Abstract
Vertebrate genomes include gene regulatory elements in protein-noncoding regions. A part of gene regulatory elements are expected to be conserved according to their functional importance, so that evolutionarily conserved noncoding sequences (CNSs) might be good candidates for those elements. In addition, paralogous CNSs, which are highly conserved among both orthologous loci and paralogous loci, have the possibility of controlling overlapping expression patterns of their adjacent paralogous protein-coding genes. The two-round whole-genome duplications (2R WGDs), which most probably occurred in the vertebrate common ancestors, generated large numbers of paralogous protein-coding genes and their regulatory elements. These events could contribute to the emergence of vertebrate features. However, the evolutionary history and influences of the 2R WGDs are still unclear, especially in noncoding regions. To address this issue, we identified paralogous CNSs. Region-focused Basic Local Alignment Search Tool (BLAST) search of each synteny block revealed 7,924 orthologous CNSs and 309 paralogous CNSs conserved among eight high-quality vertebrate genomes. Paralogous CNSs we found contained 115 previously reported ones and newly detected 194 ones. Through comparisons with VISTA Enhancer Browser and available ChIP-seq data, one-third (103) of paralogous CNSs detected in this study showed gene regulatory activity in the brain at several developmental stages. Their genomic locations are highly enriched near the transcription factor-coding regions, which are expressed in brain and neural systems. These results suggest that paralogous CNSs are conserved mainly because of maintaining gene expression in the vertebrate brain.
Keywords: noncoding sequences, genome duplication, vertebrates, gene regulation
Introduction
Regulation of gene expression in a spatial and temporal manner is crucial during the vertebrate development. Such complex transcriptional regulations are thought to be mediated by the co-ordinated binding of transcription factors known as cis-regulatory elements. They allow integration of multiple signals to regulate the expression of specific genes. These elements may not act on the physically closest gene but can act across intervening genes (Spitz et al. 2003). It was shown that certain genomic regions contain arrays of conserved noncoding sequences (CNSs), which are candidates of cis-regulatory elements, clustered around developmental regulatory genes (Bejerano et al. 2004; Woolfe et al. 2005; McEwen et al. 2006; Hufton et al. 2009; Lee et al. 2011; Takahashi and Saitou 2012). Some of them already tested have been shown to act as enhancers in transgenic reporter assays (Pennacchio et al. 2006; McEwen et al. 2006; Hufton et al. 2009; Lee et al. 2011). These genomic regions also show conserved synteny that is a prominent feature of vertebrate genomes. Moreover, these regions are conserved not only orthologously but also paralogously.
These paralogous synteny are most probably derived from ancient genome duplications. Ohno (1970) proposed that the two-round whole-genome duplications (2R WGDs) happened at the intersection of early vertebrate evolution, now called as the 2R hypothesis. It is clear that the 2R WGDs occurred early in the vertebrate evolution from phylogenetic and syntenic analysis of vertebrates and invertebrates (Dehal and Boore 2005; Nakatani et al. 2007; Putnam et al. 2008; Kuraku et al. 2009). These conserved synteny blocks are bearing paralogous genes and CNSs (Kikuta et al. 2007). They are under the strong evolutionary constraints and must have played important roles in the vertebrate evolution. However, the CNSs generated by the 2R WGDs and conserved among paralogous synteny blocks are still not completely documented. These sequences have a vertebrate-specific conservation and might be related to vertebrate morphological features such as neural crest cells and complex brains. Thus, detecting paralogous CNSs and inferring their characteristics are important in understanding the evolution of vertebrate genomes after the 2R WGDs.
The vertebrate Hox clusters are one of the most well-known examples of highly conserved paralogous syntenic regions (e.g., Garcia-Fernández 2005; Matsunami et al. 2010). Vertebrates were shown to possess at least four Hox clusters, whose genes are intimately involved in axial patterning and a strict relationship exists between respective genes and their expression limits in somitic and neural tissues. As a result of their intimate involvement in early development, a change of Hox gene expression often triggers a vertebrate morphological change (Cohn and Tickle 1999). The paralogous genes of the Hox clusters show the similar expression patterns, which suggest that there might be shared gene regulatory mechanisms among paralogous Hox clusters. Previously, we carried out the search of CNSs within these clusters, not only among orthologous clusters but also among paralogous clusters, and identified three paralogous CNSs conserved within all four Hox clusters of vertebrate species, which experienced no further genome duplication (Matsunami et al. 2010). These CNSs should contribute to the Hox cluster organization and gene expression patterns.
We used a region-focused homology search to detect weak paralogous conservations in this study. The method of paralogous CNS identification is critical for the result. Previous studies were mainly based on MegaBLAST (Zhang et al. 2000) search of whole-genome sequences to detect the paralogous CNSs (Bejerano et al. 2004; McEwen et al. 2006) on whole vertebrate species. This method is faster than conventional Basic Local Alignment Search Tool (BLAST) search (Altschul et al. 1997) and is effective to identify the paralogous CNSs showing prominent high conservation among vertebrate genomes. However, it is difficult to detect weak conservation of paralogous noncoding regions by using this strategy. The orthologous conservation of noncoding region is statistically highly significant and easy to detect. In contrast, the paralogous conservation of noncoding region is usually weaker than orthologous conservation of noncoding regions (e.g., Matsunami et al. 2010). To overcome this problem, a region-focused BLAST search of each synteny block is useful. This improved method allowed us to identify much weaker paralogous conservations derived from the 2R WGDs.
