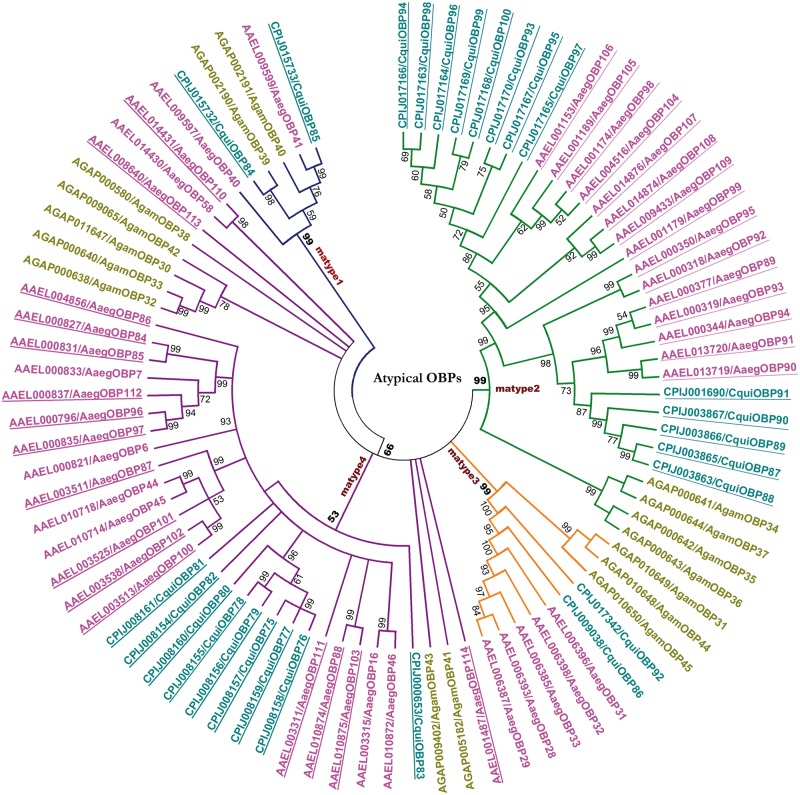
The Atypical OBPs, unlike the Classic members, formed four major clusters based on bootstrap values which are named in this study matype1–matype4 (fig. 6) and showed distinct sequence features (supplementary fig. S3c, Supplementary Material online). The matype1 forms the smallest cluster among the four subtypes with two members from each genome, and this cluster is separated from the other three subtypes with high bootstrap values. The matype2 forms a distinctive type of Atypical members holding only a total of six cysteines (C1, C3, C4, C5, C1′, and C6′) out of the 12 conserved cysteines characteristic of the other subtypes of this subfamily (fig. 1 and supplementary fig. S3c, Supplementary Material online). The matype2 still features to stand as a distinctive type with the presence of cysteines in the N-terminal domain lacking C2–C5 as previously described. The matype4 members unanimously hold a deletion of about 15 resides between the C1 and C2 which stands as the distinguishing feature of this subtype. The matype1 members are orthologous to AgamOBP39 that is located on chromosome 2R which is otherwise populated with Classic members supporting their close relation to the Classic members as observed in the phylogeny of the individual genomes. The matype2 members, intriguingly, share orthology with corresponding OBPs from A. gambiae that were mapped to chromosome X, whereas matype3 and matype4 members were sharing orthology with AgamOBPs distributed over chromosomes 3R and 3L.
Fig. 6.—
Unrooted phylogenetic tree of Atypical odorant binding proteins in the three mosquito genomes. The Anopheles gambiae, Aedes Aegypti, and Culex quinquefasciatus members are colored in mustard, pink, and turquoise, respectively. The bootstrap values of the branches are indicated on the nodes in percentage values. The names of identified clusters inside the Atypical OBPs subfamily are indicated on the branches. Detailed alignments of the members inside each cluster are provided in supplementary figure S3c, Supplementary Material online.
Discussion
Evolutionary Aspects of OBP Gene Family in Mosquitoes
C. quinquefaciatus (Arensburger et al. 2010) and Aed. aegypti genomes (Nene et al. 2007) which code for 109 and 111 OBPs, respectively, have a significantly larger OBP genes repertoire than the related A. gambiae genome which harbors only 67 OBPs. A. gambiae belongs to the Anophelinae and Aed. aegypti and C. quinquefasciatus belong to the Culicinae subfamilies. These two subfamilies of mosquitoes are estimated to have diverged around ∼120 My (Reidenbach et al. 2009). The increase in the number of genes indicates lineage-specific expansions of the OBP gene family among the mosquito species. The odorant binding gene family has been previously shown to adopt a birth and death model of evolution based on a number of factors which includes several gene gain and loss events in lineages, decrease in the number of orthology groups with increasing divergence times, and an uneven phylogenetic subfamily distribution across species (Vieira and Rozas 2011). Similar observations are made with respect to the OBP genes in Anopheles, Culex, and Aedes species, where a number of gene gain is observed in the Aedes and Culex species, uneven distribution of subfamilies is observed (where the MinusC Subfamily is absent in Anopheles), and the number of orthologous sequences are higher in the Aedes and Culex species and becomes lesser with respect to the Anopheles which is distantly related to these two species comparatively. This provides further support to the already existing fact that the OBP gene family undergoes a birth and death model of evolution. Furthermore, the appearance of new genes and subfamilies in the Aedes and Culex could relate to the requirement of these genes for environmental adaptations by these species.
