Abstract
Microsatellites, or simple sequence repeats (SSRs), are common and widespread DNA elements in genomes of many organisms. However, their dynamics in genome evolution is unclear, whereby they are thought to evolve neutrally. More available genome sequences along with dated phylogenies allowed for studying the evolution of these repetitive DNA elements along evolutionary time scales. This could be used to compare rates of genome evolution. We show that SSRs in insects can be retained for several hundred million years. Different types of microsatellites seem to be retained longer than others. By comparing Dipteran with Hymenopteran species, we found very similar patterns of SSR loss during their evolution, but both taxa differ profoundly in the rate. Relative to divergence time, Diptera lost SSRs twice as fast as Hymenoptera. The loss of SSRs on the Drosophila melanogaster X-chromosome was higher than on the other chromosomes. However, accounting for generation time, the Diptera show an 8.5-fold slower rate of SSR loss than the Hymenoptera, which, in contrast to previous studies, suggests a faster genome evolution in the latter. This shows that generation time differences can have a profound effect. A faster genome evolution in these insects could be facilitated by several factors very different to Diptera, which is discussed in light of our results on the haplodiploid D. melanogaster X-chromosome. Furthermore, large numbers of SSRs can be found to be in synteny and thus could be exploited as a tool to investigate genome structure and evolution.
Keywords: microsatellite conservation, genome evolution, social Hymenoptera, Drosophila, mosquitoes, generation time, haplodiploidy, synteny
Introduction
Large parts of eukaryotic genomes are composed of simple sequence repeats (SSRs), also called short tandem repeats (STRs) or microsatellites, are a common feature, and can account for up to 4% of genomes (Ellegren 2004; Schlötterer 2004; Molnar et al. 2012). These repeats occur throughout the genomes, the majority in noncoding regions, but they can be found also in protein coding sequences. Numerous studies showed apparent differences regarding their density, distribution, and composition (Tóth et al. 2000; Katti et al. 2001; Ross et al. 2003; Lim et al. 2004; Buschiazzo and Gemmell 2006; Galindo et al. 2009; Mayer et al. 2010; Pannebakker et al. 2010). Because of high levels of polymorphism in number of repeats, SSRs are widely used as molecular markers in a large diversity of studies. The high degree of polymorphism has been attributed to DNA slippage mutation during replication (Leclercq et al. 2010), but the process may be more complex and is still not fully understood (Li et al. 2002, 2004; Ellegren 2004; Buschiazzo and Gemmell 2006; Eckert and Hile 2009; Bhargava and Fuentes 2010; Kelkar et al. 2010; Leclercq et al. 2010). Frequent repeat number variation in SSRs at a rate of 10−2–10−6 per locus per generation (Schlötterer 2000) often follows a regular pattern which can be used as a short-term molecular clock (Sun et al. 2009) and for the inference of phylogeny (Buschiazzo and Gemmell 2009).
Traditionally, SSRs are regarded as nonfunctional and hence neutrally evolving. Consequently, these genetic elements have a higher mutation rate compared with functional or coding sequences, which are more conserved in response to selection (Schlötterer 2000). This, in combination with the polymorphic nature of SSRs, leads to the expectation of a highly dynamic system of gain, change, and loss of SSR repeats in genomes within natural populations. Nevertheless, there have been several reports of highly conserved SSRs within and across taxa. Interspecies amplification of SSR loci reveals that many SSRs are shared between closely related species (Blanquer-Maumont and Crouauroy 1995; Primmer et al. 1996; Green et al. 2001; Reber Funk et al. 2006; Barbará et al. 2007; Katada et al. 2007; Meglécz et al. 2007; Paxton et al. 2009; Stolle et al. 2009) and, for a few loci, even between species with a phylogenetic split of more than 100 Myr (Vaiman et al. 1994; FitzSimmons et al. 1995; Rico et al. 1996; Ezenwa et al. 1998; Moore et al. 1998; Green et al. 2001; Barbará et al. 2007; Buschiazzo and Gemmell 2009). Recently, Buschiazzo and Gemmell (2010) showed that a significant fraction of SSRs in vertebrates have been conserved for up to 450 Myr, but the mechanisms underlying this conservation over long evolutionary times are unknown.
Some SSRs possess biological function regarding chromosome stability, RNA folding, amino acid repeats or relations to human diseases, recombination hotspots, or transposable elements (Goldstein and Schlötterer 1999; Li et al. 2002, 2004; Brandström et al. 2008; Thomou et al. 2009; Bonen et al. 2010; Grover and Sharma 2011; Wang et al. 2012). However, the large majority of SSR repeats are located in regions without a known biological function. Nevertheless, on the basis of the flanking regions adjacent to SSRs, Stolle et al. (2011) reported a high structural conservation of the chromosomes in the honeybee Apis mellifera and the bumblebee Bombus terrestris, which diverged approximately 100 Ma. Indeed genomes of Hymenoptera have been reported to be slowly evolving compared with those of Dipteran flies or various other animal groups (Weinstock et al. 2006; Stolle et al. 2011). However, the disparate life histories within the Insecta have a considerable impact when comparing evolutionary time scales across taxa. For example, generation time and effective population size may differ by several orders of magnitude. Social insects typically have very long-lived sexual females but with a relatively small effective population size, as per generation, only one or few individuals are responsible for reproduction. In addition, other particular characteristics such as haplodiploidy, multiple mating, worker reproduction, longevity of individuals, and colonies may further obscure the actual rates of evolutionary change over generations.
Here, we investigate SSR conservation across different insect groups. Our expectation, based on the polymorphic and neutral nature of SSRs, was a fast decay of SSR loci in both Hymenoptera and Diptera. Our data suggest that high proportions of SSRs can be conserved between species. Some even can be retained for hundreds of millions of years of divergent evolution. Comparing the insect groups of Hymenoptera and Diptera, the degree of conservation differs markedly, depending upon SSR types and motif lengths, but the overall pattern is surprisingly similar. Using species with well-established phylogenies and robust divergence time estimates, we compare the rates of evolution accounting for the effect of generation time.
Materials and Methods
Genome Sequences and SSR Identification
Whole-genome sequences of 12 Drosophila, 3 mosquitoes, and 11 Hymenopteran species (fig. 1) were retrieved via GenBank (National Center for Biotechnology Information [NCBI]) and flybase (January 2011) and scanned for SSR repeats using the Phobos software (version 3.3.11, Mayer 2006–2010) with the following settings: imperfect search with minimum thresholds of 70% repeat perfection, four repetitive units of 2–5 bp motifs, and 10 bp total length, extraction of 350 bp flanking sequence at both sides. We choose these repeats because they typically account for the majority of SSRs. Further, we left out the mononucleotide repeats to avoid a bias due to differential representation in different genomes caused by the problems of sequencing homopolymers.