In this study, we characterized paralogous synteny blocks derived from the 2R WGDs by using vertebrate genome data and identified both orthologous and paralogous CNSs derived from the 2R WGDs. These paralogous CNSs are frequently located near the protein-coding regions functioned as transcription factors expressed in brain and neural system and have potential to control similar expression patterns of paralogs.
Materials and Methods
Identification of Conserved Synteny Blocks After the 2R WGDs
Nakatani et al. (2007) reported 118 conserved vertebrate linkage (CVL) regions within the human genome through comparison with medaka fish genome sequences. A total of 10,618 protein-coding genes exist in these CVL regions. Each of these regions retain four, three, or two paralogous gene sets or there is only one gene without any paralogous counterparts. These CVL regions were used as synteny block data. The Ensembl BioMart interface (http://www.ensembl.org/biomart/martview/, last accessed January 7, 2013) was used to download gene information. Paralogous genes (four, three, or two gene retentions) were used as markers of paralogous conservation to identify paralogous synteny blocks. When paralogous genes show the same combination of CVL groups, these paralogous gene sets are regarded as paralogous synteny blocks.
Identification of Paralogous CNSs
We identified paralogous CNSs within vertebrate genomes by using the paralogous synteny block data. First, the human genome sequences (Homo sapiens; NCBI36) were downloaded from the Ensembl database (http://www.ensembl.org/, last accessed January 7, 2013), and they were divided into the CVL regions following Nakatani et al. (2007). Repeat and coding regions of each block were masked based on the annotations of Ensembl database. BLAST (Altschul et al. 1997) searches were carried out between human and mouse (Mus musculus; NCBI m37) orthologous blocks to detect orthologous CNSs. The default parameter settings of BLAST search were used. The cutoff value of BLAST search directly influences the result of orthologous CNS detection. Because we effectively detected the orthologous CNSs from the Hox gene cluster regions in our previous study (Matsunami et al. 2010), the same cutoff bit score (200) was used for the human–mouse comparison. To evaluate the conservation, human–mouse CNSs were compared with six other vertebrate genomes. Species used were dog (Canis familiaris; CanFam 2.0), cow (Bos taurus; Btau_4.0), opossum (Monodelphis domestica; monDom5), chicken (Gallus gallus; WASHUC2), lizard (Anolis carolinensis; AnoCar1.0), and frog (Xenopus tropicalis; JGI 4.1). Teleosts were excluded because they underwent an additional genome duplication in their common ancestor. The cutoff e value to identify orthologous CNSs was 10−5. From these orthologous CNSs, we determined CNSs conserved among all the eight vertebrate species (see supplementary fig. S1, Supplementary Material online, for their phylogenetic relationship). Those orthologous CNSs were compared with each other with the threshold of 10−3 e value to detect paralogous CNSs. We also searched the amphioxus genome (Branchiostoma floridae v1.0; Putnam et al. 2008) to confirm whether the detected paralogous CNSs were conserved for invertebrate genomes using default BLAST settings.
Functional Validation of Paralogous CNSs
The detected paralogous CNSs were compared with previously reported sequences, which were already tested for the enhancer activities. The 1,619 human and mouse noncoding elements tested in transgenic mice at 11.5 days postcoitum (dpc) were downloaded from VISTA Enhancer Browser (http://enhancer.lbl.gov/, last accessed January 7, 2013; Visel et al. 2007). These sequences were compared with our paralogous CNSs through BLAST searches.
We also used murine ChIP-seq data produced by Shen et al. (2012) to validate gene regulatory function of paralogous CNSs. Enrichment of H3K4me3 or polII-binding signals is indicative of an active promoter, whereas the presence of H3K27ac outside promoter regions can be used as marks for enhancers (Kim et al. 2005; Heintzman et al. 2007, 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011). CTCF binding is considered as a mark for potential insulator elements (Kim et al. 2007). We downloaded the murine ChIP-seq peak data (CTCF, H3K4me3, H3K27ac, and polII) and predicted enhancer data at E14.5 brain, 8-week-old adult cortex, and 8-week-old adult cerebellum from murine ENCODE HP (http://chromosome.sdsc.edu/mouse/, last accessed January 7, 2013). Overlaps between these peak data and our paralogous CNSs were investigated. It should be noted that Shen et al. (2012) carefully examined levels of conservations of mouse genomic sequences they found. This is a clear contrast to the article of the ENCODE Project Consortium (2012) who puzzlingly assigned newly defined "biochemical functions" for 80% of the human genome.
Other possible functions of paralogous CNSs are noncoding RNA (ncRNA), such as microRNA (miRNA) or long intergenic ncRNA (lincRNA). The human ncRNA sequences (H. sapiens; NCBI37) were downloaded from the Ensembl database (http://www.ensembl.org/, last accessed January 7, 2013) and compared against paralogous CNSs to evaluate the possibility that paralogous CNSs function as ncRNA.
