Abstract
Antarctic notothenioids radiated over millions of years in subzero waters, evolving peculiar features, such as antifreeze glycoproteins and absence of heat shock response. Icefish, family Channichthyidae, also lack oxygen-binding proteins and display extreme modifications, including high mitochondrial densities in aerobic tissues. A genomic expansion accompanying the evolution of these fish was reported, but paucity of genomic information limits the understanding of notothenioid cold adaptation. We reconstructed and annotated the first skeletal muscle transcriptome of the icefish Chionodraco hamatus providing a new resource for icefish genomics (http://compgen.bio.unipd.it/chamatusbase/, last accessed December 12, 2012). We exploited deep sequencing of this energy-dependent tissue to test the hypothesis of selective duplication of genes involved in mitochondrial function. We developed a bioinformatic approach to univocally assign C. hamatus transcripts to orthology groups extracted from phylogenetic trees of five model species. Chionodraco hamatus duplicates were recorded for each orthology group allowing the identification of duplicated genes specific to the icefish lineage. Significantly more duplicates were found in the icefish when transcriptome data were compared with whole-genome data of model species. Indeed, duplicated genes were significantly enriched in proteins with mitochondrial localization, involved in mitochondrial function and biogenesis. In cold conditions and without oxygen-carrying proteins, energy production is challenging. The combination of high mitochondrial densities and the maintenance of duplicated genes involved in mitochondrial biogenesis and aerobic respiration might confer a selective advantage by improving oxygen diffusion and energy supply to aerobic tissues. Our results provide new insights into the genomic basis of icefish cold adaptation.
Keywords: deep sequencing, orthology/paralogy relationships, gene duplication, genome evolution, cold adaptation, Channichthyidae
Introduction
Chionodraco hamatus (Notothenioidei, Perciformes) is an Antarctic teleost belonging to the family Channichthyidae (icefish). All members of the family, with the exception of one species, are endemic to the Southern Ocean and are some of the most stenothermal species on Earth. They evolved in the persistently cold and oxygen-rich Antarctic waters, acquiring unique adaptations at the morphological, physiological, and biochemical level. Icefish, similar to other Antarctic notothenioids, lack a swim bladder and produce antifreeze glycoproteins (AFGPs), a key innovation preventing blood and body fluids freezing at the ambient temperature of −1.86 °C (Cheng and Detrich 2007). Icefish are a unique example of adult vertebrates lacking hemoglobin and functionally active erythrocytes, and, as a consequence, the oxygen-carrying capacity of their blood is only 10% that of red-blooded species (Ruud 1954). Fifteen of the 16 members of the family completely lack the adult β-globin gene and retain only a small 3’-fragment of the α-globin gene (Near et al. 2006). As Ruud (1954) suggested, this disadaptive phenotypic trait could have evolved only in the extreme and stable environmental conditions of Antarctic waters, where the higher oxygen solubility at low temperatures may have relaxed selection pressure for oxygen-binding proteins, allowing the successful diversification of icefish over the past 7.8–4.8 millions of years (Near et al. 2012).
Icefish do not express myoglobin in skeletal muscle, and six members of the family have lost the ability to produce the protein in cardiac myocytes either, exacerbating their hemoglobinless condition (Borley and Sidell 2010). Adequate oxygen delivery to tissues, even in the absence of respiratory pigments, is ensured in icefish thanks to the evolution of peculiar phenotypic traits such as hypertrophic heart, high cardiac output, increased blood volume, enlarged vessels lumina, low blood viscosity and pressure, well-perfused gills, improved skin and fin vascularization, cutaneous uptake of oxygen, and reduced metabolic rate (Kock 2005). One of the most remarkable icefish features is an exceptionally high mitochondrial density in the heart and skeletal muscle, which improves oxygen storage and diffusion in cells (O'Brien and Mueller 2010). These unique features make C. hamatus an excellent model to study cold adaptation. Several of the phenotypic modifications required in the constantly freezing temperatures of the Southern Ocean appear to involve stable genomic modifications (e.g., deletion of the β-globin locus), which might represent a significant limitation to adaptation to warmer environments (Patarnello et al. 2011). Understanding the genomic constraints on the evolutionary potential of Antarctic species might help to predict how well they will cope with climate change. Several molecular mechanisms ranging from point mutations of preexisting genes to large genomic rearrangements might underlie such modifications. Of all possible mechanisms, gene duplication has long been recognized to play a major role in the evolution of novel functions, a role recently confirmed by eukaryotic whole-genome sequence analysis (Ohta 1989; Lynch 2007). Cheng et al. (Cheng 1996; Cheng et al. 2003) were, to our knowledge, the first to link cold adaptation in Antarctic notothenioids to gene duplication, showing that repeated duplication of the primordial AFGP gene produced the AFGP expanded family. Duplications of other genes, with a putative role in adaptation to low temperatures, were subsequently identified, that is, metallothioneins (Bargelloni et al. 1999), pepsin enzymes (Carginale et al. 2004), and hepcidins (Xu et al. 2008). More recently, Chen et al. (2008) used an array-based comparative genomic hybridization approach to compare three high Antarctic, evolutionarily derived notothenioid species to two non-Antarctic, phylogenetically basal notothenioids, showing that 101 protein-coding genes (implicated in various biological functions including extracellular matrix remodeling, protein folding and degradation, response to stress conditions, defense from oxidative and apoptotic processes, Ras/MAPK and TGF-β signal transduction cascades, RNA transcription, binding and processing, cellular structural components, and innate immunity) were duplicated in the Antarctic species. The expression of some duplicated genes was found to be upregulated in the red-blooded Antarctic notothenioid Dissostichus mawsoni compared with temperate water teleosts. A dosage effect was thus hypothesized as the mechanism involved in the maintenance of duplicated gene copies. A recent analysis of whole-genome size in 11 red- and white-blooded notothenioid species confirmed that the evolution of phylogenetically derived notothenioid families, such as the Channichthyidae, might have been accompanied by a genome expansion (Detrich and Amemiya 2010), without further indication, however, of which genes or gene families are involved in such expansion.
The working hypothesis of this study is that targeted gene duplications occurred at loci involved in mitochondrial function and biogenesis. Energy production, in the form of ATP, is the primary function of skeletal muscle mitochondria. ATP production may be challenging in a cold environment because low temperatures typically reduce the kinetic energy of molecules and reaction rates, modify molecular interactions and diffusion rates, and affect membrane architecture. During cold acclimation, many fish enhance oxidative capacity by a variety of physiological mechanisms (Somero 2004; O'Brien 2011). Maintaining energy production at appropriate levels for physiological activities is a constant problem for Antarctic fish, which experience a narrow temperature range reaching −1.86°C, which is lethal for most ectothermic organisms. There is evidence that temperature compensatory adaptations occur at the biochemical level because key enzymes involved in energy metabolism (cytochrome c oxidase, citrate synthase, and lactate dehydrogenase) show a (proportionally) higher enzymatic activity at a common low temperature in Antarctic notothenioids than in temperate and tropical species (Crockett and Sidell 1990; Kawall et al. 2002). This may be obtained by higher enzyme concentrations and/or by increased enzyme efficiency at low temperatures. The increased activity of lactate dehydrogenase, for example, can be explained by a compensatory high catalytic rate (Fields and Somero 1998). For other enzymes, such as cytochrome c oxidase, increased enzyme concentration compensates the higher activation energies in cold-adapted notothenioids compared with temperate and tropical fish (Mark et al. 2012). Notably, an increase in aerobic enzymes concentration is observed in many fish during cold acclimation (see e.g., Lucassen et al. 2006; Orczewska et al. 2010).
Compensation of ATP production through aerobic respiration is not the only adaptation in Antarctic fish that centers on mitochondria. As mentioned earlier, higher cellular densities of these organelles have been reported to favor oxygen diffusion, especially in the hemoglobinless icefish. Therefore, a constantly higher demand for mitochondrial biogenesis might be present in these unique animals. Stably higher enzyme concentrations and increased supply for structural components of mitochondria might be achieved through different molecular mechanisms at the genetic level, for instance, mutations affecting transcriptional regulation, mRNA, and/or protein stability. Alternatively, gene amplification at target loci might selectively increase expression levels.
We tested the hypothesis of selective duplication of genes involved in mitochondrial function and biogenesis in the Antarctic fish genome through massive parallel sequencing of the skeletal muscle transcriptome of C. hamatus, which belongs to the notothenioid family showing the most extreme physiological modifications in response to cold. Icefish-specific gene duplications were then identified by comparative transcript annotation against five model fish genomes, using a dedicated bioinformatic pipeline that interrogates an existing database of orthology groups, Ensembl Compara. Finally, a functional enrichment analysis was used to evaluate the biological role of proteins encoded by duplicated loci.
Materials and Methods
Transcriptome Sequencing, Assembly, and Annotation
A normalized cDNA library, constructed from muscle tissue of four C. hamatus individuals collected at Terranova Bay, was sequenced by 454 technology (see Supplementary Material online for details). Raw sequencing data were preprocessed using LUCY (http://lucy.sourceforge.net/, last accessed December 12, 2012) to remove poly-A tails escaped to restriction and to trim adaptors and sequences smaller than 60 bases or with Phred quality lower than 30. Transcriptome assembly was performed using MIRA 3 (Chevreux et al. 1999). Two runs of assembly were carried out by MIRA in “EST” and “accurate” usage mode, respectively. Settings adopted were those defined for the 454 sequencing technology. The second run was performed on previously obtained contigs plus discarded reads, which were used as input for MIRA 3. Contigs shorter than 100 nucleotides (nt) or with average quality lower than 30 were discarded.