Evolution of MinusC Proteins in the Mosquito Genomes
The MinusC subfamily of OBPs was first identified in the Drosophila genome with some of its members lacking the second and fifth cysteine residues (Hekmat-Scafe et al. 2002) and later identified in other species which includes the Apis millefera (Forêt and Maleszka 2006) and B. mori (Gong et al. 2009 and Yoshizawa et al. 2011). In the case of mosquitoes, interestingly, MinusC OBPs are not present in the A. gambiae. But the MinusC OBPs appeared in Culicinae lineage of the mosquito OBPs. The close homology of these MinusC members with the B. mori MinusC OBPs suggests that they could have a common ancestor. Thus, it can be said that the MinusC OBPS appeared in the Endopterygota lineage of insects which is dated back to ∼300 My and suggest that these OBPs appeared earlier in the evolution and not only in the Drosophilidae, Bombyx/Tribolium, and Apis lineages as believed earlier (Vieira and Rozas 2011). However, the absence of these OBPs in the A. gambiae is intriguing and suggests that they could have species-specific expansions. This in fact supports the birth and death model of evolution observed in the OBP family of proteins. Separately, the matype2 members belonging to the two-domain OBP subfamily, which retain only six cysteines, interestingly lack the C2 and C5 cysteines in the N-terminal domain. The absence of C2 and C5 cysteines being the characteristic feature of the MinusC proteins lays an important question on the evolutionary link between these members.
Atypical OBPs are Indeed Two-Domain OBPs
The increase in the number of Atypical OBPs in the three mosquito genome revealed important facets in this subfamily of proteins. We have shown that the Atypical OBPs so far identified only in the mosquitoes are indeed two-domain OBPs. This was also reported by Vieira and Rozas (2011), based on their phylogenetic analysis that they belong to the dimer OBP clade. We further provide evidence to this by characterizing each of the Atypical OBP domains by their closest homologue in the Classic OBP subfamily in their corresponding genomes. Very interestingly, the Classic OBP members, obtained as hits by each of these domains, were mainly found among the mclassic9, mclassic8, and Obp99a members (table 2). Atypical OBPs indeed are found to share closer phylogenetic proximity to OBPs from the mclassic9, mclassic8, and OBP99a phylogenetic clusters however not with significant bootstrap value because of sequence divergence (supplementary fig. S2a–c, Supplementary Material online). Moreover, Atypical gene clusters in A. gambiae are localized in close proximity to gene clusters that contains Classic OBPs from one of these three groups at the chromosomal level. On chromosome 2R, matype1 members AgamOBP39 and AgamOBP40 are localized at the level of the same gene cluster as the mclassic9 members AgamOBP11, AgamOBP12, and AgamOBP14. Likewise, on chromosome X, the matype2 OBPs AgamOBP34 to AgamOBP37 are localized proximal to AgamOBP8 and AgamOBP9 that belong to the Obp99a phylogenetic cluster. Similarly, on chromosome 3L, the matype3 AgamOBP31, AgamOBP44, and AgamOBP45 that form a gene cluster are in close proximity to AgamOBP22 which belongs to the mclassic8 group. Another interesting observation is that OBP members from these three phylogenetic clusters (mclassic9, mclassic8, and Obp99a) are closely related to the MinusC group of proteins in the Drosophila genome (fig. 4), and it has been established that the Drosophila Dimer OBPs 83cd and 83ef (Zhou et al. 2004), which are proteins that hold two OBP domains, are related to these Drosophilidae MinusC proteins.
Table 2.
Analysis of the Two Putative OBP Domains (N-term and C-term) of Atypical OBPs from Anopheles gambiae, Aedes Aegypti, and Culex quinquefasciatus
| Mosquito Atypical OBP |
Mosquito Classic OBP Closest Homologues |
Drosophila OBP Closest Homologues |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Phylogenetic Subgroup | N-term | Phylogenetic Subgroup | E-value | C-term | Phylogenetic Subgroup | E-value | N-term | E-value | C-term | E-value |
| AAEL009597 | matype1 | AAEL005772 | Obp99a | 1e-10 | AAEL004342 | mclassic9a | 2e-14 | Obp99b | 5e-11 | Obp99a | 5e-09 |