Ontology Analysis of Paralogous CNS-Harboring Genes
The paralogous CNS-harboring genes were defined, and their features were inferred in the following manner. The closest paralogs derived from the 2R WGDs, which are conserved among both paralogous loci, were defined as paralogous CNS-harboring genes (supplementary fig. S2, Supplementary Material online). We then conducted statistical analysis by using Gene Ontology database (http://www.geneontology.org/, last accessed January 7, 2013) to find significantly enriched paralogous CNS-harboring genes. Analysis of gene function enrichment was performed using Fatigo+ web server (Al-Shahrour et al. 2007). The paralogous CNS-harboring genes were compared with the entire human genes in the Gene Ontology database to detect the overrepresented paralogous CNS-harboring genes. The expression regions and timings of paralogous CNS-harboring genes were also analyzed. The eGenetics (http://www.nhmrc.gov.au/your_health/egenetics/index.htm, last accessed January 7, 2013) database was used to investigate gene expression of paralogous CNS-harboring genes. Human anatomical system data, which give information about gene expression regions and expressed timings, were obtained from the eGenetics database by using the Ensembl Biomart (Kelso et al. 2003). We counted numbers of each gene category (paralogs derived from the 2R WGDs and paralogous CNS-harboring genes expressed in each organ and timing) and obtained their frequencies by dividing the numbers by the number of all genes.
Results
Identification of Orthologous CNSs
To identify orthologous CNSs shared among vertebrate species, we carried out comprehensive BLAST searches. We discovered 67,052 CNSs from human and mouse genome comparison under the following settings: >100 bp length and >78% similarity. Their average length was 318 bp. We compared these human–mouse CNSs with other vertebrate species. The genomes of dog, cow, opossum, chicken, lizard, and frog shared 62,611, 65,878, 44,726, 24,549, 19,724, and 10,664 CNSs with human and mouse, respectively. The number of orthologous CNSs is gradually decreased following the order of the evolutionary divergence from human and mouse. Interestingly, we identified more than 10,000 noncoding conservations from the genome of frog (X. tropicalis) in spite of the large divergence from mammals. Among the 67,052 human–mouse orthologous CNSs, 7,650 CNSs were conserved in all the eight species employed in this study; they are listed in supplementary table S1, Supplementary Material online. These CNSs may be recognized among all vertebrate species, which did not experience further genome duplications. Each synteny block contains 65 orthologous CNSs on average. The synteny blocks are scattered across the whole genome of each species except for the mammalian Y chromosome. The lack of conservation might be related to the characteristic feature of this sex chromosome. We could not find any correlations between orthologous gene density and orthologous CNS density, although important development genes such as Hox or Sox have many CNSs, other CNSs are equally distributed in the genome.
Highly Conserved Synteny Blocks
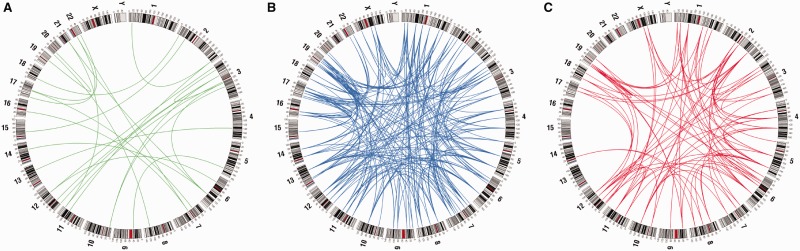
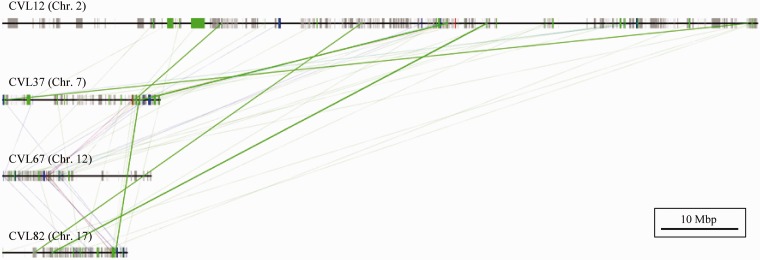
We then identified pairs of paralogous block sets (diparalogous sets), trios of paralogous block sets (triparalogous sets), and quartets of paralogous block sets (tetraparalogous sets) (tables 1 and 2). The genomic locations of paralogous synteny blocks are shown in figure 1. Lundin et al. (2003) reported that the chromosomes bearing the Hox clusters frequently include paralogous genes derived from the 2R WGDs and are organized large synteny blocks. These blocks have been considered as hallmarks of the 2R WGDs and include not only the Hox clusters but also other important genes such as Dlx, Gbx, Gli, and collagen genes. Our study also showed that Hox-linked paralogous synteny blocks had prominent paralogous conservation of not only coding regions but also noncoding regions. These Hox-linked paralogous synteny blocks were one of highly conserved synteny blocks including abundant paralogous genes. These blocks also have the high numbers of paralogous CNSs (shown with thick green lines in the fig. 2) and protein-coding genes. The paralogous gene-dense regions correspond to the dense paralogous CNS regions. Because each paralogous Hox gene showing similar expression pattern controls the vertebrate early development, CNSs in the Hox cluster might function as cis-regulatory elements such as already known paralogous conserved elements (Lehoczky et al. 2004) and control the similar expression of paralogous Hox or neighboring genes.