A first de novo functional annotation of the C. hamatus transcriptome was obtained by similarity using Basic Local Alignment Search Tool (BLAST), Blast2GO, and custom-made scripts. BLAST (ftp://ftp.ncbi.nlm.nih.gov/blast/db/, last accessed December 12, 2012) was run in local mode, and assembled contigs were compared against the National Center for Biotechnology Information (NCBI) nonredundant protein database (nr) and to the NCBI nucleotide database (nt). Alignments with an E value <10−3 were considered significant. Blast2GO suite (http://www.blast2go.org/, last accessed December 12, 2012) was used for functional annotation of transcripts through the mapping of gene ontology (GO) terms to contigs having BLAST hits, obtained by BLASTX search against nr. Only ontologies retrieved from hits with an E value <10−6, an annotation cut off >55, and a GO weight >5 were used for annotation.
Definition of Homology Relationships of C. hamatus Transcripts
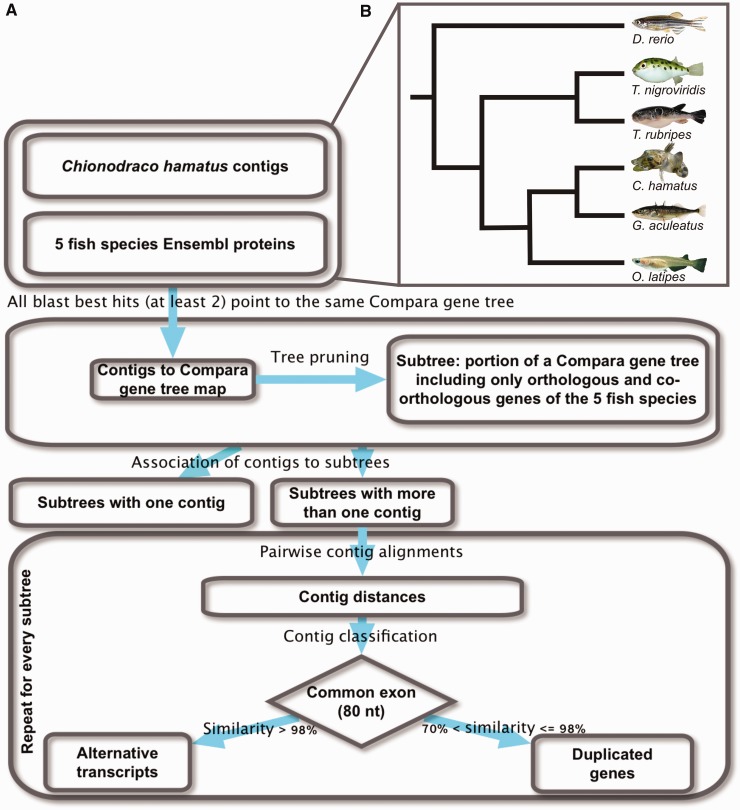
A custom-made bioinformatic pipeline based on Perl and Python scripts was developed for the definition of homology relationships of C. hamatus transcripts (fig. 1A). In particular, all queries to Ensembl database were carried out using the Ensembl Perl API version 62. Local BLAST queries were accomplished by using the BLAST+ program, version 2.2.22, made available by NCBI.
Fig. 1.—
Computational pipeline work flow and reference Ensembl Compara fish species tree. (A) Flow chart showing main steps of the computational pipeline devised to assign Chionodraco hamatus transcripts to subtrees, that is, unique sets of orthologous or co-orthologous genes in the teleost lineage, and to identify duplicated genes. (B) Schematic phylogeny of the five model species considered as reference plus C. hamatus (following Ensembl Compara species tree, based on NCBI taxonomy, and Matschiner et al. (2011) for C. hamatus branch position).
Similarity Mapping of Contigs to Ensembl Compara Trees
Contigs were used as query for local BLASTX search against Ensembl (version 62) proteins from five fully sequenced fish species: Danio rerio, zebrafish; Gasterosteus aculeatus, stickleback; Takifugu rubripes, Japanese pufferfish; Oryzias latipes, medaka; and Tetraodon nigroviridis, green spotted pufferfish (fig. 1B). BLAST hits were filtered by E value and subject coverage; in particular, only hits with an E value of 10−3 or less and covering at least 50% of the subject sequence were retained. Moreover, for each contig, a maximum of 20 BLAST hits were considered for further analysis.
BLASTX results were then used to associate C. hamatus contigs to Ensembl Compara gene trees. For each contig, and for each species, the Ensembl protein best hit was associated to the encoding transcript, to the gene, and consequently to a Compara gene tree, if any. Only nodes of the tree belonging to the five considered fish species were taken into account. If the best hit could not be associated to a Compara gene tree, the successive BLAST hit was used. This procedure was carried out in parallel for each of the five fish species. The five different contig-Compara gene tree maps were merged to obtain the mapping between contigs and Compara gene tree: A contig was considered associated to a Compara gene tree only if all BLAST best hits from different species (at least 2) pointed to the same Compara gene tree.
Identification of Transcripts Coming from Lineage-Specific Duplications
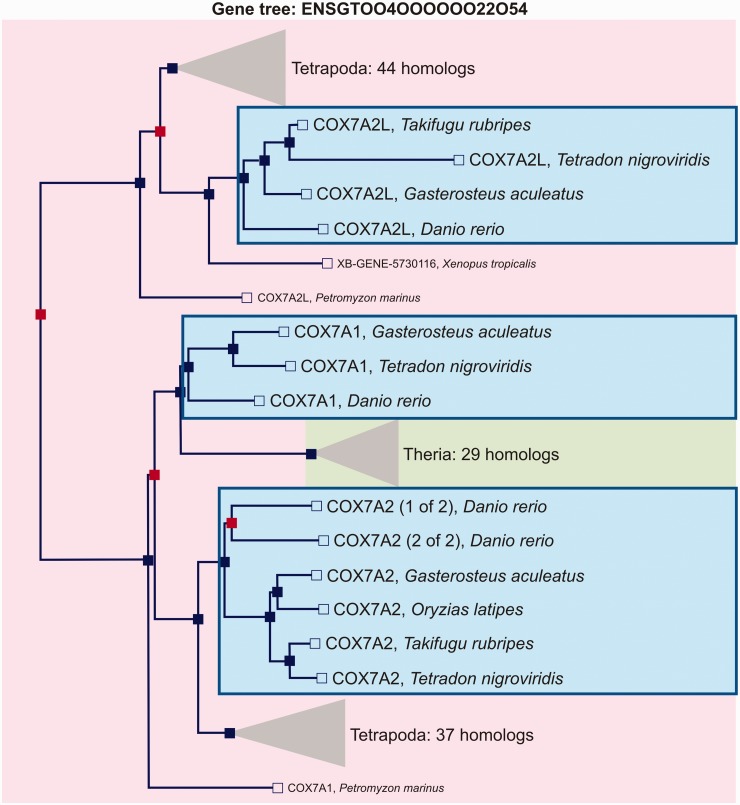
Compara trees may comprise wide groups of homologous genes, including orthologs and paralogs. To identify lineage-specific duplicates, custom subtree structures were pruned. A subtree was defined as the portion of a Compara tree including only orthologous and co-orthologous genes of the five fish model species (fig. 2).
Fig. 2.—
An example of Compara gene tree topology resulting in three subtree structures. Blue and red nodes correspond to speciation and duplication nodes, respectively. The subtree (encircled by a blue line) is defined as the portion of a Compara gene tree including orthologous and co-orthologous genes in the teleost lineage.
Sets of C. hamatus contigs mapping to the same subtree were further analyzed to discriminate between alternative transcripts of the same gene and transcripts from duplicated genes. Sequences within each set were pairwise compared using BLAST, to identify significant alignments (E value < 10−3). Pairs of contigs sharing a significant alignment of at least 80 nt (corresponding to the first quartile of exon length distribution in G. aculeatus, according to UCSC Genome Browser exon information data, i.e., 75% of exons are longer than 80 nt), with a similarity of at least 70% were considered for discrimination among alternative transcripts and duplicated loci. Contigs with a sequence similarity higher than 98% were classified as alternative transcripts of the same gene, considering that less than 2% sequence divergence might be due to allele variation and/or sequencing errors within the same locus. Alignment mismatches occurring in homopolimeric stretches longer than 3 nt were not considered for the calculation of sequence similarity between contigs, thus eliminating the confounding effect originating from the well-known error bias in 454 sequencing technology. To identify C. hamatus lineage-specific duplicates, all subtrees were selected in which the number of C. hamatus duplicates exceeded those of the closest species G. aculeatus (Matschiner et al. 2011).
Functional Enrichment Analysis of C. hamatus Putatively Duplicated Genes
Functional enrichment analysis of C. hamatus lineage-specific duplicates was performed using DAVID v6.7 (http://david.abcc.ncifcrf.gov/, last accessed December 12, 2012). The only fish species that is represented in the DAVID knowledgebase is zebrafish, therefore D. rerio Entrez gene IDs corresponding to subtrees were used as sequence identifiers to carry out the analysis. Gene functional enrichment was determined between a “Gene List,” which included the zebrafish gene IDs corresponding to the subtrees to which mapped the putatively duplicated genes in the icefish genome, and a “Background,” which contained the zebrafish gene IDs corresponding to all the considered subtrees. D. rerio gene IDs corresponding to each subtree were counted only once regardless the number of C. hamatus putative gene copies. Thresholds for Gene count and EASE score, which provides the significance of gene-term enrichment with a modified Fisher’s exact test, were set equal to 2 and 0.05, respectively.