| AaegOBP40 | AaegOBP22 | AaegOBP18 | |||||||||
| AAEL009599 | matype1 | AAEL005772 | Obp99a | 1e-10 | AAEL007014 | No group | 2e-04 | Obp99a | 9e-08 | Obp99a | 8e-04 |
| AaegOBP41 | AaegOBP22 | AaegOBP79 | |||||||||
| AGAP002190 | matype1 | AGAP000278 | OBP99a | 1e-10 | AGAP002189 | mclassic9b | 2e-15 | Obp99b | 6e-09 | Obp99a | 5e-09 |
| AgamOBP39 | AgamOBP9 | AgamOBP14 | |||||||||
| AGAP002191 | matype1 | AGAP002188 | No group | 2e-08 | — | — | — | Obp44a | 7e-07 | Obp99a | 5e-08 |
| AgamOBP40 | AgamOBP12 | ||||||||||
| AGAP011647 | matype1 | AGAP010409 | mclassic8 | 2e-11 | AGAP002025 | mclassic9b | 5e-09 | Obp99a | 7e-09 | Obp99a | 4e-03 |
| AgamOBP30 | AgamOBP22 | AgamOBP11 | |||||||||
| CPIJ015732 | matype1 | CPIJ010787 | mclassic9a | 2e-10 | CPIJ016343 | mclassic9b | 2e-17 | Obp99b | 4e-07 | Obp99a | 9e-10 |
| CquiOBP85 | CquiOBP51 | CquiOBP63 | |||||||||
| CPIJ015733 | matype1 | CPIJ010787 | mclassic9a | 1e-10 | CPIJ010782 | maclassic9b | 4e-04 | Obp44a | 3e-07 | Obp99a | 3e-07 |
| CquiOBP86 | CquiOBP51 | CquiOBP46 | |||||||||
| AAEL000318 | matype2 | AAEL007003 | No group | 5e-07 | AAEL004342 | mclassic9a | 1e-03 | Obp44a | 2e-04 | Obp99c | 1e-04 |
| AaegOBP92 | AaegOBP80 | AaegOBP18 | |||||||||
| AAEL000319 | matype2 | AAEL002617 | mclassic3a | 2e-03 | AAEL007014 | No group | 3e-04 | Obp44a | 4e-03 | — | — |
| AaegOBP93 | AaegOBP12 | AaegOBP79 | |||||||||
| AAEL000344 | matype2 | AAEL007003 | No group | 1e-04 | AAEL007014 | No group | 4e-02 | Obp44a | 4e-06 | Obp99b | 3e-03 |
| AaegOBP94 | AaegOBP80 | AaegOBP79 | |||||||||
| AAEL000350 | matype2 | AAEL011730 | mclassic8 | 3e-05 | AAEL002587 | mclassic3b | 1e-04 | Obp56d | 2e-03 | Obp99b | 5e-07 |
| AaegOBP95 | AaegOBP81 | AaegOBP11 | |||||||||
| AAEL000377 | matype2 | AAEL004343 | mclassic9a | 4e-06 | AAEL007014 | No group | 1e-03 | Obp44a | 3e-09 | Obp99b | 7e-04 |
| AaegOBP89 | AaegOBP19 | AaegOBP79 | |||||||||
| AAEL001153 | matype2 | AAEL007003 | No group | 7e-05 | AAEL013018 | OS-E/OS-F | 3e-02 | Obp99c | 1e-05 | — | — |
| AaegOBP106 | AaegOBP80 | AaegOBP3 | |||||||||
| AAEL001174 | matype2 | AAEL004343 | mclassic9a | 4e-07 | AAEL004343 | mclassic9a | 9e-07 | Obp44a | 7e-05 | Obp44a | 3e-05 |
| AaegOBP98 | AaegOBP19 | AaegOBP19 | |||||||||
| AAEL001179 | matype2 | AAEL004343 | mclassic9a | 8e-08 | AAEL007014 | No group | 1e-02 | Obp99b | 5e-07 | Obp44a | 8e-05 |
| AaegOBP99 | AaegOBP19 | AaegOBP79 | |||||||||
| AAEL001189 | matype2 | AAEL004343 | mclassic9a | 8e-07 | AAEL007003 | No group | 8e-04 | Obp44a | 1e-04 | Obp44a | 2e-03 |
| AaegOBP105 | AaegOBP19 | AaegOBP80 | |||||||||
| AAEL004516 | matype2 | AAEL004343 | mclassic9a | 2e-04 | — | — | — | Obp44a | 4e-04 | — | — |
| AaegOBP104 | AaegOBP19 | ||||||||||
| AAEL009433 | matype2 | AAEL004343 | mclassic9a | 2e-06 | — | — | — | Obp99c | 0.002 | — | — |
| AaegOBP109 | AaegOBP19 | ||||||||||
| AAEL013719 | matype2 | — | — | — | AAEL004343 | mclassic9a | 1e-03 | — | — | — | — |
| AegOBP90 | AaegOBP19 | ||||||||||
| AAEL013720 | matype2 | AAEL007003 | No group | 2e-06 | AAEL004343 | mclassic9a | 1e-04 | Obp44a | 1e-03 | — | — |
| AaegOBP91 | AaegOBP80 | AaegOBP19 | |||||||||
| AAEL014874 | matype2 | AAEL004343 | mclassic9a | 2e-06 | — | — | — | OBP99c | 2e-03 | — | — |
| AaegOBP108 | AaegOBP19 | ||||||||||
| AAEL014876 | matype2 | AAEL011730 | mclassic8 | 3e-09 | — | — | — | Obp99c | 1e-05 | — | — |
| AaegOBP107 | AaegOBP81 | ||||||||||
| AGAP000641/644 | matype2 | AGAP013182 | ND | 3e-09 | AGAP002025 | mclassic9b | 4e-10 | Pbprp2 | 1e-04 | Pbprp1 | 5e-04 |
| AgamOBP34/37 | AgamOBP59 | AgamOBP11 | |||||||||
| AGAP000642 | matype2 | AGAP013182 | ND | 2e-08 | AGAP002025 | mclassic9b | 2e-06 | Obp56d | 1e-05 | — | — |
| AgamOBP35 | AgamOBP59 | AgamOBP11 | |||||||||
| AGAP000643 | matype2 | AGAP013182 | ND | 8e-09 | AGAP002025 | mclassic9b | 2e-06 | Obp56d | 1e-05 | — | — |
| AgamOBP36 | AgamOBP59 | AgamOBP11 | |||||||||
| CPIJ001690 | matype2 | CPIJ009937 | mclassic8 | 2e-07 | — | — | — | Obp99a | 4e-07 | — | — |
| CquiOBP92 | CquiOBP44 | ||||||||||
| CPIJ003863 | matype2 | CPIJ009937 | mclassic8 | 2e-05 | CPIJ010782 | maclassic9b | 4e-02 | Obp44a | 4e-07 | — | — |