Table 1.
Number of Paralogous CNS Harboring Genes: Gene and CNS Loss Pattern of Duplicated Regions
| Conservation Level | No. of Paralogous Gene Group (No. of Genes) | No. of Paralogous CNS Group (No. of CNSs) |
|---|---|---|
| 4 | 50 (50 × 4 = 200) | 0 |
| 3 | 220 (220 × 3 = 660) | 3 (3 × 3 = 9) |
| 2 | 861 (861 × 2 = 1,722) | 150 (150 × 2 = 300) |
| 1 | 8,036 (8,036 × 1 = 8,036) | 7,341 (7,341 × 1 = 7,341) |
| Total | 9,167 (10,618) | 7,494 (7,650) |
Table 2.
Number of Paralogous CNS Harboring Genes
| No. of Paralogous CNSs | No. of CNS Harboring Genes |
|---|---|
| Quartets of paralogous genes | |
| 4 | 0 |
| 3 | 2 |
| 2 | 13 |
| 0 | 36 |
| Total | 50 |
| Trios of paralogous genes | |
| 3 | 1 |
| 2 | 29 |
| 0 | 190 |
| Total | 220 |
| Pairs of paralogous genes | |
| 2 | 31 |
| 0 | 830 |
| Total | 861 |
Fig. 1.—
Paralogous synteny blocks within the human genome. Genomic distribution of paralogous synteny blocks is shown. (A) Di-, (B) tri-, and (C) tetraparalogous blocks are identified by the gene order and homology.
Fig. 2.—
Scheme of Hox-linked paralogous block. Paralogous gene conservations and paralogous CNSs conservations are shown. Hox-linked paralogous synteny blocks also show prominent conservation of not only coding regions but also noncoding regions. Tetraparalogous, triparalogous, and diparalogous genes are represented as red, blue, and green thin lines, respectively, and diparalogous CNSs represent green thick lines. We could not identify tetraparalogous or triparalogous CNSs in these regions.
Paralogous CNSs
From the vertebrate-specific orthologous CNSs, 309 paralogous CNSs were identified, and they are listed in supplementary table S2, Supplementary Material online. Only three paralogous CNSs (SB2CNS1101, SB78CNS1119, and SB59CNS435) showed weak sequence similarities with those in the amphioxus genome. Because we could not detect clear sequence similarities between sequences of the amphioxus genome and vertebrate paralogous CNS groups, vertebrate paralogous CNSs might have emerged after amphioxus and the common vertebrate ancestor diverged. We thus focus on these vertebrate-specific CNSs in the later analyses.
We found diparalogous and triparalogous CNSs; however, tetraparalogous CNSs were not detected, probably because conservation of noncoding regions was lower than that of coding regions. Each paralogous synteny block bears several conservation levels of genes and CNSs, such as trios or pairs. We compared paralogous CNSs detected in this study with already described CNSs. Among the 309 paralogous CNSs, 194 (63%) were newly determined, whereas confirming 115 previously reported paralogous CNSs (McEwen et al. 2006). There were 15 paralogous CNSs corresponding to ncRNA (5 pairs, 1 trio, and 2 members of pair). These ncRNA included 12 miRNA and 3 lincRNA (supplementary table S3, Supplementary Material online). The paralogous CNS trio was located near the MEF family genes (MEF2A, MEF2C, and MEF2D). These paralogous CNSs function as ncRNA and are assumed to regulate the expression of neighboring genes.
According to VISTA Enhancer Browser database (Visel et al. 2007), 83 (27%) paralogous CNSs were already validated for their enhancer function through transgenic mice experiments. The 61% (51 of 83) of CNSs had positive enhancer functions, when mice were at 11.5 days postcoitum. Although the remaining 22 (39%) CNSs were not detected to have an enhancer activity, they have a possibility of silencer function. Among CNSs having positive enhancer functions, the 82% (42 of 51) of CNSs showed the prominent expression at the developmental brain region. Moreover, we used the available ChIP-seq data to confirm the enhancer function of paralogous CNSs in the brain region. From the read mapping data of ChIP-seq, 55, 16, and 17 paralogous CNSs corresponded to at least one of ChIP-seq peaks at E14.5 brain, 8-week-old adult cortex, and 8-week-old adult cerebellum, respectively (supplementary table S2, Supplementary Material online). In summary, 72 paralogous CNSs were overlapped with ChIP-seq peaks. Although these experiments covered only three time points of developmental stages, 103 paralogous CNSs (42 CNSs are positive at enhancer database and 72 CNSs are positive at ChIP-seq data) showed enhancer function in the brain region. These results suggest that paralogous CNSs may frequently regulate genes, which express brain regions at several developmental stages.
Figure 3 illustrates one of paralogous CNS pairs located in CVL11 and CVL35. These CVLs contain paralogous transcription factor genes (POU3F2 and POU3F3), and they are assigned as "paralogous CNS-harboring genes" for these CVL regions. In the multiple alignment, orthologous CNSs are obviously highly conserved. However, the conservation of paralogous CNSs is weaker than that of orthologous CNSs, so that previous studies could not detect these paralogous CNSs. These newly detected paralogous CNSs have a possibility to work as a distal enhancer for POU3F paralogous genes.