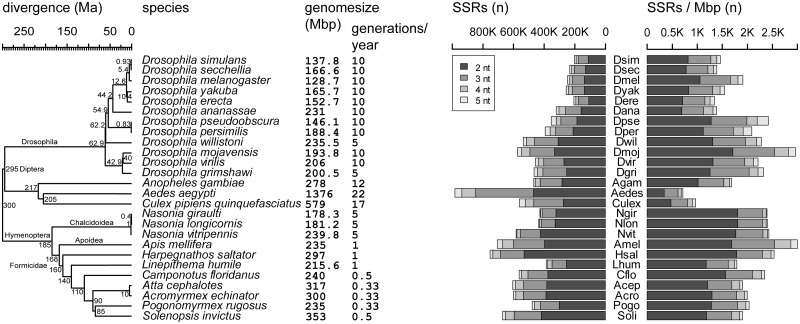
Fig. 1.—
Species overview. Summary data for species used in this study. Their phylogenetic relationships are shown at the left with divergence times at the nodes, species names with respective genome size and generation time are given in the middle part, and SSR counts and densities for each species with species abbreviations is given at the right part.
The output, with standardized SSR motifs (e.g., GA, TC, and CT are defined as AG, automatically done by Phobos), was then filtered for potential double entries, for example, if a specific imperfect SSR was found as the dinucleotide repeat AT and the trinucleotide AAT. Therefore, SSRs with a distance of 15 bp or closer to the start or end of the following SSR were discarded. This yielded initial information about the composition and genome-wide distribution of these SSRs for each species (fig. 1).
BLAST Analyses and Filtering
Libraries of SSRs flanked by 350 bp sequence were then used in pairwise Basic Local Alignment Search Tool (BLAST) analyses (NCBI BLAST 2.2.25+ [Altschul et al. 1990]), using one library (species A) as query and another library (species B) as reference. The analyses were performed using a custom-made Perl script with the SSRs sequences themselves being masked as “N.” For each query sequence, the four highest BLAST hits within the reference sequences were recorded.
The resulting BLAST hits were then processed with a second custom-made Perl script. First, those BLAST hits where the SSR motif of the query was not matching that of the reference were discarded. Second, if a query sequence yielded multiple BLAST hits on the identical reference sequence, for example, due to the gap by the masked SSR, the scores of these BLAST hits were summed up. Third, BLAST hits smaller than 100 bp and 70% or less sequence identity were excluded from further analyses.
Each query sequence, representing a SSR of species A, which passed these thresholds, was then assigned to a single sequence within the reference, representing a SSR of species B. If for a query sequence more than one BLAST hit within the reference sequences was remaining after the filtering steps, the assignment was conducted by choosing the BLAST hit with highest score. In cases where there were two or more BLAST hits with exactly same score, these entries were discarded as it could not be matched unambiguously, even if this score was the highest among the recorded BLAST hits. Similarly, we searched for multiple matches to a reference sequence. If there were more than one query sequence assigned to the same reference sequence, all were discarded but the one reference sequence which gave the highest BLAST score with the respective query. Again, we excluded those entries where two or more reference sequences had the exactly same BLAST score, even if this score represented the highest BLAST score.
Hence, the final data set contained only pairs of unique query sequences assigned to unique reference sequences, both having the same SSR motif irrespective of the number of repeat units or level of perfection. For each final data set, the result of the pairwise comparison between a query and a reference, the number of detected SSR loci was related to the number of SSR loci in the respective reference. Each query SSR locus detected in the reference is defined as a conserved SSR, although we cannot rule out the possibility that a SSR was lost during evolution within a species or lineage and independently a new, nonhomologous SSR with the same motif arose at the same or very similar position. The conserved SSR loci were determined for each analyzed species pair, the sum and the numbers for each individual SSR motif.
Validation of the Method
We validated our method by comparing the SSR libraries of Drosophila melanogaster, A. mellifera, Solenopsis invicta, Atta cephalotes, and Nasonia vitripennis with itself. The expectation was a correct recovery of each detected SSR after applying the very same thresholds, filtering, and processing steps. The result of this test is a benchmark of our approach and allows for the determination of the false-positive error rate by simply detecting erroneously assigned SSRs in the final data set.
Furthermore, we evaluated the Muller element B (chromosome 2L) of the D. melanogaster genome for synteny between D. melanogaster and D. simulans.
To proof the assumption that the BLAST analysis gives the same result irrespective which species is used as query and which as reference in a species pair, we conducted some selected reciprocal runs for the species pairs Dmel–Dsim, Dmel–Dpse, Dmel–Dvir, Amel–Soli, and Acep–Soli (for abbreviations see fig. 1).
Divergence Time and Generation Time
The generation time (here the number of generations produced per year, fig. 1) was estimated from data from the literature. The Dipteran species used in this study typically have a short generation time, and in particular, the tropical species can produce many generations per year (>20 [Keightley 2000]).
For most Drosophila species, we assumed 10 generations per year (Li and Nei 1977; Laayouni et al. 2003; Hutter et al. 2007; Cutter 2008; Barker 2011). Some Drosophila species from mountainous areas or from colder climates or such species with more extended life cycles (Begon 1976; Keightley 2000; Jennings et al. 2011) are known to have fewer generations per year, similar to D. willistoni and the Hawaiian D. grimshawi for which we assumed five generations per year.
The Hymenopteran Nasonia species are nonsocial parasites and have been reported to reproduce four to five times a year in the wild (Werren J, personal communication) (Raychoudhury et al. 2010; Powell et al. 2011). The generation times for the other, eusocial, species are typically much longer. For A. mellifera, Linepithema humile, and Harpegnathos saltator, sexual offspring is typically produced once a year. The ant species Camponotus floridanus, S. invicta, and A. cephalotes, Acromyrmex echinator, and Pogonomyrmex rugosus with larger colonies have more long living queens, and sexual offspring is only produced every 2–3 years (Hölldobler and Wilson 1990; Taber 1998, 2000; Bekkevold and Boomsma 2000; Peeters and Liebig 2000; Gadau et al. 2012).
Divergence time estimates were obtained from several phylogenetic studies based on both the fossil record and molecular clocks (Rasnitsyn and Quicke 2002; Tamura et al. 2004; Grimaldi and Engel 2005; Moreau et al. 2006; O’Grady and Desalle 2008; Werren et al. 2010; Gadau et al. 2012).
On the basis of the divergence time (in million years before present) and the number of generations, we obtained an estimate of how many generations had passed from the separation of lineage or species until the present (fig. 1 and table 1).
Table 1.