MitoCarta-Based Identification of Duplicated Genes Encoding Mitochondrial Proteins
A list of genes encoding proteins with mitochondrial localization was downloaded from MitoCarta (Pagliarini et al. 2008), an inventory of mouse genes encoding proteins with strong support for mitochondrial localization. As MitoCarta uses only gene symbols, Ensembl BioMart (http://www.ensembl.org/biomart/martview, last accessed December 12, 2012) was used to obtain Ensembl gene IDs for mouse mitochondrial proteins. A custom pipeline was developed to obtain, from Ensembl Compara, the list of fish orthologous Ensembl gene IDs for every mitochondrial mouse gene and to tag C. hamatus transcripts, and hence C. hamatus genes, as mitochondrial. The pipeline uses results from BLAST against Ensembl fish proteins to associate Ensembl gene IDs to C. hamatus transcripts and employs this information to tag C. hamatus transcripts and consequently genes as mitochondrial by using the mitochondrial fish genes obtained in the previous step. Finally, a hypergeometric distribution was used to test the enrichment for mitochondrial genes in the set of C. hamatus duplicated genes when compared with the set of all reconstructed C. hamatus genes. All computations were performed with the R statistical software.
The C. hamatus Skeletal Muscle Transcriptome Database
A database, freely available online at http://compgen.bio.unipd.it/chamatusbase/ (last accessed December 12, 2012), was implemented using SQLite and Django web framework. The database is filled with different layers of information regarding the C. hamatus transcriptome sequences and analysis results. For each contig, different data and bioinformatic analysis results are provided. All data in the database can be searched by keywords, GO terms, or by sequence using a BLAST web interface. Moreover, a query system for massive data retrieval is available.
Results
Chionodraco hamatus Skeletal Muscle Transcriptome 1.0
A total of 382,386 raw reads were obtained, with an average sequence length of 297 nt. After removal of short reads and trimming of low-quality sequence regions, the remaining 341,171 reads (SRA052291) were assembled into contigs with two runs of assembly (supplementary table S1, Supplementary Material online). After length- and quality-based filtering, a final set of 23,968 contigs (with mean length, average quality, and read coverage, respectively, 595 nt, 48, and 13) represents the C. hamatus muscle transcriptome (supplementary table S2 and fig. S1A–C, Supplementary Material online).
For 9,556 transcripts (40%), BLAST identified significant similarity with known proteins. In total, 109,307 nr hits were found, with an average of 11.4 hits per contig (94% of best hits E value <10−6 and 89% E value <10−9). In parallel, 7,155 contigs (30%) had significant hits among known nucleotide sequences (94% of best hits E value <10−6 and 88% E value <10−9). Comprehensively, 48% of the C. hamatus transcriptome (11,509 contigs) resulted to be similar to known protein or DNA sequences.
In total, 7,293 contigs were associated to one or more of 5,805 unique GO terms, for a total of 61,170 term occurrences (5.9 ± 1.8 terms per contig). Supplementary figure S1D, Supplementary Material online, reports the slim-based term classification of GO terms.
Identification of Lineage-Specific Duplicated Genes in C. hamatus
Using the bioinformatics pipeline depicted in figure 1A, assembled contigs were first univocally assigned to one Ensembl Compara gene tree. To this end, results of contigs BLAST against five fish species Ensembl proteins were used (table 1). Each tree corresponds to a homology group. Then, the structure of each gene tree was split to obtain one or more new entities, called subtrees, defined as a portion of a gene tree including orthologous or co-orthologous genes of five fish model species (D. rerio, G. aculeatus, T. rubripes, O. latipes and Tet. nigroviridis) (fig. 2). A total of 2,401 C. hamatus contigs were assigned to 1,712 subtrees, that is, unique sets of orthologous or co-orthologous genes in the teleost lineage. For 1,232 subtrees, a single icefish contig was found, whereas 480 subtrees were associated with two or more C. hamatus transcripts for a total of 1,169 transcripts.
Table 1.
Results of Chionodraco hamatus Contigs BLASTX Comparison against Ensembl Proteins of the Five Model Teleosts
| Species | Contigs with Significant Hits (E value <10−3) | Contigs with Significant Hits Covering At Least 50% of the Subject Sequence | Total Number of Protein Hits |
|---|---|---|---|
| Danio rerio | 9,203 | 5,084 | 165,094 |
| Gasterosteus aculeatus | 9,411 | 4,776 | 113,220 |
| Takifugu rubripes | 8,958 | 4,485 | 148,743 |
| Oryzias latipes | 9,070 | 4,740 | 106,126 |
| Tetraodon nigroviridis | 8,764 | 4,627 | 98,293 |
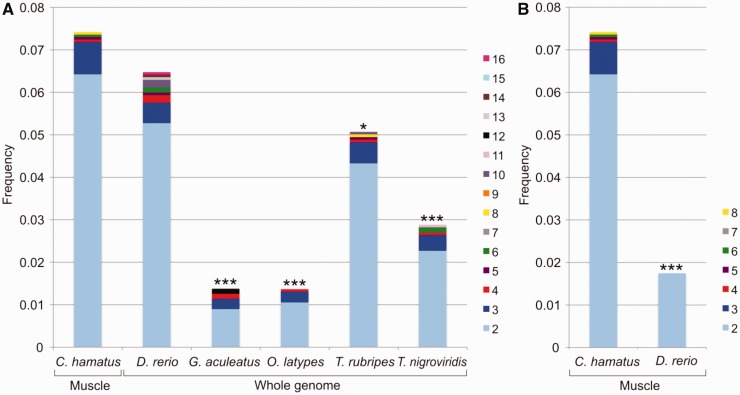
Subsequently, a highly stringent filtering process to exclude potential alternative transcripts/alleles from the same locus was applied (fig. 1A), yielding 353 transcripts, which could be considered the product of distinct genetic loci and were associated with 127 subtrees. In 124 subtrees, the number of C. hamatus gene copies exceeded that observed in the closest species, G. aculeatus, suggesting that for each orthology group, one or more gene duplication events occurred after the separation between the stickleback and icefish lineages. The number of gene copies ranged from 2 to 8 (2.26 on average), for a total of 276 putatively duplicated genes (supplementary table S3, Supplementary Material online). For each subtree, the proportion of C. hamatus duplicated genes was compared with the proportion of duplicated genes in each of the five considered fish genomes. Because duplicated genes of each species were counted in each subtree, only lineage-specific duplications were considered. Using a test of equal proportion, C. hamatus showed a significantly higher proportion (P value with Bonferroni correction for multiple testing < 0.05) of duplicated genes in all comparisons with the other species, except against D. rerio (fig. 3A). The evidence obtained for a significantly higher occurrence of gene duplications in C. hamatus is likely underestimated, because gene copy number was inferred based on transcriptome data from a single tissue and compared with whole-genome information for fish model species. In fact, limiting the comparison to zebrafish genes expressed in the skeletal muscle (expression data are available for D. rerio only and provided by ZFIN) produced a significant difference in the proportion of duplicated loci (fig. 3B).
Fig. 3.—
Comparison of duplicated gene frequencies in different fish species. (A) Bar plots represent, for each species, the fraction of analyzed subtrees with at least one gene represented, including two or more duplicated genes. The proportion of Chionodraco hamatus duplicated genes, inferred in this study from the analysis of the muscle transcriptome, is pairwise compared with the proportion of duplicated genes of the five fish species for which the whole genome is available. Test of equal proportions: ***P value <0.001, **P value <0.01, and *P value <0.05, after Bonferroni correction for multiple tests. (B) Comparison of the proportion of duplicated genes expressed in muscle in C. hamatus and D. rerio.
Functional Enrichment Analysis of Duplicated Loci
Out of 124 subtrees/orthology groups that showed at least one icefish-specific gene duplication, 6 subtrees could not be associated to any zebrafish gene ID (five of these subtrees did not include any D. rerio gene) and one additional subtree was not associated with any functional information in DAVID. Using zebrafish functional annotations for each subtree, a total of 11 terms resulted to be significantly over-represented (P value <0.05), which include 2 KEGG pathways, 1 SP-PIR keyword, 1 biological process (BP) GO term, 6 molecular function (MF) GO terms, and 1 cellular component (CC) GO term (table 2). Overall, most enriched terms are related to two key cellular processes, namely protein translation and oxidative phosphorylation.
Table 2.