Fig. 3.—
Paralogous CNSs shared between POU3F2 and POU3F3 genes. Genomic locations of each orthologous CNS in the human genome and the alignment of paralogous CNS are shown. This paralogous CNS pair is located nearby POU3 paralogs, POU3F2 (BRN2), and POU3F3 (BRN1), that is derived from the 2R WGDs. These are strong candidates of gene regulatory sequences of these paralogs.
Location of CNSs and Paralogous CNS-Harboring Genes
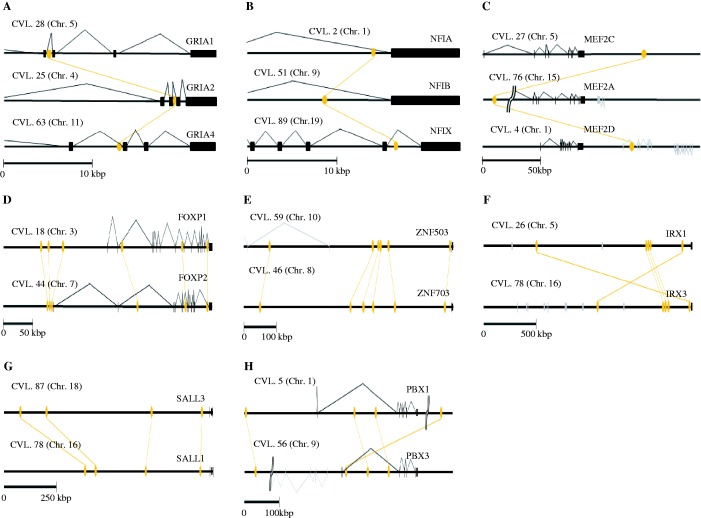
We searched paralogous CNS-harboring genes of each paralogous CNS (tables 1 and 2). We then inferred the functional bias of paralogous CNS-harboring genes based on the Gene Ontology database and the gene expression database. Tables 3 and 4 present paralogous CNS-harboring genes with particularly abundant paralogous CNSs. The majority of paralogous CNSs are located at intron, upstream region or downstream region of the genes encoding transcription factor. We identified three gene families each of which retained paralogous CNS trios (fig. 4A–C) and five gene families each of which retained more than three paralogous CNS pairs (fig. 4D–H). The functions of paralogous CNSs located near well-studied transcription factors such as FoxP1/P2, Sox14/21, and the Irx cluster are already known (de la Calle-Mustienes et al. 2005; McEwen et al. 2006). These paralogous CNSs function as distal enhancers and partially share their gene expression regions between paralogous pairs. Among other paralogous CNSs, some paralogous CNSs also showed experimentally validated distal enhancer functions in the database or function as ncRNA (supplementary tables S2 and S3, Supplementary Material online). This result strongly suggests that the remaining paralogous CNS pairs whose function is not yet tested have also enhancer or ncRNA function.
Table 3.
List of Paralogous CNSs Harboring Genes: Pair of Paralogous CNSs
| Number of Pairs | Pair of Harboring Gene |
|---|---|
| 6 | FOXP1&FOXP2, ZNF503&ZNF703 |
| 5 | IRX1&IRX3 |
| 4 | PBX1&PBX3, SALL1&SALL3 |
| 3 | EBF1&EBF3, EVX1&EVX2, NR2F1&NR2F2, POU4F1&POU4F2, SOX5&SOX6 |
| 2 | ESRP1&ESRP2, FOXB1&FOXB2, HOXA5&HOXB5, LMO1&LMO3, LRBA&NBEA, LRP3&LRP12, NEUROD1&NEUROD2, NRXN1&NRXN3, OTX1&OTX2, POU3F1&POU3F2, POU3F2&POU3F3, PRDM16&MECOM, SLIT2&SLIT3, SOX14&SOX21, TCF4&TCF12, TFAP2A&TFAP2B, TOX&TOX3, TSHZ1&TSHZ2, VRK1&VRK2 |
| 1 | ACTL6A&ACTL6B, ARL5A&ARL5C, ARSB&ARSI, ARSJ&ARSB, BACE1&BACE2, BMP3&GDF10, CCNL1&CCNL2, CPA1&CPA2, CUX1&CUX2, DNM1&DNM3, ENPP2&ENPP3, FOXO1&FOXO3, FOXP2&FOXP4, GNB2&GNB4, GPC2&GPC6, GPC3&GPC5, GPM6A&PLP1, GRIA2&GRIA3, HMGB1&HMGB3, HOXA4&HOXD4, HSF2&HSF4, ING1&ING2, INPP5D&SH2D1A, IRX2&IRX5, KANK1&KANK4, KCNK9&KCNK15, KHDRBS2&KHDRBS3, LASS3&LASS6, MACF1&DST, MBNL1&MBNL2, MCTP1&MCTP2, MEF2A&MEF2C, MEIS1&MEIS2, NFIA&NFIB, NPNT&EGFL6, ODZ2&ODZ3, P4HA1&P4HA2, PDE4B&PDE4C, PIK3C2A&PIK3C2B, PLS1&PLS3, PTCH1&PTCH2, QSOX1&QSOX2, R3HCC1&c10orf28, RALA&RALB, RHAG&RHCG, RNF38&RNF44, SALL1&SALL4, SEC24C&SEC24D, SEPT6&SEPT10, SGMS1&SGMS2, SH3RF3&SORBS2, SHH&IHH, SLC12A1&SLC12A3, SLC12A2&SLC12A3, SLC4A4&SLC4A10, SLC6A15&SLC6A18, SLC9A2&SLC9A3, SLIT1&SLIT2, SMAD2&SMAD3, SOX1&SOX2, SOX2&SOX3, ST8SIA3&ST8SIA4, SULF1&GNS, TFAP2A&TFAP2C, ZEB2&KIAA0087, ZFHX3&ZFHX4, ZIC2&ZIC3, ZNF423&ZNF521 |
Table 4.