Pairwise Comparisons for SSR Conservation
| Query | Reference | Conserved SSRs (n) | SSRs in Reference (n) | Conserved (%) | Divergence Time (Ma) | Generations per Year | Generations (Million) |
|---|---|---|---|---|---|---|---|
| Dper | Dpse | 232,926 | 353,383 | 65.91 | 0.85 | 10 | 8.5 |
| Dsec | Dsim | 129,022 | 201,053 | 64.17 | 0.93 | 10 | 9.3 |
| Dsim | Dmel | 115,053 | 246,106 | 46.75 | 5.4 | 10 | 54 |
| Dsec | Dmel | 117,217 | 246,106 | 47.63 | 5.4 | 10 | 54 |
| Dere | Dyak | 93,489 | 256,427 | 36.46 | 10.4 | 10 | 104 |
| Dere | Dmel | 88,213 | 246,106 | 35.84 | 12.6 | 10 | 126 |
| Dyak | Dmel | 90,661 | 246,106 | 36.84 | 12.6 | 10 | 126 |
| Dmoj | Dvir | 104,551 | 456,107 | 22.92 | 40 | 10 | 400 |
| Dgri | Dvir | 86,405 | 456,107 | 18.94 | 42.9 | 7.5 | 321.75 |
| Dana | Dyak | 36,375 | 256,427 | 14.19 | 44.2 | 10 | 442 |
| Dana | Dmel | 36,511 | 246,106 | 14.84 | 44.2 | 10 | 442 |
| Dana | Dsim | 34,477 | 201,053 | 17.15 | 44.2 | 10 | 442 |
| Dper | Dmel | 26,608 | 246,106 | 10.81 | 54.9 | 10 | 549 |
| Dpse | Dmel | 27,504 | 246,106 | 11.18 | 54.9 | 10 | 549 |
| Dwil | Dmel | 14,855 | 246,106 | 6.04 | 62.2 | 7.5 | 466.5 |
| Dwil | Dsim | 13,619 | 201,053 | 6.77 | 62.2 | 7.5 | 466.5 |
| Dwil | Dvir | 22,130 | 456,107 | 4.85 | 62.9 | 7.5 | 471.75 |
| Dgri | Dmel | 13,618 | 246,106 | 5.53 | 62.9 | 7.5 | 471.75 |
| Dmoj | Dmel | 14,099 | 246,106 | 5.73 | 62.9 | 10 | 629 |
| Dvir | Dmel | 14,742 | 246,106 | 5.99 | 62.9 | 10 | 629 |
| Aedes | Culex | 8,689 | 561,135 | 1.55 | 205 | 21 | 4,305 |
| Agam | Culex | 4,311 | 561,135 | 0.77 | 217 | 16 | 3,472 |
| Pogo | Soli | 104,911 | 671,437 | 15.62 | 85 | 0.42 | 35.42 |
| Acro | Soli | 127,832 | 671,437 | 19.04 | 90 | 0.42 | 37.5 |
| Acep | Soli | 120,761 | 671,437 | 17.99 | 90 | 0.42 | 37.5 |
| Cflo | Soli | 77,896 | 671,437 | 11.6 | 110 | 0.5 | 55 |
| Lhum | Soli | 73,464 | 671,437 | 10.94 | 140 | 0.75 | 105 |
| Hsal | Soli | 66,510 | 671,437 | 9.91 | 160 | 0.75 | 120 |
| Amel | Soli | 19,323 | 671,437 | 2.88 | 168 | 0.75 | 126 |
| Nvit | Soli | 5,933 | 671,437 | 0.88 | 185 | 2.25 | 416.25 |
| Acro | Acep | 285,148 | 603,455 | 47.25 | 10 | 0.33 | 3.33 |
| Lhum | Acep | 66,203 | 603,455 | 10.97 | 140 | 0.67 | 93.33 |
| Hsal | Cflo | 59,205 | 562,525 | 10.52 | 160 | 0.75 | 120 |
| Cflo | Amel | 26,633 | 704,546 | 3.78 | 168 | 0.75 | 126 |
| Hsal | Amel | 20,134 | 704,546 | 2.86 | 168 | 1 | 168 |
| Nvit | Amel | 6,393 | 704,546 | 0.91 | 185 | 2 | 370 |
| Nlon | Ngir | 322,594 | 426,704 | 75.6 | 0.41 | 5 | 2.05 |
| Nvit | Ngir | 317,256 | 426,704 | 74.35 | 1 | 5 | 5 |
Note.—The analyzed species pairs (query vs. reference) are shown with the detected number of SSRs (conserved between both species), the number of used SSRs (number of SSRs in the reference), the proportion found to be conserved, the time when both species split (divergence time), the generation time as the average of the number of generations produced per year by each species in this pair, and the number of million generation potentially produced since divergence.
Conservation of SSR Loci in Genomes of Species Pairs
Each node in the phylogeny represents the time at which the most recent common ancestor species separated into two different lineages or species. Drosophila melanogaster (Dmel) was selected as the reference genome because it is an intensely studied model species. Hence, all other Drosophila species were compared with Dmel. In addition, some additional pairwise comparisons were chosen to cover nodes that provided additional phylogenetic time points (e.g., D. secchellia–D. simulans or D. mojavense–D. virilis). We analogously proceeded within the Hymenoptera, with the S. invicta (Soli) as the main reference genome to cover most nodes on the phylogenetic tree.
Rate of Decay of SSR Loci
An exponential decay function was fitted to our data to determine the rate of decay of SSR conservation using R (Team 2011). This was achieved by minimizing the square of the deviance of our data points to the decay function, searching the parameter space with the assumption of a constant rate of decay.
Conservation SSR Types and Motifs
Pairwise comparisons were used to analyze the conservation of specific SSR types, di-, tri-, tetra-, and pentanucleotide repeats, and their motifs. First, counts for each SSR type and motif were determined in the reference species. The same was done for the data set resulting from the pairwise comparison, the conserved SSRs loci. The relationship between the total numbers of SSRs shared between both species represents the total decay of the SSRs or the proportion of all SSRs which are conserved between both species. This analysis was repeated for each of the SSR types and motifs. The decay of each different type of SSRs and the repeat motif length (di-, tri-, tetra-, and pentanucleotide repeats) between both species was related to the decay of total number of SSRs. Comparing the four SSR types, we can determine whether the decay of a specific type of SSR is slower (less decay) than the overall decay of all SSRs. Analogously, the specific SSR sequence motifs were analyzed within each SSR type, that is, the decay of a certain dinucleotide repeat motif was compared with the decay of all dinucleotide repeats. Therefore, if certain motifs decay slower than others, it infers that they are more stable than others over evolutionary time scales. Differences across motifs and types of SSRs were tested by comparing within (including correction for multiple testing) and between the Hymenoptera and Diptera using a two-tailed Mann–Whitney U test.
Results
Genomic SSR Content
SSRs with repeat units of two to five base pairs were identified in 12 Drosophila, 3 mosquitoes, 3 Nasonia, 1 bee, and 7 ant genomes. The total numbers, the density, and the composition vary among the genomes of different species, sometimes even between closely related species (fig. 1 and supplementary file S1, Supplementary Material online). There was a positive linear relation of genome size and SSR count (supplementary file S1, Supplementary Material online).