Functional Enrichment Analysis of 124 Chionodraco hamatus Putatively Duplicated Genes Using DAVID Web Tool
| Category | Term | Gene Count | P | FE |
|---|---|---|---|---|
| GOTERM_MF | GO:0003735 structural constituent of ribosome | 16 | 0.0003 | 2,6 |
| GOTERM_MF | GO:0005198 structural molecule activity | 17 | 0.0006 | 2,4 |
| SP_PIR_KEYWORDS | Ribosomal protein | 15 | 0.0008 | 2,6 |
| GOTERM_BP | GO:0006412 translation | 17 | 0.0017 | 2,2 |
| KEGG_PATHWAY | dre03010 ribosome | 12 | 0.0076 | 2,3 |
| KEGG_PATHWAY | dre00190 oxidative phosphorylation | 12 | 0.0250 | 2,0 |
| GOTERM_CC | GO:0005840 ribosome | 17 | 0.0280 | 1,6 |
| GOTERM_MF | GO:0016675 oxidoreductase activity, acting on heme group of donors | 4 | 0.0440 | 4,7 |
| GOTERM_MF | GO:0015002 heme-copper terminal oxidase activity | 4 | 0.0440 | 4,7 |
| GOTERM_MF | GO:0016676 oxidoreductase activity, acting on heme group of donors, oxygen as acceptor | 4 | 0.0440 | 4,7 |
| GOTERM_MF | GO:0004129 cytochrome-c oxidase activity | 4 | 0.0440 | 4,7 |
Note.—Reported are GO terms, KEGG pathways, and SP-PIR keywords significantly over-represented (P value <0.05) in the set of duplicates. For each enriched term, the number of genes involved in the term (Gene count), enrichment P value, and fold enrichment (FE) are indicated.
A total of 1,098 mouse genes encoding proteins with strong support for mitochondrial localization were downloaded from MitoCarta. When converting gene symbols given by MitoCarta to Ensembl gene IDs, a total of 1,057 (96%) genes were obtained. A custom pipeline was developed to obtain, from Ensembl Compara, the list of 5,826 fish orthologs to mitochondrial mouse genes. The set of 5,826 fish mitochondrial orthologs was composed by 1,378 D. rerio genes, 1,115 G. aculeatus genes, 1,114 T. rubripes genes, 1,061 O. latipes genes, and 1,158 Tet. nigroviridis genes. Subsequently, a mini pipeline was developed to tag C. hamatus contigs as mitochondrial. Using results of BLAST against Ensembl fish proteins, C. hamatus contigs were associated to an Ensembl gene ID and tagged as mitochondrial if the corresponding Ensembl gene ID was present among the 5,826 fish mitochondrial genes. As a result, 34 of 124 putative duplicated genes and 274 of 1,588 nonduplicated genes were tagged with mitochondrial localization (table 3 reports the set of 34 C. hamatus putative duplicated genes encoding proteins with mitochondrial localization). A hypergeometric distribution was then used to test the enrichment for mitochondrial genes in the set of 124 putative duplicated C. hamatus genes when compared with the set of all reconstructed C. hamatus genes. A P value of 0.0235 indicates a significant enrichment of mitochondrial genes in the group of duplicated C. hamatus genes.
Table 3.
Thirty-Four Chionodraco hamatus Putative Duplicated Genes Encoding Proteins with Mitochondrial Localization
| Gene Symbol | Gene Description | Number of C. hamatus Duplicated Gene Copies |
|---|---|---|
| acn9 | ACN9 homolog (Saccharomyces cerevisiae) (Source: ZFIN; Acc: ZDB-GENE-041010-94) | 2 |
| atp5s | ATP synthase, H+ transporting, mitochondrial F0 complex, subunits (Source: ZFIN; Acc: ZDB-GENE-040426-959) | 2 |
| bola1 | bolA homolog 1 (Escherichia coli) (Source: HGNC Symbol; Acc:24263) | 2 |
| C13H6orf57 | Chromosome 6 open reading frame 57 (Source: HGNC Symbol; Acc:20957) | 2 |
| C4H1orf31 | chromosome 1 open reading frame 31 (Source: HGNC Symbol; Acc:18025) | 2 |
| coa5 | Cytochrome C oxidase assembly factor 5 (Source: HGNC Symbol; Acc:33848) | 3 |
| cox17 | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) (Source: ZFIN; Acc: ZDB-GENE-040912-91) | 8 |
| cox4i1 | Cytochrome c oxidase subunit IV isoform 1 (Source: ZFIN; Acc: ZDB-GENE-030131-5175) | 2 |
| cox6a2 | Cytochrome c oxidase subunit VIa polypeptide 2 (Source: HGNC Symbol; Acc:2279) | 3 |
| cox7a2 | Cytochrome c oxidase, subunit VIIa 2 (Source: ZFIN; Acc: ZDB-GENE-050522-153) | 2 |
| etfa | Electron-transfer-flavoprotein, alpha polypeptide (Source: ZFIN; Acc: ZDB-GENE-030131-4449) | 2 |
| fdx1 | Ferredoxin 1 (Source: ZFIN; Acc: ZDB-GENE-071015-2) | 2 |
| glrx2 | Glutaredoxin 2 (Source: ZFIN; Acc: ZDB-GENE-040718-101) | 2 |
| mrpl-13 | Mitochondrial ribosomal protein L13 (Source: ZFIN; Acc: ZDB-GENE-050522-167) | 2 |
| mrpl-17 | Mitochondrial ribosomal protein L17 (Source: ZFIN; Acc: ZDB-GENE-040912-133) | 2 |
| mrpl-2 | Mitochondrial ribosomal protein L2 (Source: HGNC Symbol; Acc:14056) | 2 |
| mrpl-48 | Mitochondrial ribosomal protein L48 (Source: ZFIN; Acc: ZDB-GENE-041008-125) | 2 |
| mrpl-52 | Mitochondrial ribosomal protein L52 (Source: HGNC Symbol; Acc:16655) | 2 |
| mrpl-55 | Mitochondrial ribosomal protein L55 (Source: HGNC Symbol; Acc:16686) | 2 |
| mterfd1 | MTERF domain containing 1 (Source: ZFIN; Acc: ZDB-GENE-040718-359) | 2 |
| ndufa11 | zgc:66391 (Source: ZFIN; Acc: ZDB-GENE-040426-1631) | 2 |
| ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13 kDa (Source: HGNC Symbol; Acc: 7688) | 2 |
| ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (Source: ZFIN; Acc: ZDB-GENE-040426-1124) | 2 |
| ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 (Source: ZFIN; Acc: ZDB-GENE-011010-1) | 2 |
| ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 (Source: ZFIN; Acc: ZDB-GENE-040426-1858) | 2 |
| ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) (Source: ZFIN; Acc: ZDB-GENE-041111-261) | 2 |
| oxa1l | Oxidase (cytochrome c) assembly 1-like (Source: ZFIN; Acc: ZDB-GENE-071004-49) | 2 |
| romo1 | Reactive oxygen species modulator 1 (Source: HGNC Symbol; Acc:16185) | 3 |
| sdhc | Succinate dehydrogenase complex, subunit C, integral membrane protein (Source: ZFIN; Acc: ZDB-GENE-040801-26) | 2 |
| timm9 | Translocase of inner mitochondrial membrane 9 homolog (Source: ZFIN; Acc: ZDB-GENE-021206-14) | 2 |
| tmem14c | Transmembrane protein 14C (Source: ZFIN; Acc: ZDB-GENE-060825-156) | 3 |
| uqcrb | Ubiquinol-cytochrome c reductase binding protein (Source: ZFIN; Acc: ZDB-GENE-050522-542) | 2 |
| uqcrc2a | ubiquinol-cytochrome c reductase core protein IIa (Source: ZFIN; Acc: ZDB-GENE-040718-405) | 2 |
| zgc:56493 | zgc:56493 (Source: ZFIN; Acc: ZDB-GENE-030131-8581) | 5 |
Note.—The number of C. hamatus putative duplicated gene copies is reported for each gene, identified by a gene symbol, and a gene description (provided by Ensembl BioMart using Danio rerio Ensembl gene ID of the related subtree).
Discussion
This study describes the C. hamatus skeletal muscle transcriptome. Deep sequencing information, by means of a dedicated bioinformatic pipeline, disclosed putative duplicated genes specific to the C. hamatus lineage. This unique investigation licensed exploration of the genetic and genomic adaptations to extreme cold conditions. A global functional enrichment analysis of putative duplicated genes and the localization and biological role of single-encoded proteins supported our hypothesis that duplicated genes were involved in mitochondrial function and biogenesis.
Gene and genome duplications are thought to have played a major role in genome evolution and in the generation of the enormous phenotypic variability that exists among living species. In the teleost lineage, duplicated genes were shown to have significantly impacted the evolution of key traits, including immune response (Van der Aa et al. 2009), development (Zou et al. 2009), sensory systems (Rennison et al. 2012), adaptation to different biological environments (Nomiyama et al. 2008), endocrine system (Arterbery et al. 2011), and body plans (Guo et al. 2010). DNA sequences generated by duplication increase genome redundancy, a distinctive trait of eukaryotic genomes, which offers the raw genetic material for the evolution of genetic novelties. Four major outcomes of gene duplication have been proposed: 1) both duplicates may be preserved in a genome with one copy evolving new functional properties and the other copy retaining the original function (“neofunctionalization”) (Ohno 1970); 2) the two copies may be maintained for a selective advantage in increasing the synthesis of the ancestral gene product (“dosage effect”) (Ohno 1970); 3) one copy may accumulate degenerative mutations and be silenced (“nonfunctionalization”) (Ohno 1972); and 4) both copies may be partially impaired by deleterious mutations, so that the ancestral function is divided between duplicated loci (“subfunctionalization”) (Force et al. 1999).