List of Paralogous CNSs Harboring Genes: Trio of Paralogous CNSs
| Number of Trio | Trio of Harboring Gene |
|---|---|
| 1 | MEF2A&MEF2C&MEF2D, NFIA&NFIB&NFIX, and GRIA1&GRIA2&GRIA4 |
Fig. 4.—
The locations of paralogous CNS. The triparalogous CNSs were identified only near (A) GRIA gene family, (B) NFI gene family, and (C) MEF2 gene family. The paralogous CNS-harboring genes having more than three pair-paralogous CNS pairs are only five gene families. These are (D) FOXP1/P2, (E) ZNF503/703, (F) IRX1/3, (G) SALL1/3, and (H) PBX1/3. Protein-coding regions of paralogous CNS-harboring genes are represented by black boxes. The orange ellipse is paralogous CNSs. The connected lines show paralogous conservation of each CNS.
Table 5 is the result of the gene ontology analysis. The paralogous CNS-harboring genes were compared with the entire human genes in the Gene Ontology database. We found that paralogous CNSs were frequently located near genes which function as sequence-specific DNA binding (i.e., transcription factors). These results are consistent with previous studies and suggest that, after the 2R WGDs, genes which function as gene regulation were more conservative than genes having other functions.
Table 5.
Overrepresented Gene Functions of Host Genes
| GO Term | P |
|---|---|
| Sequence-specific DNA binding (GO:0043565) | 3.39E–15 |
| Ionotropic glutamate receptor activity (GO:0004970) | 7.69E–05 |
| Phosphoinositide binding (GO:0035091) | 6.05E–05 |
| Lipid kinase activity (GO:0001727) | 5.33E–04 |
| 1-Phosphatidylinositol-3-kinase activity (GO:0016303) | 8.92E–06 |
| Follicle-stimulating hormone receptor activity (GO:0004963) | 1.77E–04 |
| Low-density lipoprotein receptor activity (GO:0005041) | 3.41E–04 |
Note.—Adjusted P values are calculated by comparing the distribution of the host genes with that of human genes.
The expression regions and stages of paralogous CNS-harboring genes were investigated by examining the eGenetics database. We found that paralogous CNS-harboring genes frequently include genes expressed in the brain at early developmental stages (supplementary fig. S3, Supplementary Material online). The proportions of paralogous CNSs enhancer activities were compared with the proportions of the entire enhancer database (table 6 and supplementary table S2, Supplementary Material online). Majority of paralogous CNSs in the enhancer database show expression in the brain at an early developmental stage (P < 0.01, χ2 test). These results imply that existing paralogous CNSs may contribute to the vertebrate-specific complex brain morphology at their early developmental stages.
Table 6.
Proportion of Enhancer Activities
| Expression | Paralogous CNSs | All Sequences in Database |
|---|---|---|
| No expression | 22 (26.51%) | 815 (50.34%) |
| At brain region | 42 (50.60%) | 416 (25.69%) |
| At other region | 19 (22.89%) | 388 (23.97%) |
| Total | 83 | 1,619 |
Discussion
In this study, we identified vertebrate-specific CNSs, and they may be related to the vertebrate-specific features. Previously, CNSs were described by several criteria. Bejerano et al. (2004) defined 483 ultra-conserved elements (UCEs). These included coding regions and ≥200 bp length with 100% identity among human, rat, and mouse. Woolfe et al. (2007) defined 6,957 CNEs that were ≥40 bp length and 65% identity between human and fugu. These genome-wide studies used very stringent criteria and often missed rather weak paralogous conservations. It is difficult to compare those results with our present study, partly because compared genome sequences are different. Our results covered 308 human–rat–mouse UCEs and 3,388 human–fugu CNEs, whereas other conserved elements previously detected are not overlapped with our CNSs. We defined vertebrate-specific CNSs as conserved among all the eight vertebrate species in this study. Because we used genome sequences of many species, other conserved elements missing in some species were not included in our results. These missed conserved elements may be overlapped with long gap regions. Because some CNSs are missed because of low genome quality, our results may underestimate the number of orthologous CNSs. However, it is clear that orthologous CNSs we detected are significantly conserved throughout the vertebrate evolution.