Conservation of SSRs between Pairs of Species
Each pairwise comparison of the SSR libraries with Blast identifies potentially homologous SSR loci between species, which were retained since divergence of both species from a common ancestor. As expected, SSRs conservation decreases over phylogenetic time scales (table 1 and supplementary file S2, Supplementary Material online). Species that separated within the last 1 Myr retained more than 60% of the SSR loci. The Drosophila species of the subgenus Sophophora retained still more than 5% of the SSR loci during their more than 60 Myr of separate evolution; the ants and the honeybee retained approximately 3% since 185 Myr and Aedes and Culex more than 1.5% since more than 200 Myr. Even between the Diptera and the Hymenoptera, separated for approximately 300 Myr (Grimaldi and Engel 2005), approximately 0.1% of the SSR loci were conserved.
Validation of the Method
As a benchmark of our method, we compared the genomes of several species with themselves, using identical processing and filtering. For D. melanogaster, we detected 80.84%, for A. mellifera 88.06%, for S. invicta 84.36%, for A. cephalotes 91.49%, and for N. vitripennis 83%.
When checked for the correct assignment of the identical SSRs, we found 0.8% of the SSRs in D. melanogaster to be incorrectly assigned. This measure represents the rate of false positives detected with our method and filtering thresholds. For A. mellifera, this rate was 1.87%, for S. invicta 2.46%, for A. cephalotes 1.1%, and for N. vitripennis 1.29%, giving an average of 1.68% for the tested Hymenoptera. Approximately a quarter of these false positives are SSRs close by the correct SSR, within the 350 bp flanking sequence and with the same motif, thus this fraction could potentially be corrected by manual inspection.
Another indication of the validity of our approach is the comparison of genome structure between the closely related D. melanogaster and D. simulans using the detected conserved SSRs. Using more than 19,000 SSRs from Muller element B (chromosome 2L) from both species, we found this element to be highly similar in terms of the order and distances of the SSRs, which indicated that the majority of this chromosome is in synteny. This agrees largely with the previous findings using gene locations (Bhutkar et al. 2008). The syntenic relationship of the first 9,030 SSRs corresponding to the first 10 Mbp from Muller element B are visualized with AutoGRAPH (Derrien et al. 2007) (supplementary file S3, Supplementary Material online).
Reciprocal BLAST analysis in some selected species pairs yielded very similar numbers of conserved SSRs. The difference in proportion of conserved SSRs caused by slightly different absolute numbers in reciprocal runs are for Dmel–Dsim 0%, Dmel–Dvir 0.14%, Dmel–Dpse 0.24%, Amel–Soli 0.34%, and Acep–Soli 1.01%, thus neglectable in our analysis.
Rate of Decay of SSRs Loci
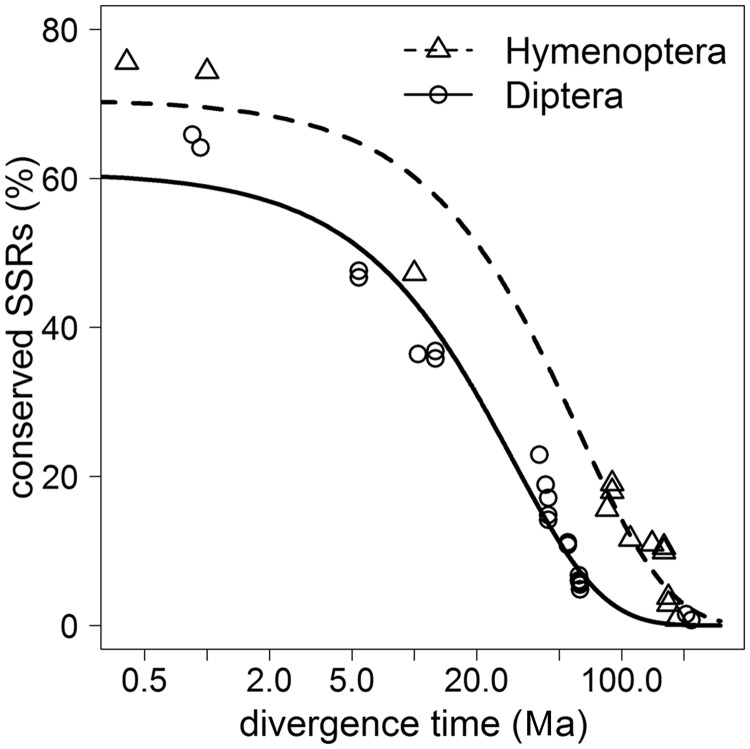
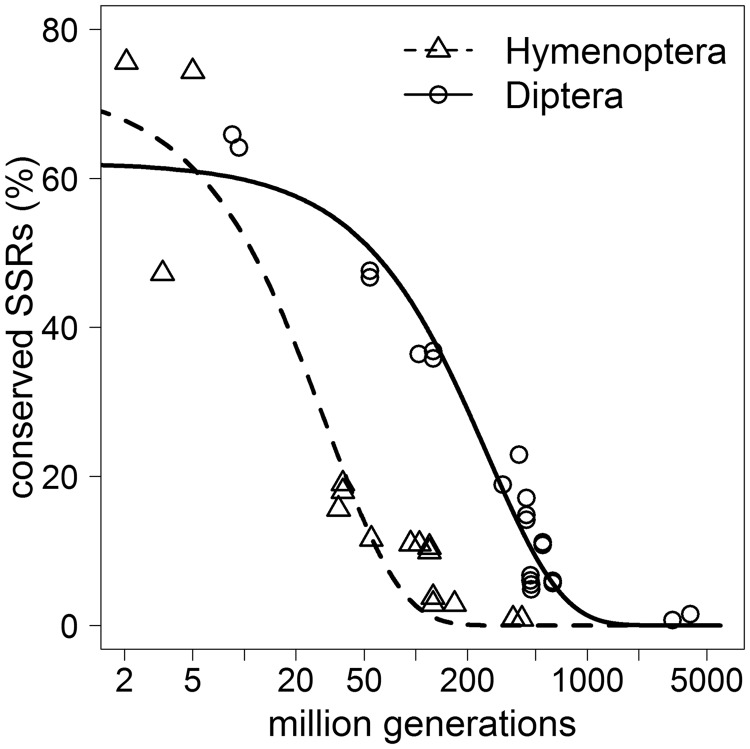
Fitting an exponential decay function to the proportion of conserved SSR loci, we were able to determine the rate of decay for both Dipteran and Hymenopteran SSRs (table 2). The decay rates were related to the time of divergence between two species (fig. 2) and to the estimated number of generations passed since then (fig. 3). In both cases, this fit was highly significant with low standard errors. Although the Dipteran SSR decay rate is two times faster than in the Hymenoptera, the Hymenoptera show an 8.5 times faster decay of SSR loci than the Diptera in relation to the number of generations.
Table 2.