| CquiOBP89 | CquiOBP44 | CquiOBP46 | |||||||||
| CPIJ003865 | matype2 | CPIJ009937 | mclassic8 | 4e-06 | CPIJ010782 | maclassic9b | 2e-02 | Obp44a | 7e-07 | — | — |
| CquiOBP88 | CquiOBP44 | CquiOBP46 | |||||||||
| CPIJ003866 | matype2 | CPIJ009937 | mclassic8 | 3e-07 | CPIJ010782 | maclassic9b | 1e-04 | Obp44a | 1e-09 | Obp99b | 1e-03 |
| CquiOBP90 | CquiOBP44 | CquiOBP46 | |||||||||
| CPIJ003867 | matype2 | CPIJ009937 | mclassic8 | 1e-09 | CPIJ010782 | maclassic9b | 4e-02 | OBP99a | 9e-07 | ||
| CquiOBP91 | CquiOBP44 | CquiOBP46 | |||||||||
| CPIJ017163 | matype2 | CPIJ009937 | mclassic8 | 2e-06 | CPIJ017326 | Obp99a | 6e-02 | Obp44a | 4e-04 | Obp56g | 4e-03 |
| CquiOBP99 | CquiOBP44 | CquiOBP43 | |||||||||
| CPIJ017164 | matype2 | CPIJ009937 | mclassic8 | 6e-06 | CPIJ010789 | mclassic7 | 2e-02 | Obp44a | 2e-04 | — | — |
| CquiOBP97 | CquiOBP44 | CquiOBP53 | |||||||||
| CPIJ017165 | matype2 | CPIJ009937 | mclassic8 | 7e-10 | CPIJ010782 | maclassic9b | 1e-02 | Obp99c | 2e-04 | — | — |
| CquiOBP98 | CquiOBP44 | CquiOBP46 | |||||||||
| CPIJ017166 | matype2 | CPIJ009937 | mclassic8 | 1e-03 | CPIJ016343 | mclassic9b | 3e-02 | Obp44a | 1e-04 | — | — |
| CquiOBP95 | CquiOBP44 | CquiOBP63 | |||||||||
| CPIJ017167 | matype2 | CPIJ009937 | mclassic8 | 1e-03 | CPIJ001365 | Pbprp1 | 1e-02 | Obp44a | 4e-04 | Obp56d | 7e-05 |
| CquiOBP96 | CquiOBP44 | CquiOBP7 | |||||||||
| CPIJ017168 | matype2 | CPIJ016951 | Obp19a | 6e-04 | CPIJ016343 | mclassic9b | 3e-02 | — | — | — | — |
| CquiOBP101 | CquiOBP57 | CquiOBP63 | |||||||||
| CPIJ017169 | matype2 | CPIJ009937 | mclassic8 | 8e-03 | CPIJ010789 | mclassic7 | 2e-02 | Obp44a | 3e-03 | Obp99b | 2e-03 |
| CquiOBP100 | CquiOBP44 | CquiOBP53 | |||||||||
| CPIJ017170 | matype2 | CPIJ009937 | mclassic8 | 3e-02 | CPIJ016343 | mclassic9b | 6e-03 | Obp44a | 6e-03 | — | — |
| CquiOBP94 | CquiOBP44 | CquiOBP63 | |||||||||
| AAEL006385 | matype3 | AAEL002596 | mclassic3a | 8e-04 | AAEL004343 | mclassic9a | 2e-06 | Obp56d | 8e-04 | Obp99a | 9e-08 |
| AaegOBP33 | AAEgOBP9 | AaegOBP19 | |||||||||
| AAEL006387 | matype3 | AAEL002617 | mclassic3a | 1e-05 | AAEL004343 | mclassic9a | 3e-06 | Obp56d | 3e-05 | Obp99a | 5e-07 |
| AaegOBP29 | AaegOBP12 | AaegOBP19 | |||||||||
| AAEL006393 | matype3 | AAEL002617 | mclassic3a | 1e-05 | AAEL004343 | mclassic9a | 3e-06 | Obp56d | 3e-05 | Obp99a | 5e-07 |
| AaegOBP28 | AaegOBP12 | AaegOBP19 | |||||||||
| AAEL006396 | matype3 | AAEL002617 | mclassic3a | 1e-05 | AAEL004342 | mclassic9a | 2e-03 | Obp56d | 1e-05 | Obp99a | 4e-05 |
| AaegOBP31 | AaegOBP12 | AaegOBP18 | |||||||||
| AAEL006398 | matype3 | AAEL011730 | mclassic8 | 2e-02 | AAEL011730 | mclassic8 | 1e-06 | Obp56d | 9e-03 | Obp99a | 1e-06 |
| AaegOBP32 | AaegOBP81 | AaegOBP81 | |||||||||
| AGAP010648 | matype3 | AGAP002025 | mclassic9b | 1e-10 | AGAP002025 | mclassic9b | 1e-08 | Obp99a | 2e-05 | Obp99b | 5e-04 |
| AgamOBP44 | AgamOBP11 | AgamOBP11 | |||||||||
| AGAP010649 | matype3 | AGAP013182 | ND | 5e-09 | AGAP002025 | mclassic9b | 1e-12 | Obp99a | 8e-07 | Obp99b | 6e-08 |
| AgamOBP31 | AgamOBP59 | AgamOBP11 | |||||||||
| AGAP010650 | matype3 | AGAP013182 | ND | 1e-11 | AGAP002189 | mclassic9b | 3e-11 | Obp99b | 9e-05 | Obp99a | 3e-10 |
| AgamOBP45 | AgamOBP59 | AgamOBP14 | |||||||||
| CPIJ009038 | matype3 | CPIJ009937 | mclassic8 | 9e-08 | CPIJ009937 | mclassic8 | 7e-08 | Obp56d | 2e-03 | Obp99a | 1e-10 |
| CquiOBP87 | CquiOBP44 | CquiOBP44 | |||||||||
| CPIJ017342 | matype3 | CPIJ009937 | mclassic8 | 2e-08 | CPIJ006551 | Obp19a | 2e-05 | Obp56c | 7e-03 | Obp99b | 1e-08 |
| CquiOBP93 | CquiOBP44 | CquiOBP11 | |||||||||
| AAEL000796 | matype4 | AAEL011730 | mclassic8 | 8e-04 | AAEL004343 | mclassic9a | 6e-07 | — | — | Obp56i | 1e-05 |
| AaegOBP96 | AaegOBP81 | AaegOBP19 | |||||||||
| AAEL000821 | matype4 | AAEL011730 | mclassic8 | 1e-05 | AAEL005770 | Obp99a | 1e-06 | — | — | — | — |
| AaegOBP6 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL000827 | matype4 | AAEL007014 | No group | 4e-03 | AAEL004342 | mclassic9a | 4e-05 | — | — | Obp99a | 5e-05 |
| AaegOBP84 | AaegOBP79 | AaegOBP18 | |||||||||