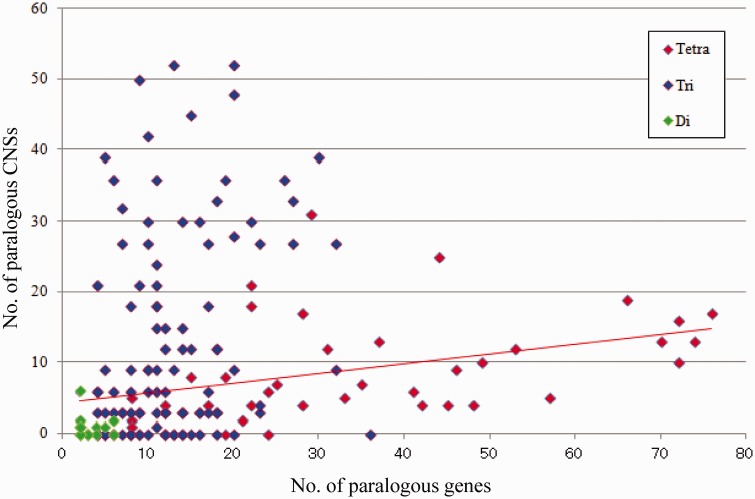
Why the paralogous synteny block is conserved remains elusive. The genomic regulatory block hypothesis is proposed to explain this enigmatic synteny blocks (Becker and Lenhard 2007; Kikuta et al. 2007). This hypothesis suggests that CNSs scattered across each synteny block prevent each block from breakage of the synteny. Under this hypothesis, paralogous CNSs maintain paralogous conserved synteny. We inferred the relationship between the distribution of paralogous CNSs and the distribution of paralogous genes. As the result, paralogous gene orders (synteny) and paralogous CNS conservations are weakly correlated, especially for tetraparalogous synteny blocks (Pearson's product–moment correlation coefficient = 0.485; fig. 5). This means that paralogous synteny blocks bearing many paralogs also include abundant paralogous CNSs. In other words, highly conserved syntenic regions have more paralogous CNSs. This implies that these paralogous CNSs may constrain the synteny blocks from the breakage and play a key role in the genomic regulatory block hypothesis.
Fig. 5.—
Relationship between numbers of paralogs and paralogous CNSs derived from the 2R WGDs. Paralogs and paralogous CNSs within the synteny blocks were counted and were plot to a scatter plot. The horizontal axis is the number of conserved paralogous genes. Vertical axis is the number of conserved paralogs CNSs. The red line is an approximate linear regression of tetraparalogous block points. There is a clear positive correlation.
The majority of vertebrate CNSs may have been generated before the divergence of the major extant vertebrate lineages. The sequencing of the genome of a cephalochordate, amphioxus (Branchiostoma floridae), has uncovered traces of the origins of very small number of vertebrate CNSs (Holland et al. 2008; Putnam et al. 2008). Although we also found few weak conservations in noncoding regions between the amphioxus genome and vertebrate orthologous CNSs, we could not detect conservations that were shared among each vertebrate paralogous loci and amphioxus noncoding regions. Invertebrate groups have been found to possess their own sets of CNSs (Glazov et al. 2005; Vavouri et al. 2007), and interestingly, there are similarities between the functions of genes around with both vertebrate and invertebrate CNSs cluster. This suggests parallel evolution of CNS networks (Vavouri et al. 2007). Consequently, although the slow evolution of coding sequences can be charted readily across the invertebrate/vertebrate boundary, the CNSs changed very quickly during the vertebrate evolution. Recently, noncoding sequences that are conserved from several basal vertebrates were reported. The elephant shark (Callorhinchus milii) is a cartilaginous fish and a basal jawed vertebrate. Its genome contains a few thousand vertebrate CNSs in spite of their low-coverage genome information (Lee et al. 2011). However, the number of CNSs retained in the sea lamprey (Petromyzon marinus), one of extant jawless vertebrates, is much smaller than that of other vertebrates (McEwen et al. 2009). The lamprey CNSs show remarkably shorter lengths than those of vertebrates and low homology with vertebrate CNSs. Whether the jawless vertebrate genomes experience the 2R WGDs is still unclear, so that the orthologies of sequences are difficult to assign (Kuraku et al. 2009). Nevertheless, these observations suggest that vertebrate CNSs have not constantly evolved. We can interpret that lamprey noncoding sequences are extremely changed such as their coding regions (Qiu et al. 2011), or the evolutionary rate of jawed vertebrate noncoding region has become slower after the jawless vertebrate lineages branched off. To answer these issues, we should analyze the high-quality basal vertebrate genomes. In either case, the existence of massive CNSs is not an usual situation compared with other invertebrate species closer to vertebrates. These CNSs might contribute to vertebrate-specific features.