Comparison of SSR Decay in Hymenoptera and Diptera in Relation to Divergence Time or Generation Time
| Decay (Slope) Estimate | Decay (Slope) SE | Origin (Intercept) Estimate | Origin (Intercept) SE | F | P | |
|---|---|---|---|---|---|---|
| Hymenoptera, divergence time | 1.59 | 0.0183 | 70.64 | 2.4658 | 185.7523 | 8.33E − 11 |
| Diptera, divergence time | 3.32 | 0.0143 | 60.9 | 1.0954 | 285.2023 | 6.77E − 15 |
| Hymenoptera, generations | 3.24 | 0.0167 | 72.4 | 2.6891 | 62.5389 | 1.05E − 07 |
| Diptera, generations | 0.38 | 9.45E − 04 | 62.14 | 1.0857 | 211.7773 | 1.03E − 13 |
Note.—Comparison between the decay of SSRs in Hymenoptera and Diptera in relation to divergence time (split in Ma) and to the estimated number of million generations passed using an exponential decay function. Score and P value from a general regression statistics (F test) are given as well as standard errors (SE) for the slope estimate (decay) and the intercept.
Fig. 2.—
Conserved SSR proportions by divergence time. Proportions of SSRs conserved in species pairs of Hymenoptera and Diptera relative to their phylogenetic divergence time (split in Ma, log scale).
Fig. 3.—
Conserved SSR proportions by generation time. Proportions of SSRs conserved in species-pairs of Hymenoptera and Diptera relative to the estimated number of million generations since their divergence (log scale).
A more stringent analysis, in which Dmel or Soli SSRs were only considered to be conserved if they were found in species from subsequent branches in the phylogeny, gave much lower proportion of conserved SSRs but showed essentially the very same pattern of decay (supplementary file S4, Supplementary Material online). Another additional analysis was performed using only those SSRs, which are located on the Dmel X-chromosome in comparison to the other Dmel chromosomes. The haplodiploid X-chromosome showed a slightly faster loss of SSR loci compared with the diploid chromosomes (supplementary file S5, Supplementary Material online, Wilcoxon matched pairs test: P = 0.0077).
Conservation of SSR Types and Motifs
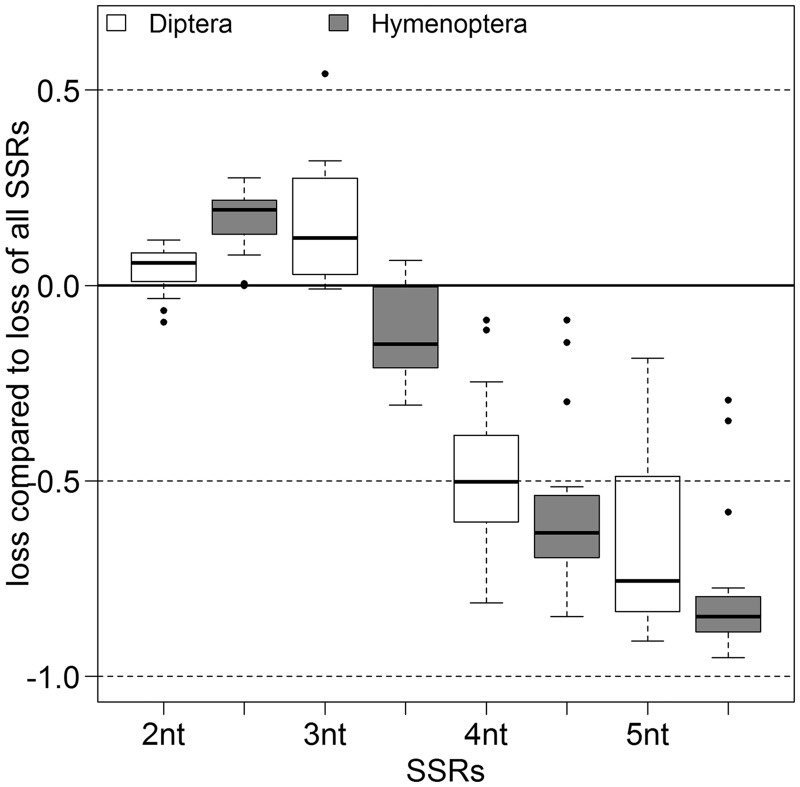
From each pairwise comparison, we separately analyzed the different types of SSRs: di-, tri-, tetra-, and pentanucleotide repeats and their motifs. For the Hymenoptera, we found a distinct relationship between the length of the repeat motif and its conservation. Dinucleotide repeats were found to decay more slowly than the overall rate (set to zero), indicated by a positive value of relative SSR loss, trinucleotide repeats slightly faster, and tetra- and pentanucleotide repeats significantly faster, indicated by a negative value of relative SSR loss (fig. 4 and supplementary file S2, Supplementary Material online). In Diptera, the pattern is similar but the trinucleotide repeats decay was slower than SSRs in general.
Fig. 4.—
Relative loss of SSRs by their motif length. The loss of di-, tri-, tetra-, and pentanucleotide SSRs compared with the loss of all SSRs (y = 0, indicated by a black line). The Diptera are shown in white and the Hymenoptera with gray filling. The black bar within the box shows the median. Black dots represent outliers. All groups are significantly different.
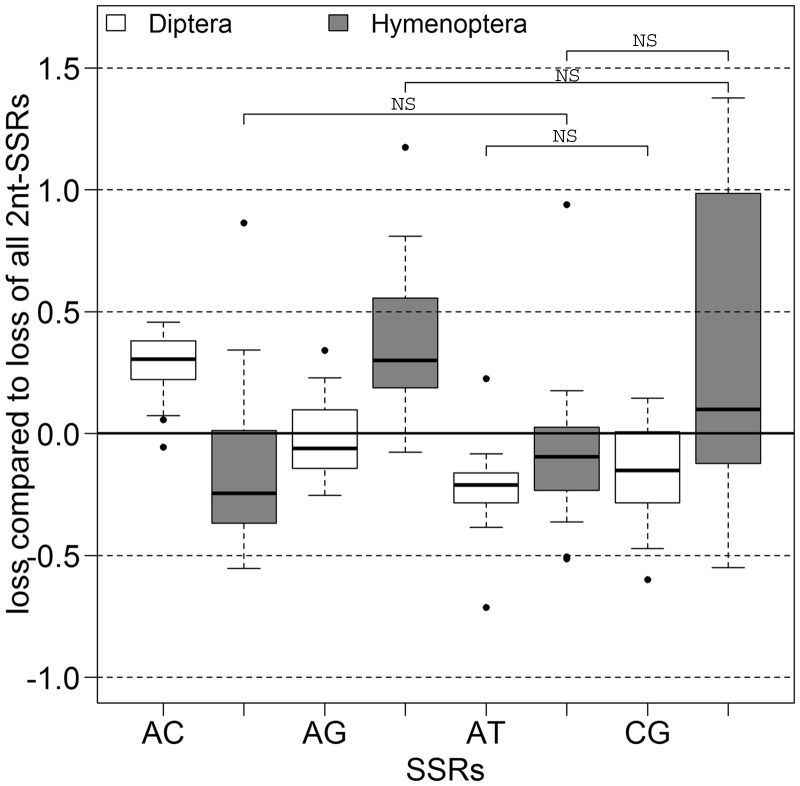
Dinucleotide repeats, although slower decaying than SSRs altogether, show significant differences among their four motifs (fig. 5 and supplementary file S2, Supplementary Material online). In Hymenoptera, AC and AT repeats are very similar and decay slightly faster than dinucleotide repeats altogether, whereas AG and CG repeats similarly decay slower. Differing in Diptera, AC repeats decay slowest of all the dinucleotide repeats, and AG and CG repeats decay slightly faster.