| AAEL000831 | matype4 | AAEL011730 | mclassic8 | 1e-02 | AAEL002596 | mclassic3a | 3e-05 | — | — | Obp56g | 1e-04 |
| AaegOBP85 | AaegOBP81 | AaegOBP9 | |||||||||
| AAEL000833 | matype4 | AAEL004339 | mclassic7 | 4e-03 | AAEL004343 | mclassic9a | 8e-05 | — | — | Obp99d | 5e-05 |
| AaegOBP7 | AaegOBP17 | AaegOBP19 | |||||||||
| AAEL000835 | matype4 | AAEL011730 | mclassic8 | 8e-03 | AAEL004343 | mclassic9a | 6e-07 | — | — | Obp56i | 1e-05 |
| AaegOBP97 | AaegOBP81 | AaegOBP19 | |||||||||
| AAEL000837 | matype4 | — | — | — | AAEL011730 | mclassic8 | 6e-08 | — | — | Pbprp2 | 7e-04 |
| AaegOBP112 | AaegOBP81 | ||||||||||
| AAEL001487 | matype4 | AAEL011730 | mclassic8 | 4e-03 | — | — | — | Obp51a | 1e-02 | — | — |
| AaegOBP114 | AaegOBP81 | ||||||||||
| AAEL003311 | matype4 | AAEL011730 | mclassic8 | 7e-05 | AAEL002596 | mclassic3a | 2e-04 | — | — | Obp99b | 1e-03 |
| AaegOBP111 | AaegOBP81 | AaegOBP9 | |||||||||
| AAEL003315 | matype4 | AAEL011730 | mclassic8 | 2e-08 | AAEL005770 | Obp99a | 2e-04 | — | — | Obp99c | 4e-03 |
| AaegOBP16 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL003511 | matype4 | AAEL011730 | mclassic8 | 2e-04 | AAEL004343 | mclassic9a | 1e-06 | — | — | Obp99a | 2e-05 |
| AaegOBP87 | AaegOBP81 | AaegOBP19 | |||||||||
| AAEL003513 | matype4 | AAEL011730 | mclassic8 | 5e-07 | AAEL005770 | Obp99a | 1e-07 | — | — | Obp99a | 2e-05 |
| AaegOBP100 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL003525 | matype4 | AAEL011730 | mclassic8 | 2e-03 | AAEL005770 | Obp99a | 4e-07 | — | — | Obp99a | 2e-04 |
| AaegOBP101 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL003538 | matype4 | AAEL011730 | mclassic8 | 5e-07 | AAEL005770 | Obp99a | 1e-07 | — | — | OBP99a | 2e-05 |
| AaegOBP102 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL004856 | matype4 | AAEL011730 | mclassic8 | 3e-05 | AAEL007014 | No group | 6e-06 | — | — | Obp99a | 1e-04 |
| AaegOBP86 | AaegOBP81 | AaegOBP79 | |||||||||
| AAEL010714 | matype4 | AAEL011730 | mclassic8 | 3e-05 | AAEL005770 | Obp99a | 2e-07 | — | — | Obp99a | 8e-06 |
| AaegOBP45 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL010718 | matype4 | AAEL011730 | mclassic8 | 5e-07 | AAEL005770 | Obp99a | 2e-06 | Obp56g | 6e-03 | Obp99a | 1e-04 |
| AaegOBP44 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL010872 | matype4 | AAEL011730 | mclassic8 | 9e-05 | AAEL004342 | mclassic9a | 5e-05 | — | — | Pbprp5 | 8e-04 |
| AaegOBP46 | AaegOBP81 | AaegOBP18 | |||||||||
| AAEL010874 | matype4 | AAEL011730 | mclassic8 | 3e-06 | AAEL005770 | Obp99a | 3e-06 | — | — | Obp99b | 3e-05 |
| AaegOBP88 | AaegOBP81 | AaegOBP21 | |||||||||
| AAEL010875 | matype4 | AAEL011730 | mclassic8 | 3e-06 | AAEL005770 | Obp99a | 2e-05 | — | — | Obp99d | 7e-05 |
| AaegOBP103 | AaegOBP81 | AaegOBP21 | |||||||||
| CPIJ000653 | matype4 | — | — | — | CPIJ016343 | maclassic9b | 2e-06 | — | — | Obp99b | 3e-04 |
| CquiOBP84 | CquiOBP63 | ||||||||||
| CPIJ008154 | matype4 | CPIJ014525 | maclassic6 | 1e-02 | CPIJ016343 | mclassic9b | 7e-05 | — | — | — | — |
| CquiOBP83 | CquiOBP24 | CquiOBP63 | |||||||||
| CPIJ008155 | matype4 | CPIJ010789 | mclassic7 | 2e-03 | CPIJ016343 | mclassic9b | 1e-06 | — | — | Obp99a | 2e-04 |
| CquiOBP79 | CquiOBP53 | CquiOBP63 | |||||||||
| CPIJ008156 | matype4 | CPIJ010789 | mclassic7 | 4e-02 | CPIJ010782 | maclassic9b | 2e-11 | — | — | Obp99a | 2e-08 |
| CquiOBP80 | CquiOBP53 | CquiOBP46 | |||||||||
| CPIJ008157 | matype4 | CPIJ010787 | mclassic9a | 1e-02 | CPIJ010782 | maclassic9b | 1e-09 | — | — | Obp99a | 6e-08 |
| CquiOBP76 | CquiOBP51 | CquiOBP46 | |||||||||
| CPIJ008158 | matype4 | CPIJ016343 | mclassic9b | 1e-08 | CPIJ016343 | mclassic9b | 8e-07 | Obp99a | 1e-05 | Obp99a | 3e-08 |
| CquiOBP77 | CquiOBP63 | CquiOBP63 | |||||||||
| CPIJ008159 | matype4 | CPIJ009937 | mclassic8 | 6e-03 | CPIJ016343 | mclassic9b | 4e-10 | — | — | Obp99a | 6e-09 |
| CquiOBP78 | CquiOBP44 | CquiOBP63 | |||||||||
| CPIJ008160 | matype4 | CPIJ009937 | mclassic8 | 1e-02 | CPIJ016343 | mclassic9b | 1e-09 | — | — | Obp44a | 3e-03 |
| CquiOBP81 | CquiOBP44 | CquiOBP63 | |||||||||
| CPIJ008161 | matype4 | CPIJ010789 | mclassic7 | 2e-02 | CPIJ016343 | mclassic9b | 2e-09 | — | — | Obp99b | 3e-06 |
| CquiOBP82 | CquiOBP53 | CquiOBP63 | |||||||||