Duplication events may involve either the whole genome (whole-genome duplication [WGD]) or single genes or genomic regions (segmental duplication). WGDs occurred in many lineages during evolution (Van de Peer et al. 2009). Two WGD events appear to have occurred early in the vertebrate evolution, whereas the third round of WGD involved the teleost lineage soon after its divergence from tetrapods (Dehal and Boore 2005; Meyer and Van de Peer 2005). A vast literature exists regarding WGD events and their evolutionary implications in vertebrates. Much less attention has been paid to recent gene duplicates and their evolutionary consequences, which may reveal specific adaptation processes in individual species or lineages, as in C. hamatus. A genome-wide analysis of four mammalian genomes (human, macaque, mouse, and rat), focusing on recent gene duplicates, revealed that 10% of all lineage-specific duplicated pairs evolved under positive selection, which is a higher percentage than a comparable set of single-copy genes (Han et al. 2009), suggesting that analysis of recent duplicates may reveal significant footprints of adaptive change. Human-specific duplicates identified in the study were involved in the development of neural and cognitive functions (Han et al. 2009).
The prediction of orthology and paralogy relationships between genes is a central issue in comparative genomics. Typically, orthologs are defined as homologous sequences, which diverge as a consequence of a speciation event, whereas paralogs are homologous sequences originating from a duplication node. Various methods have been proposed for the inference of reliable orthology/paralogy relationships at the genome-wide level. Some methods rely on pairwise sequence comparison between genomes, whereas an increasing number of web tools are based on phylogenetic approaches, following the evolutionary basis of the original orthology/paralogy definition (Altenhoff and Dessimoz 2012). In Ensembl Compara, gene phylogenies are computed using the TreeBest method, which reconciles calculated gene tree topologies with known species trees to determine whether a node corresponds to a duplication or a speciation event (Vilella et al. 2009). Ensembl Compara is a wide-ranging collection of gene trees, spanning 91% of genes across vertebrates and including five well-annotated fish species: D. rerio, G. aculeatus, T. rubripes, O. latipes, and Tet. nigroviridis.
The final aim of this study was to distinguish between duplicated genes originating from WGD events and C. hamatus lineage-specific duplications, which may offer new insights into the genetic/genomic mechanisms of cold adaptation in icefish. We used a targeted computational pipeline relying on Ensembl Compara phylogenetic resources and exploiting deep sequencing information of the C. hamatus muscle transcriptome. Using a quite stringent similarity-based approach (all BLAST best hits from different species pointing the same Compara gene tree), a subset of 2,401 C. hamatus transcripts was univocally assigned to custom subtree structures, which are portions of an Ensembl Compara gene tree including unique sets of orthologous or co-orthologous genes in the teleost lineage. By definition, duplicated genes resulted from WGD events should end up in different subtrees, with the latter including only duplicated genes arisen independently in each of the model species lineages. This is true also for C. hamatus mapped transcripts, because duplicated genes were identified and counted inside each subtree.
We found 124 subtrees with C. hamatus lineage-specific duplications, with the number of duplicates per gene ranging from 2 to 8. Moreover, C. hamatus displayed a significantly higher proportion of lineage-specific duplicates than the other model fish species, excepting D. rerio, which, however, is known to have a highly duplicated genome. This difference is striking considering that it was obtained comparing whole-genome data from model species (all transcripts, no matter of expression characteristics), with C. hamatus duplicated genes expressed in one tissue (skeletal muscle). Limiting the comparison to muscle expressed genes in D. rerio, C. hamatus displayed a significantly higher proportion of duplicated genes. It is worth noting that we are not comparing the level of retention of duplicated genes resulted from WGD events but the amount of duplicated genes arisen thereafter and independently in each lineage.
We reasoned that identified duplicated genes could be simply genes free to duplicate with minimal negative outcomes. In this case, we would expect that the same genes might be duplicated in other fish. From a qualitative point of view, the overlap between the set of C. hamatus duplicated genes and that of zebrafish is narrow (out of 124 subtrees with C. hamatus lineage-specific duplications, only 6 also include zebrafish duplicates) showing that these two sets are different in composition. Moreover, in none of these six subtrees, we observed more than one zebrafish gene copy expressed in muscle. Thus, although zebrafish duplicated genes evolved as tissue-specific isoforms, C. hamatus duplicates might play a role in muscle.
Functional enrichment analyses, based on GO terms, pathways and MitoCarta information, showed that C. hamatus duplicated loci are enriched in mitochondrial function and respiratory activities. In fact, “oxidative phosphorylation” was one of the most represented KEGG pathways identified by functional enrichment analysis of duplicates. Moreover, enrichment analysis for mitochondrial proteins using MitoCarta information confirmed that at least 34 out of 124 duplicated genes encode proteins with mitochondrial localization, leading to a significant enrichment of mitochondrial function in duplicated genes. Additionally, gene-by-gene manual annotation based on relevant literature showed that many proteins encoded by duplicated loci are involved in mitochondrial biogenesis and function. Thus, these results not only support a trend for genome expansion accompanying the evolution of phylogenetically derived notothenioid families reported by Chen et al. (2008) and Detrich and Amemiya (2010) but also confirm the working hypothesis of targeted gene duplications at loci involved in mitochondrial function and biogenesis. This observation might explain, at least in part, the high mitochondrial density documented in Antarctic notothenioids. These fish possess an increased mitochondrial abundance compared with Perciformes from warmer climates (Johnston et al. 1998), with an extreme value recorded for the active and pelagic Pleuragramma antarcticum (Johnston et al. 1988).
There are three lines of evidence showing that a high mitochondrial density is advantageous at low temperatures: 1) it facilitates ATP production thanks to higher concentrations of enzymes involved in aerobic metabolism; 2) it enhances intracellular oxygen diffusion through the mitochondrial lipid membrane as oxygen solubility in phospholipids is approximately four times higher than in water; and 3) it reduces the mean diffusion distance for oxygen transfer from capillaries to mitochondria (O'Brien 2011). The lack of myoglobin expression in skeletal muscle is displayed by both white- and red-blooded notothenioids (Sidell et al. 1997; Moylan and Sidell 2000). All icefish completely lack hemoglobin (Ruud 1954), and six species do not express myoglobin in cardiac myocytes (Borley and Sidell 2010). Thus, oxygen supply and ATP generation are extremely challenging in white-blooded icefish. As reviewed by O'Brien and Mueller (2010), mitochondrial density in icefish muscle reaches values that largely exceed those reported for red-blooded notothenioids, both in skeletal muscle and in the myocardium, with a particular increase in nonexpressing myoglobin icefish. Icefish mitochondria were indeed found to be structurally different from that of red-blooded notothenioids, displaying a particularly high lipid-to-protein ratio (O'Brien and Mueller 2010), suggesting a major role of lipids in facilitating oxygen storage and diffusion in cells in the absence of oxygen-carrying proteins (Sidell 1998).
Together, the progressive expansion of the mitochondrial compartment in Antarctic notothenioids, the enrichment analyses performed at the gene category level, and a close examination of recent literature regarding single protein functions indicate a role of duplicated genes in mitochondrial function and biogenesis. Among identified mitochondrial proteins, 12 are structural subunits of the electron transport chain complexes localized in the mitochondrial inner membrane. In addition, multiple gene copies (from 3 to 8) were detected for COX17 cytochrome c oxidase assembly homolog (cox17), COX19 cytochrome c oxidase assembly homolog (cox19), and cytochrome c oxidase assembly factor 5 (coa5), which are involved in the assembly of complex IV (Horng et al. 2004; Rigby et al. 2007; Huigsloot et al. 2011). MTERF domain containing 1 (Mterfd1) is an important regulator of mitochondrial transcription and is required for the normal assembly of mitochondrial respiratory complexes (Park et al. 2007). The duplicated electron-transfer-flavoprotein alpha polypeptide (efta) gene codes for one of the two subunits of an electron-transferring flavoprotein, a mitochondrial matrix enzyme located at a key metabolic branch point, which shuttles electrons from various primary dehydrogenases to the main respiratory chain (Toogood et al. 2007). Translocase of inner mitochondrial membrane 9 homolog (timm9) encodes a subunit of the Tim9-Tim10 complex (or Tim9-Tim10-Tim12 complex), a mitochondrial intermembrane chaperone that mediates the translocation of nuclear-encoded hydrophobic precursor proteins into the intermembrane space and is thus essential for the import of inner membrane proteins (Schmidt et al. 2010). Moreover, oxidase (cytochrome c) assembly 1-like (oxa1l) codes for an integral protein of the mitochondrial inner membrane and represents the key component of the machinery that mediates insertion of both nuclear and mitochondrial translation products into the inner membrane (Hell et al. 1998; Ott and Herrmann 2010). In mammals, Oxa1l cross-links to six mitochondrial ribosomal proteins (MRPs) (Mrpl-13, Mrpl-20, Mrpl-28, Mrpl-48, Mrpl-49 and Mrpl-51) (Haque et al. 2010); notably, mrpl-13 and mrpl-48 genes were duplicated in our analysis.
Mitochondrial ribosomes seem to have diverged substantially during eukaryotic evolution because protein composition is highly variable between different species, both in the number and in the type of contained MRPs (Smits et al. 2007). Newly acquired MRPs, which were shown to have originated by several gene duplication events (Smits et al. 2007), may have contributed to the introduction of novel and/or more specialized functions. Mrpl-13 and mrpl-48, which originated by duplication of mrps-10 in the metazoan lineage, and four additional genes encoding MRPs were found to be duplicated in our analysis. All of them are components of the large ribosomal subunit involved in peptide bonds formation and thus more prone to cooperate with co- and/or post-translational interacting factors. As Gruschke and Ott (2010) suggested, modifications at this level may result in novel ways to organize the biogenesis of mitochondrially encoded proteins, that is, the major components of the mitochondrial respiratory complexes.