Fig. 5.—
Relative loss of 2 nt SSRs by their motif sequence. The loss of the different 2 nt SSRs compared with the loss of all 2 nt SSRs (y = 0, indicated by a black line). The Diptera are shown in white, the Hymenoptera with gray filling. The black bar within the box shows the median. Black dots represent outliers. All groups are significantly different, except those indicated with “NS.” NS, not significant.
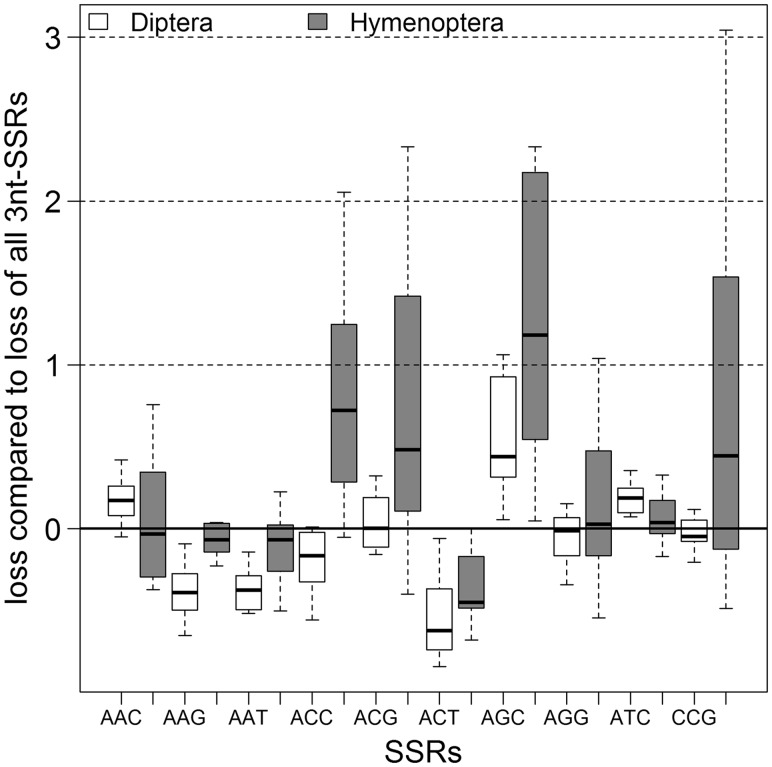
Trinucleotide repeats show significant differences in both groups as well within the groups (fig. 6 and supplementary file S2, Supplementary Material online). In Hymenoptera, a slower decay was detected for ACC, ACG, CCG, and especially AGC repeats and a faster decay for AAG and especially ACT repeats; the other motifs are close to zero, so their decay is very similar to the overall decay 3 nt SSRs. In Diptera, AAC, ATC, and especially AGC decay slower than the trinucleotide repeats altogether, and ACG, AGG and CCG are close to zero. The remaining motifs, and especially ACT, were found to have a faster decay. So despite some variance, the strongest deviation from the overall decay of all trinucleotide repeats in both insect orders was found for AGC and ACT repeats (fig. 6 and supplementary file S2, Supplementary Material online).
Fig. 6.—
Relative loss of 3 nt SSRs by their motif sequence. The loss of the different 3 nt SSRs compared with the loss of all 3 nt SSRs (y = 0, indicated by a black line). The Diptera are shown in white and the Hymenoptera with gray filling. The black bar within the box shows the median, outliers not shown.
The numbers of tetra- and pentanucleotide repeats and the proportion detected as conserved were much lower than in the previous SSR types. Therefore, the data show higher variability (supplementary file S2, Supplementary Material online). Consistently, ACTG is the slowest decaying motif in both insect orders. In contrast, ACCT is lost slowest in Hymenoptera but relatively rapid in Diptera. Between closely related species, the relative losses of specific SSRs were usually very similar.
Interestingly, in the AT-rich genomes of the Hymenoptera, AT-rich SSRs are common (AT as well as AAT, AAAT, AATT, AAAAT, and AATAT). Similarly, frequencies of AG might be somewhat correlated with the similar motifs AAG, AAAG, and AAAAG; CG with CCG, CCCG, CCGG; and AC with AAC, AAAC, and AAAAC. In the Dipteran genomes, such potential correlations apparently do not occur except for AT with AAT, AAAT, and AAAAT (supplementary file S1, Supplementary Material online).
Discussion
We show that SSRs can be conserved for many millions of years in the genomes of Hymenoptera and Diptera. Unlike previous work on vertebrates (Buschiazzo and Gemmell 2010), our data are not based on whole-genome alignments and subsequent selection of homologous regions to extract conserved SSRs. We used a BLAST-based approach to find a homologous SSR in pairwise genome comparisons. For both approaches, there is a risk to erroneously detect SSRs in the other genomes as a homolog because of its proximity to the correct locus, which might have lost the SSR during evolution. We tested our method by analyzing a genome with itself. Overall, our methodology correctly recovered 99.2% and 98.3% of loci in Drosophila and the Hymenoptera, respectively. The erroneously assigned SSRs were mainly located toward the ends of chromosomes or scaffolds. Although some studies show that SSR loci related to transposable elements can influence and bias SSR detection (Smýkal et al. 2009; Tay et al. 2010), our recovery rates and low error rates suggest that these cases are not relevant at the phylogenetic level. Another advantage of our approach is that it is not dependent on any previous alignment of homologous regions conserved for many species, which might introduce a bias toward more conserved loci resulting in a reduced sample size. In our method, each locus is analyzed independently for each pairwise comparison, this way we can include many more SSRs independent of possible differences of chromosome structures. Furthermore, the analysis is independent of the quality of the assembly in terms of misassembled sequences or assembly gaps.
As predicted, we found that the number of shared SSRs between two species decreases with increasing phylogenetic distance. Nevertheless, high numbers of conserved SSRs are still present many million years after divergence of two species.
In support of vertebrate data (Buschiazzo and Gemmell 2010), a very small fraction of below 0.1% of SSRs were even retained over more than 300 Myr of separate evolution of Diptera and Hymenoptera. Interestingly, Janes et al. (2011) discovered additional noncoding DNA sequences that were retained for long times and in differential proportions in both reptiles and mammals. This suggests that, in general, noncoding DNA elements can be conserved for many millions of years and/or generations.