| AAEL008640 | — | AAEL011730 | mclassic8 | 2e-08 | AAEL011730 | mclassic8 | 1e-05 | — | — | — | — |
| AaegOBP113 | AaegOBP81 | AaegOBP81 | |||||||||
| AAEL014430 | — | AAEL007003 | No group | 2e-08 | AAEL011730 | mclassic8 | 2e-05 | Obp99c | 3e-07 | Obp44a | 1e-06 |
| AaegOBP58 | AaegOBP80 | AaegOBP81 | |||||||||
| AAEL014431 | — | AAEL011730 | mclassic8 | 4e-12 | AAEL004342 | mclassic9a | 9e-09 | Obp99b | 1e-07 | Obp99b | 3e-03 |
| AaegOBP110 | AaegOBP81 | AaegOBP18 | |||||||||
| AGAP000580 | — | AGAP002189 | mclassic9b | 2e-06 | AGAP002025 | mclassic9b | 4e-05 | Obp99b | 6e-05 | Obp99c | 2e-06 |
| AgamOBP38 | AgamOBP14 | AgamOBP11 | |||||||||
| AGAP000638 | — | AGAP010409 | mclassic8 | 2e-11 | AGAP002025 | mclassic9b | 8e-07 | Obp99a | 1e-06 | Obp99c | 1e-03 |
| AgamOBP32 | AgamOBP22 | AgamOBP11 | |||||||||
| AGAP000640 | — | AGAP010409 | mclassic8 | 2e-11 | AGAP002025 | mclassic9b | 8e-07 | Obp99a | 1e-06 | Obp99c | 1e-03 |
| AgamOBP33 | AgamOBP22 | AgamOBP11 | |||||||||
| AGAP005182 | — | AGAP013182 | ND | 8e-07 | AGAP002025 | mclassic9b | 7e-11 | — | — | Obp44a | 2e-04 |
| AgamOBP41 | AgamOBP59 | AgamOBP11 | |||||||||
| AGAP009065 | — | AGAP013182 | ND | 6e-10 | AGAP002025 | mclassic9b | 2e-08 | Obp99a | 5e-05 | Obp99c | 7e-05 |
| AgamOBP42 | AgamOBP59 | AgamOBP11 | |||||||||
| AGAP009402 | — | AGAP010409 | mclassic8 | 5e-11 | AGAP002189 | mclassic9b | 6e-16 | Obp99a | 4e-06 | Obp99a | 2e-09 |
| AgamOBP43 | AgamOBP22 | AgamOBP14 | |||||||||
Note.—The table shows top hits results of the BLAST search among all mosquito Classic OBPs and Drosophila OBPs after splitting the Atypical proteins into their two respective putative domains.
The recent publication of a functional dimer in the C. quinquefasciatus genome (Mao et al. 2010) supports the current important speculations on Atypical members, indicating the importance of the presence of two-domain proteins in the binding of relatively large ligands. Thus, it is confirmed that the Atypical OBP members are indeed two-domain OBPs which were previously observed in Drosophila as Dimer OBPs and that they no more stand specific to the mosquito genomes as reported earlier (Xu et al. 2003). Furthermore, the matype2 members which carry a presence of only 6 cysteines in the place of 12 cysteines as in the other two-domain OBPs is suggestive of a possible adaptation in the fold with 3 disulphide bonds in place of 6 disulphide bonds in the other types. The astound distribution of these matype2 OBP genes from A. gambiae on the X chromosome further increases the speculative importance of these proteins in the blood feeding mechanism by female mosquitoes. Interestingly, most of the members of the two-domain OBP subfamily are reported as differentially expressed with respect to blood time series which adds to the importance of these proteins in host recognition (Dissanayake et al. 2010).
Ecological adaptations might have driven the need for the observed expansion in two-domain OBP gene repertoire in the Aed. aegyptii and C. quinquefasciatus genome when compared with A. gambiae. Our observations indicate that this expansion most probably occurred through gene duplication events in localized genome regions which lead to the observed gene clusters. We hence hypothesize that two distinct mechanisms could underlie the emergence of Atypical genes in mosquitoes. The observations made in A. gambiae genome sustain the first hypothesis that two-domain OBPs might have originated from gene duplicates of mclassic9, mclassic8, or Obp99a related members and their subsequent gene fusion leading to Atypical genes coclustered with their Classic counterparts. The observations made in Aed. aegyptii and C. quinquefasciatus support the second complementary hypothesis whereby the Atypical genes have undergone further gene duplications probably in response to ecological constraints in these mosquito lineages.
Our analysis hence sustains the proposition that the Atypical OBP genes to be renamed two-domain OBP proteins. Their future structural characterization and ligand binding profiling would be of significant importance in deciphering their contribution in olfaction in mosquitoes.