Another interesting duplication is reactive oxygen species modulator 1 (romo1), encoding the first mitochondrial inner membrane protein known to be involved in the regulation of mitochondrial fission in mammalian cells (Zhao et al. 2009). Enhanced expression of romo1 was shown to inhibit nuclear DNA synthesis and mitosis and, in particular, to induce mitochondrial fragmentation, resulting in numerous sphere-shaped organelles with no noticeable internal structural alterations (Zhao et al. 2009). The duplication of romo1 may be associated with the need to coordinate cell cycle with mitochondrial proliferation. Increasing mitochondrial abundance without a concomitant increase in cell division would be a valuable strategy to obtain higher mitochondrial densities in cells.
An additional putative duplicated gene, possibly involved in the regulation of mitochondrial morphology, encodes the ATP synthase, H+ transporting, mitochondrial F0 complex, and subunit s (Atp5s). Atp5s is an essential subunit of the ATP synthase complex that couples proton translocation through the F0 subunit with ATP synthesis at F1 subunit (Belogrudov and Hatefi 2002). The protein is also responsible for preventing passive proton diffusion through F0 and for regulating proton electrochemical gradient across the inner membrane (Belogrudov 2008, 2010). Atp5s may enhance ATP production under proton-limited conditions and improve the efficiency of oxidative phosphorylation in animal mitochondria (Belogrudov 2009). Icefish mitochondria display an increased coupling of proton transport and ATP synthesis compared with red-blooded notothenioids, which, in turn, show a higher coupling than temperate teleosts (Mueller et al. 2011). Belogrudov (2010) showed that overexpression of atp5s affects mitochondria morphology in cultured animal cells, with thicker and more packaged cristae of the inner membrane, likely as a consequence of enhanced protein–protein interactions. This structural modification is supposed to increase the rate of exchange of ATP synthasome substrates across the inner membrane and/or the rate of substrate conduction along the respiratory chain. This might explain the 2-folds higher respiratory control ratio found in atp5s-overexpressing cells, suggesting increased coupling between respiration and ATP production (Belogrudov 2010).
The duplication of dynactin 6 (dctn6) also supports the hypothesis of increased mitochondrial biogenesis coupled with enhanced lipid synthesis in the icefish lineage. Dynactin is a major component of the activator complex that stimulates dynein-mediated vesicle transport, a highly ATP-dependent and key cellular biological process. Upregulation of this gene, which is not present in MitoCarta, is associated with the dramatic proliferation of mitochondria in skeletal muscle of mice following the genetic inactivation of the mitochondrial adenine nucleotide translocator-1, which exports mitochondrial ATP to the cytosol (Murdock et al. 1999). Mouse Dctn6 is understood to play a role in mitochondrial lipid synthesis due to protein sequence similarity to an enzyme involved in bacterial lipid A biosynthesis (Murdock et al. 1999). The mitochondrial fatty acid synthetic pathway is present in many eukaryotic organisms and has been shown to be of particular importance for normal mitochondrial morphology and function (Hiltunen et al. 2010). Duplication of dctn6 may be relevant for cold adaptation because it is probably a multifunctional protein involved in vesicle transport regulation, vasopressin-regulated water reabsorption, and in mitochondrial biogenesis, via lipid biosynthesis. However, it should be emphasized here that very little is known about how the biosynthesis of mitochondrial phospholipids is integrated into the process of mitochondrial biogenesis (O'Brien and Mueller 2010).
Oxygen solubility increases at lower temperatures, thus Antarctic fish are constantly exposed to high oxygen tensions, which enhance reactive oxygen species (ROS) production and levels of oxidative stress. Chen et al. (2008) have documented a potential evolutionary response in Antarctic notothenioids using an array-based comparative genomic hybridization technique. Nine genes involved in stress response, anti-ROS, and antiapoptotic processes were found to be duplicated in three Antarctic notothenioids when comparing the hybridization intensity of their DNA with that of two non-Antarctic and phylogenetically basal notothenioids (Chen et al. 2008). These nine genes were not detected as duplicated in our analysis, possibly because of their missed transcription in skeletal muscle, transcriptome incompleteness, the conservative approach followed by our pipeline, or because duplicated gene copies were not divergent enough to be detected by sequence analysis. However, several genes associated with anti-ROS activities or upregulated in the presence of high ROS levels were found to be duplicated. Among them, BCL2/adenovirus E1B interacting protein 3 (bnip3) is activated in ventricular myocytes in response to stress when mitochondria are severely damaged, inducing mitochondrial dysfunction and subsequent cell death (Regula 2002; Gustafsson 2011). Recently, a bnip3 apoptosis-independent function was discovered in the myocardium, where high mitochondrial volume densities are required to satisfy the large ATP demand. Bnip3 expression seems to be essential to regulating mitochondrial turnover by selective autophagy (Gustafsson 2011). Long-term studies in aged bnip3-knockout mice showed that Bnip3 deficiency promotes the accumulation of dysfunctional mitochondria and the development of cardiac disease (Dorn 2010). Given that bnip3 functions as a redox sensor activated in response to localized accumulation of oxidative stress (Kubli et al. 2008), clearly a constitutively increased expression of this gene in Antarctic notothenioids, via gene duplication, may improve mitochondrial turnover efficiency in extremely oxidizing conditions.
Another duplicated gene with anti-ROS function is glutaredoxin 2 (glrx2), encoding a glutathione-dependent oxidoreductase activated by oxidative stress, which facilitates the maintenance of mitochondrial redox homeostasis, protects cells from oxidative damage, and inhibits mitochondrially mediated apoptosis (Enoksson et al. 2005; Lillig et al. 2005). Additionally, zgc:56493 (for which five gene copies were detected) and zgc:92254, which code for a thioredoxin and a glutathione S-transferase, respectively, are multifunctional proteins displaying antioxidative and antiapoptotic properties (Arner and Holmgren 2000; Burmeister et al. 2008; Piaggi et al. 2010). Microsomal glutathione S-transferase 3 (mgst3) encodes an enzyme displaying glutathione-dependent peroxidase activity toward lipid hydroperoxides (Jakobsson et al. 1997). This function may be essential to preserve icefish mitochondrial membranes which, because of their increased phospholipidic component, are particularly susceptible to lipid peroxidation (Mueller et al. 2011). Mgst3 was also found overexpressed in carp in response to cold stress (Gracey et al. 2004).
Other noteworthy duplicated genes are: 1) bolA homolog 1 (Escherichia coli) (bola1), which is well studied in E. coli and is involved in the response and adaptation to various stress conditions, among which are pH variation and oxidative stress (Adnan et al. 2011); 2) cystatin A (csta), encoding a cysteine proteinase that restrains UVB-induced apoptosis of keratinocytes and confers enhanced resistance to high salt concentrations, drought, oxidative, and cold stresses in plants (Takahashi et al. 2007; Zhang et al. 2008); 3) nuclear protein 1 (nupr1), encoding a key player in cellular stress response (Goruppi and Iovanna 2010), which is upregulated in the liver of rainbow trout during stress response and recovery (Momoda et al. 2007); 4) ring finger protein 7 (rnf7), encoding a redox-inducible and apoptosis-protective antioxidant protein, which also decreases endogenous ROS production and oxidative DNA damage after exposure to heat shock when overexpressed (Lee et al. 2008; Sun 2008); 5) defender against cell death 1 (dad1), which codes for a subunit of the oligosaccharyltransferase complex and is upregulated in zebrafish embryonic cell line by XBP-1S, a key transcriptional regulator of the unfolded protein response that allows the recovery of endoplasmic reticulum (ER) function (Hu et al. 2007); and 6) chaC, cation transport regulator-like 1 (chac1), which encodes a component of the mammalian unfolded protein response pathway (Mungrue et al. 2009). Duplicated genes were found for four additional proteins involved in protein folding: Prefoldin subunit 2 (Pfdn2), which is a subunit of the molecular chaperone prefoldin involved in the folding of newly synthesized proteins, mainly actins and tubulins (Pfdn2 subunit is thought to facilitate the formation of the prefoldin complex and also contain a DNA binding motif) (Martin-Benito et al. 2002; Miyazawa et al. 2011); Thioredoxin domain containing 9 (Txndc9), which, curiously, has an action antagonistic to that of prefoldin and the combined action of the two proteins is thought to be crucial to establishing a functional cytoskeleton (Stirling et al. 2006); Ubiquitin fusion degradation 1-like (Ufd1l), which is part of a multiprotein complex involved in the recognition and export of polyubiquitin-tagged misfolded proteins from the ER to the cytoplasm, where they are degraded by the 26S proteasome (Bays and Hampton 2002); Ufd1l subunit is the one that directly interacts and stimulates the activity of ubiquitin ligase gp78, which mediates the degradation of misfolded ER proteins (Cao et al. 2007) and is also induced in yeast in response to cold (Schade et al. 2004); ubiquitin-like 5 (ubl5), which is upregulated in response to mitochondrial stress, is required for mounting mitochondrial unfolded protein response, leading to the induction and subsequent import into mitochondria of mitochondrial chaperones (Haynes and Ron 2010).