There might be a balance between SSR length and probability of a mutation event, the longer the SSR, the greater the probability it will be "broken" by a point mutation, which might impair further slippage mutation. Thus a higher rate of decay would be expected if the mutation rate is high. This point of view is also supported by Sun et al. (2009). Under the assumption that that the majority of SSRs do not exhibit any relevant function and are thus neutrally evolving, SSR decay could be interpreted as a measure of the rate of genome evolution.
We detected slower rates of genome evolution in bees, wasps, and ants relative to the flies. This supports earlier reports where a high degree of conservation of structural chromosomal organization was observed between the bumble bee B. terrestris and the honeybee A. mellifera despite diverging approximately 100 Ma (Stolle et al. 2011) or where higher sequence identities in orthologous genes in A. mellifera than in other insects were found (Weinstock et al. 2006).
However, estimating rates of evolution solely based on mutations over time has been repeatedly criticized (Kimura 1983; Easteal 1985). Two compared organisms might comprise very different characteristics in many aspects, so that sequence differences can be achieved in very different time scales, potentially leading to false conclusions regarding relative rates evolution. Mutation rates can be affected by life history traits such as metabolism or body size (Mooers and Harvey 1994; Bromham et al. 1996) and can be linked to diversification rate or environmental energy (Davies et al. 2004; Lanfear et al. 2010). Furthermore, population structure can be an important factor, especially effective population size (Kimura and Ohta 1971; Woolfit and Bromham 2005), which determines the level of genetic drift. Traits such as fecundity, longevity, or ploidy can also covary with rates of molecular evolution and could influence on population genetic structure.
The comparison of SSR decay in our study showed a 2-fold slower decay over phylogenetic time in the Hymenopterans than in the Dipterans. Numerous studies in plants and vertebrates highlighted the importance of the generation time for the rate of evolution (Sarich and Wilson 1973; Kimura 1983; Easteal 1985; Laroche and Bousquet 1999; Gissi et al. 2000; Andreasen and Baldwin 2001; Nabholz et al. 2008; Welch et al. 2008). Species that produce more generations per unit time tend to have faster evolutionary rates, presumably due to more meiotic DNA replication errors, as observed within the invertebrates (Thomas et al. 2010). The species used in our study differ in the number of generation produced per year. Some social Hymenoptera produce reproductive individuals only after several years (Hölldobler and Wilson 1990), whereas the Drosophila species have many generation each year (Keightley 2000). We corrected for this discrepancy by relating our data to the number of generations, the Hymenopteran SSRs decay 8.5 times faster than the Dipteran SSRs. This striking difference might be explained by several factors.
We find differences of several orders of magnitude on examining the population sizes of the species studied here. Compared with Drosophila and mosquitoes, the Hymenopteran species represented in this study are parasitic or social and both have very small effective population sizes (Moran 1984; Owen and Owen 1989; Peeters and Liebig 2000; Zayed 2004; Nolte and Schlötterer 2008; Petit and Barbadilla 2009; Alves et al. 2010; Elias et al. 2010; Jaffé et al. 2010; Andolfatto et al. 2011), although some species have reproductive females with very high fecundity and longevity (Nabholz et al. 2008; Welch et al. 2008). Small effective population sizes enhance the loss of genetic diversity through drift and hence could cause smaller SSR polymorphism.
Furthermore, social Hymenoptera have been shown to have a much higher genomic recombination rate (11.15 cM/Mb) compared with that of Drosophila (1.59 cM/Mb) (Wilfert et al. 2007; Lattorff and Moritz 2008; Stolle et al. 2011). There is no sufficient data for all species to investigate this relationship further, but recombination rate could influence genome evolution and thus SSR loss.
Finally, in Hymenopterans, males are haploid, whereas Dipterans are diploid. The haploid male sex further decreases the effective population size and thus could have some influence on the rates of evolution. Interestingly, we found a slightly faster rate of SSR loss for the D. melanogaster haplodiploid X-chromosome than in the diploid Dmel chromosomes. The X-chromosome has only 75% effective population size than the other chromosomes (males are haploid for the X-chromosome, which means it has 50% of the effective population size, and females are diploid for the X-chromosome, which means 100% of the effective population size). Because of stronger genetic drift, one could expect a lower degree of polymorphism, which was confirmed by previous studies (Begun and Whitley 2000; Betancourt et al. 2002; Andolfatto et al. 2011). However, because a loss of polymorphism due to genetic drift has probably no influence on mutation rate as such, differences in effective population size might have little effect on the pattern we found in the Hymenoptera and Diptera. A possible explanation could be differences in the number of cell divisions in the germ cells between both sexes, whereby although detected, the difference was found to be weak in D. melanogaster (Bauer and Aquadro 1997). However, if D. melanogaster females would reproduce early in their life, the weak female bias in the number of germ-cell divisions could enhance the SSR turnover in the X chromosome and thus cause a slightly faster SSR loss. If such differing numbers of germ-cell divisions between sexes would play a role in other species as well, it might explain a faster loss of SSRs in the Hymenoptera, in which all chromosomes are haplodiploid. And this further could be enhanced by the longevity of queens of the social Hymenoptera in comparison to the short living males. On the other hand, this would be detectable by enhanced evolutionary rates, for which previous studies (Bauer and Aquadro 1997; Begun and Whitley 2000; Betancourt et al. 2002) found no evidence in Drosophila, and is also opposed by the finding of a faster mutation rate on the male Y chromosome versus the X-chromosome (Bachtrog 2008).
Although distinct patterns relating to motif composition within and between insect orders are lacking, differences in the frequency and conservation of particular motifs were observed between Hymenoptera and Diptera. This constraint could indicate that some motifs are more stable than others or actually might be somehow selected. Our data suggest at least a constraint of the length of a motif which might be related to probabilities of point mutations disrupting the slippage-mutational process. There also might be a relationship between frequency and conservation of a motif, and the frequencies of related motifs which could give some indications for the turnover (birth and death rate) of specific motifs. However, other conclusions for the different patterns within and between each insect order, especially for specific repeat motifs, are hard to draw, especially as the process of birth and death of a SSR, potentially from SSRs changed by mutations, is poorly understood.
The functional implications of the conservation or frequency of SSRs, if there are any, also unfortunately must remain unclear at this stage. Opposing the general view of functionless DNA elements, some SSRs could play some functional roles, although this would not explain the whole pattern of the large number of SSRs. Palindromic repeats, such as AT and CG, could be involved in formation of DNA hairpin structures, some trinucleotide repeats could be constrained by functions within coding regions or on chromosomal level. Thus far, only a few specific SSRs are known to be involved in some biological processes (for further reading see Goldstein and Schlötterer 1999; Li et al. 2002, 2004; Buschiazzo and Gemmell 2010; Grover and Sharma 2011) or other relevant impact (Auer et al. 2001; Kerrest et al. 2009; Blackwood et al. 2010; Bonen et al. 2010; Mueller et al. 2011). Some SSRs were also related to recombination hotspots (Brandström et al. 2008) and transposable elements (Smýkal et al. 2009; Tay et al. 2010).