The existences of paralogous CNSs detected in this study are difficult to explain by previous duplication models. Classical models predicted that the most likely fate of duplicated genes is the degeneration of one of the pair to a pseudogene (or completely lost from the genome) or less frequently the acquisition of novel gene functions as a result of alterations in coding or regulatory sequences in a process known as neo-functionalization. Force et al. (1999) proposed the duplication–degeneration–complementation (DDC) model in which duplicated genes undergo complementary deleterious mutations in independent subfunctions, so that both genes are required to share the functions of the ancestral gene. These models are difficult to explain the existence of paralogous CNSs. An alternative model is the gene balance hypothesis proposed by Papp et al. (2003). It postulates that selection against gene dosage imbalances will promote the retention of particular types of genes, though this model has not explicitly been applied to evolutionary fates of noncoding sequences (Papp et al. 2003). Immediately after a WGD event, genome-wide relative gene dosage is maintained, but subsequent step-wise mutation or deletion of duplicated genes can lead to deleterious dosage imbalances. Genes whose proteins have many interaction partners may be more sensitive to these dosage changes, possibly leading to an over-retention of highly connected gene functions, such as transcriptional regulators and signaling complexes. Conversely, small-scale duplications immediately disrupt relative dosage, so highly connected genes should avoid this type of duplication during evolution. Duplicated genes derived from the 2R WGDs are more sensitive to dosage change than other duplicated genes and frequently associated with disease in human genome (Makino and McLysaght 2010). This different correlation between gene retention after WGD and small-scale duplication is a key distinction between the gene balance hypothesis and the DDC model; DDC model should promote the same patterns of gene retention for all types of gene duplication. In support of the gene balance hypothesis, vertebrate genes that function in transcription regulation or signal transduction are over-retained after the 2R WGD events but not after small-scale duplications (Blomme et al. 2007). We also found that the paralogous CNSs are frequently retained near the transcription factors. The transcription and developmental genes have more complex function than other genes, such as pleiotropic expressions, highly connected protein networks, and dosage sensitivity. These characters may allow greater subfunctionalization. They often share gene expression regions and have similar functions among paralogous genes. However, the existence of paralogous CNSs is difficult to be explained by the DDC model, because this model does not assume same enhancer functions among paralogous loci. The one possible explanation of the existence of paralogous CNSs is the gene balance hypothesis (Papp et al. 2003). These paralogous CNSs have possibility to control similar expression patterns of paralogs and dosage compensation of paralogs through the highly conserved sequences.
The alternative possible function of paralogous CNSs is ncRNA. Rinn et al. (2007) reported interchromosomal interactions between paralogous regions through ncRNA. Some enhancers act not only cis but also trans via ncRNA transcription (Ørom et al. 2010). Paralogous CNSs may be related to these interchromosomal interactions of duplicated genome regions.
Supplementary Material
Supplementary figures S1–S3 and table S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Drs. Shigehiro Kuraku, Akatsuki Kimura, Toshiyuki Takano, Hiroshi Akashi, and Toshihiko Shiroishi for their many helpful discussions, suggestions, and comments. This work was supported partly by the Graduate University for Advanced Studies (SOKENDAI) and SOKENDAI Short-term Study-abroad Program to M.M. and by a Grant-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to M.M. and N.S.
Literature Cited
- Al-Shahrour F, et al. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TS, Lenhard B. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genomics. 2007;278:487–491. doi: 10.1007/s00438-007-0287-0. [DOI] [PubMed] [Google Scholar]
- Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Blomme T, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2007;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:1700–1708. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernández J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton AL, et al. Deeply conserved chordate noncoding sequences preserve genome synteny but not drive gene duplication retention. Genome Res. 2009;19:2036–2051. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso J, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Fredman D, Rinkwitz S, Lenhard B, Becker TS. Retroviral enhancer detection insertions in zebrafish combined with comparative genomics reveal genomic regulatory blocks—a fundamental feature of vertebrate genomes. Genome Biol. 2007;8(1 Suppl):S4. doi: 10.1186/gb-2007-8-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 26. 2009:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 2011;28:1205–1215. doi: 10.1093/molbev/msq304. [DOI] [PubMed] [Google Scholar]
- Lehoczky JA, Williams ME, Innis JW. Conserved expression domains for genes upstream and within the HoxA and HoxD clusters suggests a long-range enhancer existed before cluster duplication. Evol Dev. 2004;6:423–430. doi: 10.1111/j.1525-142X.2004.04050.x. [DOI] [PubMed] [Google Scholar]
- Lundin LG, Larhammar D, Hallböök F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics. 2003;3:53–63. [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami M, Sumiyama K, Saitou N. Evolution of conserved non-coding sequences within the vertebrate Hox clusters through the two-round whole duplications revealed by phylogenetic footprinting analysis. J Mol Evol. 2010;71:427–436. doi: 10.1007/s00239-010-9396-1. [DOI] [PubMed] [Google Scholar]
- McEwen GK, et al. Ancient duplicated conserved noncoding elements in vertebrates: a genomic and functional analysis. Genome Res. 2006;16:451–465. doi: 10.1101/gr.4143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen GK, Goode DK, Parker HJ, Woolfe A, Callaway H, et al. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS Genet. 2009;12:e1000762. doi: 10.1371/journal.pgen.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;43:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Qiu H, Hildebrand F, Kuraku S, Meyer A. Unresolved orthology and peculiar coding sequence properties of lamprey genes: the KCNA gene family as test case. BMC Genomics. 2011;12:325. doi: 10.1186/1471-2164-12-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Saitou N. Identification and characterization of lineage-specific highly conserved noncoding sequences in mammalian genomes. Genome Biol Evol. 2012;4:641–657. doi: 10.1093/gbe/evs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Walter K, Gilks WR, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:116–130. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. CONDOR: a database resource of developmentally-associated conserved non-coding elements. BMC Dev Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.