Duplicated genes were found for the heat shock factor binding protein 1 (Hsbp1), a negative regulator of the heat shock response (HSR) (Satyal et al. 1998). The HSR typically consists in the upregulation of heat shock proteins (HSPs), after induction by the heat shock factors in response to elevated temperatures or other stress. Hsbp1 is a nuclear-localized protein that converts the active trimeric state of Heat shock factor 1 (Hsf1) to the inert monomeric state. Hsbp1 prevents both Hsf1 DNA-binding and transactivation activity and, consequently, blocks HSR activation (Satyal et al. 1998). In Antarctic notothenioids, both Hsp70 mRNA and Hsf1 with DNA-binding activity were found, but no increased expression of Hsp70, or of any other HSP size class, was detected after stress exposure (Buckley et al. 2004; Buckley and Somero 2009). Hsbp1 might have a role in preventing HSPs induction in Antarctic notothenioids. Interestingly, another putative duplicated gene is BCL2-associated athanogene 2 (bag2), encoding a regulator of Hsp70 family chaperones through the inhibition of their chaperone activity by promoting substrate release (Takayama et al. 1999). Bag2 specifically inhibits ubiquitinylation of misfolded proteins by CHIP, an Hsp70-associated ubiquitin ligase, thus providing a regulatory mechanism for preventing uncontrolled degradation of chaperone substrates (Dai et al. 2005).
Finally, in addition to the six MRPs already discussed, 14 cytoplasmic ribosomal proteins were identified among duplicated genes, and, consistently, “structural constituent of ribosome” and “ribosome” were the most represented MF term and KEGG pathway, respectively, found by the enrichment analysis of duplicates in DAVID. Duplication of ribosomal protein genes might reflect the need to enhance protein synthesis capacity at low temperatures. A similar response, the upregulation of ribosomal proteins, was observed in Saccharomices cerevisiae after cold acclimation (Tai et al. 2007) and in the head kidney of a red-blooded notothenioid, Dis. mawsoni, compared with temperate and tropical teleosts (Chen et al. 2008).
Our results identified duplicated genes specific to the icefish lineage and showed that they are mainly involved in mitochondrial biogenesis and aerobic respiration. Thus, our findings confirm the importance of genomic expansions in the evolution of Antarctic notothenioids (Chen et al. 2008; Detrich and Amemiya 2010) and provide new indications about the functional role of gene duplicates. Energy production might be exceptionally challenging in extreme cold conditions and icefish, in the absence of oxygen-carrying proteins, rely on high mitochondrial densities to achieve high concentrations of aerobic enzymes, enhanced lipid-mediated oxygen diffusion, and short oxygen route from capillaries to mitochondria, thus ensuring energy supply and oxygen diffusion to aerobic tissues (O'Brien and Mueller 2010). Therefore, the preferential maintenance of duplicated genes involved in mitochondrial biogenesis and aerobic respiration, which implies a strong selective pressure for increased mitochondrial function acting for millions of years during the evolutionary history of Antarctic notothenioids, seems to underlie icefish adaptation to the cold. Gene duplications specific to the icefish lineage might have provided the raw genetic material for the natural selection of cold-adapted phenotypes able to survive at subzero temperatures. The acquisition of new functional properties or fitness benefits due to an increased synthesis of the ancestral gene product might explain the preservation of mitochondrial-related duplicates, providing an updated framework for the experimental investigation of the genomic and physiological bases of cold adaptation.
Supplementary Material
Supplementary methods, figure S1, and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Vittorio Varotto (Department of Biology, University of Padova, Padova, Italy) for collecting C. hamatus samples. This work was supported by the Italian National Program for Antarctic Research (PNRA) to T.P., L.B., and L.Z. and by the University of Padova to S.B. A.C. and I.A.M.M. are recipients of post-doc grants from the University of Padova (grant numbers GRIC11Z79P and CPDR084151/08). C.A. is a PhD student in Evolutionary Biology at the University of Padova, with a program partially supported under NSF grant 0741348.
Literature Cited
- Adnan M, Morton G, Hadi S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2-DDCT method. Mol Cell Biochem. 2011;357:275–282. doi: 10.1007/s11010-011-0898-y. [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. Inferring orthology and paralogy. Methods Mol Biol. 2012;855:259–279. doi: 10.1007/978-1-61779-582-4_9. [DOI] [PubMed] [Google Scholar]
- Arner ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Arterbery AS, et al. Evolution of ligand specificity in vertebrate corticosteroid receptors. BMC Evol Biol. 2011;11:14. doi: 10.1186/1471-2148-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargelloni L, et al. Metallothioneins in Antarctic fish: evidence for independent duplication and gene conversion. Mol Biol Evol. 1999;16:885–897. doi: 10.1093/oxfordjournals.molbev.a026178. [DOI] [PubMed] [Google Scholar]
- Bays NW, Hampton RY. Cdc48-Ufd1-NpI4: stuck in the middle with Ub. Curr Biol. 2002;12:R366–R371. doi: 10.1016/s0960-9822(02)00862-x. [DOI] [PubMed] [Google Scholar]
- Belogrudov GI. The proximal N-terminal amino acid residues are required for the coupling activity of the bovine heart mitochondrial factor B. Arch Biochem Biophys. 2008;473:76–87. doi: 10.1016/j.abb.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogrudov GI. Recent advances in structure-functional studies of mitochondrial factor B. J Bioenerg Biomembr. 2009;41:137–143. doi: 10.1007/s10863-009-9210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogrudov GI. Coupling factor B affects the morphology of mitochondria. J Bioenerg Biomembr. 2010;42:29–35. doi: 10.1007/s10863-009-9263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogrudov GI, Hatefi Y. Factor B and the mitochondrial ATP synthase complex. J Biol Chem. 2002;277:6097–6103. doi: 10.1074/jbc.M111256200. [DOI] [PubMed] [Google Scholar]
- Borley KA, Sidell BD. Evolution of the myoglobin gene in Antarctic icefishes (Channichthyidae) Polar Biol. 2010;34:659–665. [Google Scholar]
- Buckley BA, Place SP, Hofmann GE. Regulation of heat shock genes in isolated hepatocytes from an Antarctic fish, Trematomus bernacchii. J Exp Biol. 2004;207:3649–3656. doi: 10.1242/jeb.01219. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Somero GN. cDNA microarray analysis reveals the capacity of the cold-adapted Antarctic fish Trematomus bernacchii to alter gene expression in response to heat stress. Polar Biol. 2009;32:403–415. [Google Scholar]
- Burmeister C, et al. Oxidative stress in Caenorhabditis elegans: protective effects of the omega class glutathione transferase (GSTO-1) FASEB J. 2008;22:343–354. doi: 10.1096/fj.06-7426com. [DOI] [PubMed] [Google Scholar]
- Cao J, et al. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Carginale V, Trinchella F, Capasso C, Scudiero R, Parisi E. Gene amplification and cold adaptation of pepsin in Antarctic fish: A possible strategy for food digestion at low temperature. Gene. 2004;336:195–205. doi: 10.1016/j.gene.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Chen Z, et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 2008;105:12944–12949. doi: 10.1073/pnas.0802432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-HC. Genomic basis for antifreeze glycopeptide heterogeneity and abundance in Antarctic notothenioid fishes. In: Ennion SJ, Goldspink G, editors. Gene expression and manipulation in aquatic organisms. Cambridge: Cambridge University Press; 1996. pp. 1–20. [Google Scholar]
- Cheng C-HC, Chen L, Near TJ, Jin Y. Functional antifreeze glycoprotein genes in temperate-water New Zealand nototheniid fish infer an Antarctic evolutionary origin. Mol Biol Evol. 2003;20:1897–1908. doi: 10.1093/molbev/msg208. [DOI] [PubMed] [Google Scholar]
- Cheng C-HC, Detrich HW ., III Molecular ecophysiology of Antarctic notothenioid fishes. Philos Trans R Soc Lond B Biol Sci. 2007;362:2215–2232. doi: 10.1098/rstb.2006.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. German Conference on Bioinformatics. Hanover, Germany: 1999. Genome sequence assembly using trace signals and additional sequence information; pp. 45–56. [Google Scholar]
- Crockett EL, Sidell BD. Some pathways of energy metabolism are cold adapted in Antarctic fishes. Physiol Zool. 1990;63:472–488. [Google Scholar]
- Dai Q, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:1700–1708. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW, III, Amemiya CT. Antarctic notothenioid fishes: genomic resources and strategies for analyzing an adaptive radiation. Integr Comp Biol. 2010;50:1009–1017. doi: 10.1093/icb/icq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW., 2nd Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoksson M, et al. Overexpression of glutaredoxin 2 attenuates apoptosis by preventing cytochrome c release. Biochem Biophys Res Commun. 2005;327:774–779. doi: 10.1016/j.bbrc.2004.12.067. [DOI] [PubMed] [Google Scholar]
- Fields PA, Somero GN. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci U S A. 1998;95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. J Biol Chem. 2010;285:1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey AY, et al. Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci U S A. 2004;101:16970–16975. doi: 10.1073/pnas.0403627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Ott M. The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle. Bioessays. 2010;32:1050–1057. doi: 10.1002/bies.201000081. [DOI] [PubMed] [Google Scholar]
- Guo B, Gan X, He S. Hox genes of the Japanese eel Anguilla japonica and Hox cluster evolution in teleosts. J Exp Zool B Mol Dev Evol. 2010;314:135–147. doi: 10.1002/jez.b.21318. [DOI] [PubMed] [Google Scholar]