Materials and Methods
Sequence Searches
The database of the predicted protein sequences of the three mosquito genomes A. gambiae (A. gambiae annotation, AgamP3.4), Aed. aegypti (Aed. aegypti annotation, AaegL1.1), and C. quinquefasciatus (C. quinquefasciatus annotation, CpipJ1.2) were downloaded from the VectorBase (Lawson et al. 2009) version 3.4 (http://www.vectorbase.org, last accessed January 9, 2013) and Ensembl Genomes (Hubbard et al. 2009). The putative OBPs in the three mosquito species were identified using 10 Drosophila query sequences which belong to three different subfamilies Classic/General OBPs, PlusC, and MinusC OBPs using a PSI-BLAST (Altschul et al. 1997) run of 10 query sequences with an E-value cutoff of 3e−10 (Vieira et al. 2007) and an alignment length cutoff of 75% with respect to the query sequence. At this level, all of the previously identified members in the three genomes were identified with identification of a few additional members. A second run of PSI-BLAST was initiated with the hits from the previous runs. Using this protocol it was possible to not only pick up all the members of OBPs reported so far (Vogt 2002; Xu et al. 2003; Zhou et al. 2004, 2008; Pelletier and Leal 2009, 2011; Vieira and Rozas 2011) but also a remarkable number of additional members. The additional sequences were checked for the presence of a signal peptide using the SignalP server (Petersen et al. 2011), PBP/GOBP domain using CD-Search (Marchler-Bauer and Bryant 2004) in the case of classic OBPs, and alignment of the new sequences with their subfamily members in case of Atypical and PlusC proteins. The D7 proteins which were identified using this method but which are considered as a distinct family of proteins related to the OBPs were also retained for further analysis and used as an outgroup in the construction of phylogenetic trees. The orthologous sequences were identified based on the reciprocal best hit approach using BLAST (Moreno-Hagelsieb and Latimer 2008). The newly added sequences were named according to the naming conventions used in the earlier reports (Vogt 2002; Xu et al. 2003; Zhou et al. 2004, 2008; Pelletier and Leal 2009).
Multiple Sequence Alignment
The multiple sequence alignment forms the basis for any analysis of a family of proteins and it is highly necessary to obtain an accurate alignment. The error rate in the alignment increases with the increase in divergence of the proteins. Structure-based alignments in turn are considered to be the most accurate forms of alignments and hence, in this study, the structure alignment was used in constructing the alignments. The structure alignment was constructed using 10 OPBs in the OBP gene family using COMPARER (Sali and Blundell 1990). However, the use of the structure alignment as profiles was restricted to seven members in the case of OBPs and two members for the D7 family due to the limited number of structural data (data not shown). The OBPs and the D7 sequences were aligned to their respective structure alignments as profiles, and a combined alignment of the two family of proteins was constructed using the profile–profile alignment option using ClustalX (Thompson et al. 1994, 1997; Jeanmougin et al. 1998). The alignments were truncated based on the structure alignment on the N-terminal end which corresponds to the signal peptide region that has a high substitution rate; however, the C-terminal ends were retained due to the presence of an extended C-terminal in the case of Atypical subfamily members of the OBP family. This method was applied for aligning the sequences in all the three different genomes. Alignments for the different subclasses were constructed with sequences from all the three mosquito genomes and in the case of Classic subfamily, along with Drosophila sequences. The alignment of the Atypical and PlusC subclasses of OBPs were however not based on the structure alignment.
Phylogenetic Analysis
The phylogenetic trees were inferred using the Neighbor-Joining method (Saitou and Nei 1987) in MEGA 4.0 (Tamura et al. 2007). The percentage of replicate trees in which the associated sequences cluster together in the bootstrap test (1000 replicates) are shown next to the branches of the bootstrap consensus trees (Felsenstein 1985) and branches with <50% bootstrap cutoff were collapsed. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of number of amino acid substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). The trees were rooted at the branches of the D7 family of proteins which was considered as an outgroup (supplementary fig. S2a–c, Supplementary Material online).
The trees of the different subclasses (figs. 4–6) used for the comparative analysis of the different genomes were analyzed as unrooted trees. The phylogenetic trees were inferred using the Neighbor-Joining method (Saitou and Nei 1987) in MEGA 4.0 (Tamura et al. 2007). The percentage of replicate trees in which the associated sequences cluster together in the bootstrap test (1000 replicates) are shown next to the branches of the bootstrap consensus trees (Felsenstein 1985) and branches with <50% bootstrap cutoff were collapsed for the PlusC and Atypical OBP trees. The branches were not collapsed for the Classic OBP tree, however the subtype definition was still based on 50% bootstrap cutoff.
Orthology, Paralogy, Chromosomal Mapping, and Tentative Syntenic Analysis
OBP orthologs have been identified using the reciprocal BLAST hit approach (Moreno-Hagelsieb and Latimer 2008) which is widely used in the detection of orthologs. The inParanoid database (O’Brien et al. 2005) was used to examine the inparalogous relationship between OBPs. Assembled genome data was only available for A. gambiae at the date of this work in the Ensembl Genome (Hubbard et al. 2009) and VectorBase (Lawson et al. 2009). The chromosomal locations of OBPs from A. gambiae were identified using this data. The genome data of Aed. aegypti and C. quinquefasciatus as featured todate in Ensembl Genomes and VectorBase are not yet assembled and were used to map the OBP genes in these genomes at the supercontigs level. The exact chromosomal locations are known for only about 10% of their supercontigs among which very few harbor OBP genes. Orthologous OBP genes identified as described above were used to establish putative synteny between chromosomal segments from A. gambiae and supercontigs from the other two Culicinae species. The genes were mapped to their respective location on the chromosome or supercontigs (supplementary fig. S1a–e, Supplementary Material online). The chromosomes of A. gambiae was used as reference and were represented as a yellow bar and the contigs of Aedes and Culex are represented in purple and green, respectively. The direct three-way (1:1:1) orthology relationships among the three genomes are represented as green lines. The two-way (1:1) orthology relationships between two species are represented as black lines, and the inparalogy relationships are represented as red lines. The figures of the chromosomal mapping were drawn to scale using Adobe illustrator CS5.