- Gustafsson AB. Bnip3 as a dual regulator of mitochondrial turnover and cell death in the myocardium. Pediatr Cardiol. 2011;32:267–274. doi: 10.1007/s00246-010-9876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Spremulli LL, Fecko CJ. Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem. 2010;285:34991–34998. doi: 10.1074/jbc.M110.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Ron D. The mitochondrial UPR—protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci U S A. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ. Mitochondrial fatty acid synthesis—an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog Lipid Res. 2010;49:27–45. doi: 10.1016/j.plipres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- Hu MC, et al. XBP-1, a key regulator of unfolded protein response, activates transcription of IGF1 and Akt phosphorylation in zebrafish embryonic cell line. Biochem Biophys Res Commun. 2007;359:778–783. doi: 10.1016/j.bbrc.2007.05.183. [DOI] [PubMed] [Google Scholar]
- Huigsloot M, et al. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am J Hum Genet. 2011;88:488–493. doi: 10.1016/j.ajhg.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson PJ, Mancini JA, Riendeau D, Ford-Hutchinson AW. Identification and characterization of a novel microsomal enzyme with glutathione-dependent transferase and peroxidase activities. J Biol Chem. 1997;272:22934–22939. doi: 10.1074/jbc.272.36.22934. [DOI] [PubMed] [Google Scholar]
- Johnston IA, Calvo J, Guderley H, Fernandez D, Palmer L. Latitudinal variation in the abundance and oxidative capacities of muscle mitochondria in perciform fishes. J Exp Biol. 1998;201:1–12. doi: 10.1242/jeb.201.1.1. [DOI] [PubMed] [Google Scholar]
- Johnston IA, Camm JP, White M. Specializations of swimming muscles in the pelagic Antarctic fish Pleuragramma antarcticum. Mar Biol. 1988;100:3–12. [Google Scholar]
- Kawall H, Torres J, Sidell B, Somero G. Metabolic cold adaptation in Antarctic fishes: evidence from enzymatic activities of brain. Mar Biol. 2002;140:279–286. [Google Scholar]
- Kock K-H. Antarctic icefishes (Channichthyidae): a unique family of fishes: A review, Part I. Polar Biol. 2005;28:862–895. [Google Scholar]
- Kubli DA, Quinsay MN, Huang C, Lee Y, Gustafsson AB. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am J Physiol Heart Circ Physiol. 2008;295:H2025–H2031. doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, et al. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167–176. doi: 10.1016/j.freeradbiomed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- Lillig CH, et al. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci U S A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen M, Koschnick N, Eckerle LG, Portner HO. Mitochondrial mechanisms of cold adaptation in cod (Gadus morhua L.) populations from different climatic zones. J Exp Biol. 2006;209:2462–2471. doi: 10.1242/jeb.02268. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- Mark FC, et al. Mitochondrial function in Antarctic nototheniids with ND6 translocation. PLoS One. 2012;7:e31860. doi: 10.1371/journal.pone.0031860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J, et al. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 2002;21:6377–6386. doi: 10.1093/emboj/cdf640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, Hanel R, Salzburger W. On the origin and trigger of the notothenioid adaptive radiation. PLoS One. 2011;6:e18911. doi: 10.1371/journal.pone.0018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Miyazawa M, et al. Prefoldin subunits are protected from ubiquitin-proteasome system-mediated degradation by forming complex with other constituent subunits. J Biol Chem. 2011;286:19191–19203. doi: 10.1074/jbc.M110.216259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momoda TS, et al. Gene expression in the liver of rainbow trout, Oncorhynchus mykiss, during the stress response. Comp Biochem Physiol Part D Genomics Proteomics. 2007;2:303–315. doi: 10.1016/j.cbd.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Moylan TJ, Sidell BD. Concentrations of myoglobin and myoglobin mRNA in heart ventricles from Antarctic fishes. J Exp Biol. 2000;203:1277–1286. doi: 10.1242/jeb.203.8.1277. [DOI] [PubMed] [Google Scholar]
- Mueller IA, Grim JM, Beers JM, Crockett EL, O'Brien KM. Inter-relationship between mitochondrial function and susceptibility to oxidative stress in red- and white-blooded Antarctic notothenioid fishes. J Exp Biol. 2011;214:3732–3741. doi: 10.1242/jeb.062042. [DOI] [PubMed] [Google Scholar]
- Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart muscle isoform of the adenine nucleotide translocator. J Biol Chem. 1999;274:14429–14433. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- Near TJ, Parker SK, Detrich HW., III A genomic fossil reveals key steps in hemoglobin loss by the antarctic icefishes. Mol Biol Evol. 2006;23:2008–2016. doi: 10.1093/molbev/msl071. [DOI] [PubMed] [Google Scholar]
- Near TJ, et al. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci U S A. 2012;109:3434–3439. doi: 10.1073/pnas.1115169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H, et al. Extensive expansion and diversification of the chemokine gene family in zebrafish: identification of a novel chemokine subfamily CX. BMC Genomics. 2008;9:222. doi: 10.1186/1471-2164-9-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KM. Mitochondrial biogenesis in cold-bodied fishes. J Exp Biol. 2011;214:275–285. doi: 10.1242/jeb.046854. [DOI] [PubMed] [Google Scholar]
- O'Brien KM, Mueller IA. The unique mitochondrial form and function of Antarctic channichthyid icefishes. Integr Comp Biol. 2010;50:993–1008. doi: 10.1093/icb/icq038. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- Orczewska JI, Hartleben G, O'Brien KM. The molecular basis of aerobic metabolic remodeling differs between oxidative muscle and liver of threespine sticklebacks in response to cold acclimation. Am J Physiol Regul Integr Comp Physiol. 2010;299:R352–R364. doi: 10.1152/ajpregu.00189.2010. [DOI] [PubMed] [Google Scholar]
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767–775. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CB, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Patarnello T, Verde C, di Prisco G, Bargelloni L, Zane L. How will fish that evolved at constant sub-zero temperatures cope with global warming? Notothenioids as a case study. Bioessays. 2011;33:260–268. doi: 10.1002/bies.201000124. [DOI] [PubMed] [Google Scholar]
- Piaggi S, et al. Glutathione transferase omega 1-1 (GSTO1-1) plays an anti-apoptotic role in cell resistance to cisplatin toxicity. Carcinogenesis. 2010;31:804–811. doi: 10.1093/carcin/bgq031. [DOI] [PubMed] [Google Scholar]
- Regula KM. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- Rennison DJ, Owens GL, Taylor JS. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 2012;62:986–1008. doi: 10.1016/j.ympev.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Rigby K, Zhang L, Cobine PA, George GN, Winge DR. Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J Biol Chem. 2007;282:10233–10242. doi: 10.1074/jbc.M610082200. [DOI] [PubMed] [Google Scholar]
- Ruud JT. Vertebrates without erythrocytes and blood pigment. Nature. 1954;173:848–850. doi: 10.1038/173848a0. [DOI] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY. Cold adaptation in budding yeast. Mol Biol Cell. 2004;15:5492–5502. doi: 10.1091/mbc.E04-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Sidell BD. Intracellular oxygen diffusion: the roles of myoglobin and lipid at cold body temperature. J Exp Biol. 1998;201:1119–1128. doi: 10.1242/jeb.201.8.1119. [DOI] [PubMed] [Google Scholar]
- Sidell BD, et al. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proc Natl Acad Sci U S A. 1997;94:3420–3424. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN. Adaptation of enzymes to temperature: searching for basic “strategies.”. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:321–333. doi: 10.1016/j.cbpc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Stirling PC, et al. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J Biol Chem. 2006;281:7012–7021. doi: 10.1074/jbc.M513235200. [DOI] [PubMed] [Google Scholar]
- Sun Y. RNF7 (RING finger protein-7) Atlas Genet Cytogenet Oncol Haematol. 2008;12:289–291. [Google Scholar]
- Tai SL, Daran-Lapujade P, Walsh MC, Pronk JT, Daran JM. Acclimation of Saccharomyces cerevisiae to low temperature: a chemostat-based transcriptome analysis. Mol Biol Cell. 2007;18:5100–5112. doi: 10.1091/mbc.E07-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, et al. Cystatin A suppresses ultraviolet B-induced apoptosis of keratinocytes. J Dermatol Sci. 2007;46:179–187. doi: 10.1016/j.jdermsci.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Takayama S, Xie ZH, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- Toogood HS, Leys D, Scrutton NS. Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes. FEBS J. 2007;274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Van der Aa LM, et al. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, et al. Adaptive evolution of hepcidin genes in Antarctic notothenioid fishes. Mol Biol Evol. 2008;25:1099–1112. doi: 10.1093/molbev/msn056. [DOI] [PubMed] [Google Scholar]
- Zhang XX, Liu SK, Takano T. Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 2008;68:131–143. doi: 10.1007/s11103-008-9357-x. [DOI] [PubMed] [Google Scholar]
- Zhao J, et al. The novel conserved mitochondrial inner-membrane protein MTGM regulates mitochondrial morphology and cell proliferation. J Cell Sci. 2009;122:2252–2262. doi: 10.1242/jcs.038513. [DOI] [PubMed] [Google Scholar]
- Zou S, Kamei H, Modi Z, Duan C. Zebrafish IGF genes: gene duplication, conservation and divergence, and novel roles in midline and notochord development. PLoS One. 2009;4:e7026. doi: 10.1371/journal.pone.0007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.