Atypical Domain Analysis
The two constitutive PBP/GOBP OBP domains of Atypical OBPs were further characterized for their relationship with Classic or PlusC OBPs. For each Atypical OBP, the boundary between the N-term and C-term PGP/GOBP domains was manually delimited. This was performed by subjecting the full-length sequence to Pfam (Finn et al. 2010) and Conserved Domain Database (Marchler-Bauer et al. 2011) and was further validated by analyzing their cysteine profiles. Each N-term and C-term domain hence delimited was then subjected to a PSI-BLAST search (E-value cutoff value of 10−2) against a database that contains all OBPs from the same mosquito species in an attempt to find their putative distantly related single-domain OBPs. A similar search was performed against a database of Drosophila OBPs.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
R.S. is thankful to University of La Reunion for supporting this collaborative research through continuous invited professorship support since 2005. The authors are thankful to Manipal University, NCBS (TIFR), and the University of La Reunion for bioinformatics infrastructural support. The authors thank Prof. N. Srinivasan (Indian Institute of Science), Prof. Matthew (NCBS), Prof. Frédéric Cadet (University of La Réunion), Prof. Vinh Tran (University of Nantes), and Prof. Sankararamakrishnan (IIT, Kanpur) for useful discussions regarding this work. The authors are also thankful to Olivier Cadet, Swapnil Mahajan, and Nicolas Fontaine for technical assistance toward setting up the mOBPdb database. This work was in part supported by a grant from Conseil Régional de La Réunion, the French Ministry of Research, and the European Union in the framework of the GRI Phase III project. M.M. was supported by an international PhD fellowship from Conseil Regional de La Reunion in the framework of the joint dual-studentship program between Manipal University and University of La Réunion.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, deBianchi AG, James AA, Marinotti O. The major acid soluble proteins of adult female Anopheles darlingi salivary glands include a member of the D7-related family of proteins. Insect Biochem Mol Biol. 2002;32:1419–1427. doi: 10.1016/s0965-1748(02)00062-0. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci U S A. 2009;106:3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choumet V, et al. The salivary glands and saliva of Anopheles gambiae as an essential step in the Plasmodium life cycle: a global proteomic study. Proteomics. 2007;7:3384–3394. doi: 10.1002/pmic.200700334. [DOI] [PubMed] [Google Scholar]
- Dissanayake S, et al. aeGEPUCI: a database of gene expression in the dengue vector mosquito, Aedes aegypti. BMC Res Notes. 2010;3:248. doi: 10.1186/1756-0500-3-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera) Genome Res. 2006;16:1404–1413. doi: 10.1101/gr.5075706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D-P, Zhang H-J, Zhao P, Xia Q-Y, Xiang Z-H. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10:332. doi: 10.1186/1471-2164-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Davies PL. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene. 2002;292:43–55. doi: 10.1016/s0378-1119(02)00672-8. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, et al. Intriguing olfactory proteins from the yellow fever mosquito, Aedes aegypti. Naturwissenschaften. 2004;91:426–431. doi: 10.1007/s00114-004-0551-7. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Kalume DE, et al. A proteomic analysis of salivary glands of female Anopheles gambiae mosquito. Proteomics. 2005;5:3765–3777. doi: 10.1002/pmic.200401210. [DOI] [PubMed] [Google Scholar]
- Lawson D, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZX, Pickett JA, Field LM, Zhou JJ. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. Arch Insect Biochem Physiol. 2005;58:175–189. doi: 10.1002/arch.20047. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Ribeiro JM, Andersen JF. Structure, function, and evolution of biogenic amine-binding proteins in soft ticks. J Biol Chem. 2008;283:18721–18733. doi: 10.1074/jbc.M800188200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, et al. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci U S A. 2010;107:19102–19107. doi: 10.1073/pnas.1012274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J Biol Chem. 1994;269:16340–16347. [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Leal WS. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS One. 2009;4:e6237. doi: 10.1371/journal.pone.0006237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Leal WS. Characterization of olfactory genes in the antennae of the Southern house mosquito, Culex quinquefasciatus. J Insect Physiol. 2011;57:915–929. doi: 10.1016/j.jinsphys.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol B Biochem Mol Biol. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Plettner E, Lazar J, Prestwich EG, Prestwich GD. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochemistry. 2000;39:8953–8962. doi: 10.1021/bi000461x. [DOI] [PubMed] [Google Scholar]
- Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Definition of general topological equivalence in protein structures. A procedure involving comparison of properties and relationships through simulated annealing and dynamic programming. J Mol Biol. 1990;212:403–428. doi: 10.1016/0022-2836(90)90134-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thangudu RR, et al. Analysis on conservation of disulphide bonds and their structural features in homologous protein domain families. BMC Struct Biol. 2008;8:55. doi: 10.1186/1472-6807-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangudu RR, et al. Native and modeled disulfide bonds in proteins: knowledge-based approaches toward structure prediction of disulfide-rich polypeptides. Proteins. 2005;58:866–879. doi: 10.1002/prot.20369. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JM. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 2011;3:476–490. doi: 10.1093/gbe/evr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Sanchez-Gracia A, Rozas J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 2007;8:R235. doi: 10.1186/gb-2007-8-11-r235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG. Odorant binding proteins of the malaria mosquito Anopheles gambiae; possible orthologues of the OS-E and OS-F OBPs of Drosophila melanogaster. J Chem Ecol. 2002;28:2371–2376. doi: 10.1023/a:1021009311977. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hasan G, Pikielny CW. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J Biol Chem. 1999;274:10309–10315. doi: 10.1074/jbc.274.15.10309. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003;12:549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- Yoshizawa Y, et al. Ligand carrier protein genes expressed in larval chemosensory organs of Bombyx mori. Insect Biochem Mol Biol. 2011;41:545–562. doi: 10.1016/j.ibmb.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol. 2008;17:147–163. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Huang W, Zhang GA, Pickett JA, Field LM. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004;327:117–129. doi: 10.1016/j.gene.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.