Irrespective of the actual mechanisms that drive the evolutionary changes in SSRs, we show that they allow for a comparison of rates of genome evolution. We find that the rate of decay of SSRs, and, therefore, the rate of genome evolution, is not 2-fold slower in the Hymenoptera compared with the Diptera as indicated by absolute substitution rates but is 8.5 times faster when correcting for generation time. Thus, previous studies on structural conservation (Stolle et al. 2011) and sequence similarity (Weinstock et al. 2006) based on absolute time should be re-evaluated regarding generation time and future studies need to account for it.
Conserved SSRs can potentially also be exploited for a rapid, cost-efficient, and yet comprehensive development of markers for arrays of even distantly related species. They can also be a powerful tool to investigate genome structure and synteny between genomic regions with a resolution, which can be orders of magnitude higher than using genes.
Supplementary Material
Supplementary files S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
E.S. and R.F.A.M. designed the study and wrote the manuscript; E.S. conducted the analyses; the custom PERL scripts were written by J.K.; and statistical analyses were done by E.S. and J.K. The authors thank Wee Tek Tay, David Nash, Yannick Wurm, and Jack Werren for valuable comments. Furthermore, they are indebted to Tomás Murray and four anonymous referees for constructive comments on an earlier version of the manuscript. This work was supported by the German Science Foundation DFG.
Literature Cited
- Altschul S, Gish W, Miller W, Myers E. Basic local alignment search tool. J Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alves DA, et al. Successful maintenance of a stingless bee population despite a severe genetic bottleneck. Conserv Genet. 2010;12:647–658. [Google Scholar]
- Andolfatto P, Wong KM, Bachtrog D. Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species. Genome Biol Evol. 2011;3:114–128. doi: 10.1093/gbe/evq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen K, Baldwin BG. Unequal evolutionary rates between annual and perennial lineages of checker mallows (Sidalcea, Malvaceae): evidence from 18S–26S rDNA internal and external transcribed spacers. Mol Biol Evol. 2001;18:936–944. doi: 10.1093/oxfordjournals.molbev.a003894. [DOI] [PubMed] [Google Scholar]
- Auer RL, et al. Role for CCG-trinucleotide repeats in the pathogenesis of chronic lymphocytic leukemia. Blood. 2001;97:509–515. doi: 10.1182/blood.v97.2.509. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Evidence for male-driven evolution in Drosophila. Mol Biol Evol. 2008;25:617–619. doi: 10.1093/molbev/msn020. [DOI] [PubMed] [Google Scholar]
- Barbará T, et al. Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol. 2007;16:3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x. [DOI] [PubMed] [Google Scholar]
- Barker JSF. Effective population size of natural populations of Drosophila buzzatii, with a comparative evaluation of nine methods of estimation. Mol Ecol. 2011;20:4452–4471. doi: 10.1111/j.1365-294X.2011.05324.x. [DOI] [PubMed] [Google Scholar]
- Bauer VL, Aquadro CF. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol Biol Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- Begon M. Temporal variations in the reproductive condition of Drosophila obscura Fallén and D. subobscura Collin. Oecologia. 1976;23:31–47. doi: 10.1007/BF00351213. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc Natl Acad Sci U S A. 2000;97:5960–5965. doi: 10.1073/pnas.97.11.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkevold D, Boomsma JJ. Evolutionary transition to a semelparous life history in the socially parasitic ant Acromyrmex insinuator. J Evol Biol. 2000;13:615–623. [Google Scholar]
- Betancourt AJ, Presgraves DC, Swanson WJ. A test for faster X evolution in Drosophila. Mol Biol Evol. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- Bhargava A, Fuentes FF. Mutational dynamics of microsatellites. Mol Biotechnol. 2010;44:250–266. doi: 10.1007/s12033-009-9230-4. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood JK, Okely EA, Zahra R, Eykelenboom JK, Leach DRF. DNA tandem repeat instability in the Escherichia coli chromosome is stimulated by mismatch repair at an adjacent CAG · CTG trinucleotide repeat. Proc Natl Acad Sci U S A. 2010;107:22582–22586. doi: 10.1073/pnas.1012906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquer-Maumont A, Crouauroy B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J Mol Evol. 1995;41:492–497. doi: 10.1007/BF00160321. [DOI] [PubMed] [Google Scholar]
- Bonen L, Haerty W, Golding GB. Low-complexity sequences and single amino acid repeats: not just “junk” peptide sequences. Genome. 2010;53:753–762. doi: 10.1139/g10-063. [DOI] [PubMed] [Google Scholar]
- Brandström M, Bagshaw AT, Gemmell NJ, Ellegren H. The relationship between microsatellite polymorphism and recombination hot spots in the human genome. Mol Biol Evol. 2008;25:2579–2587. doi: 10.1093/molbev/msn201. [DOI] [PubMed] [Google Scholar]
- Bromham L, Rambaut A, Harvey PH. Determinants of rate variation in mammalian DNA sequence evolution. J Mol Evol. 1996;43:610–621. doi: 10.1007/BF02202109. [DOI] [PubMed] [Google Scholar]
- Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006;28:1040–1050. doi: 10.1002/bies.20470. [DOI] [PubMed] [Google Scholar]
- Buschiazzo E, Gemmell NJ. Evolutionary and phylogenetic significance of platypus microsatellites conserved in mammalian and other vertebrate genomes. Aust J Zool. 2009;57:175. [Google Scholar]
- Buschiazzo E, Gemmell NJ. Conservation of human microsatellites across 450 million years of evolution. Genome Biol Evol. 2010;2:153–165. doi: 10.1093/gbe/evq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG. Environmental energy and evolutionary rates in flowering plants. Proc Biol Sci. 2004;271:2195–2200. doi: 10.1098/rspb.2004.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Andre C, Galibert F, Hitte C. AutoGRAPH: an interactive web server for automating and visualizing comparative genome maps. Bioinformatics. 2007;23:498–499. doi: 10.1093/bioinformatics/btl618. [DOI] [PubMed] [Google Scholar]
- Easteal S. Generation time and the rate of molecular evolution. Mol Biol Evol. 1985;2:450–453. doi: 10.1093/oxfordjournals.molbev.a040361. [DOI] [PubMed] [Google Scholar]
- Eckert K, Hile SE. Every microsatellite is different: intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J, Dorn S, Mazzi D. No evidence for increased extinction proneness with decreasing effective population size in a parasitoid with complementary sex determination and fertile diploid males. BMC Evol Biol. 2010;10:366. doi: 10.1186/1471-2148-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Ezenwa VO, et al. Ancient conservation of trinucleotide microsatellite loci in polistine wasps. Mol Phylogenet Evol. 1998;10:168–177. doi: 10.1006/mpev.1998.0528. [DOI] [PubMed] [Google Scholar]
- FitzSimmons NN, Moritz C, Moore SS. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol Biol Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- Gadau J, et al. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 2012;28:14–21. doi: 10.1016/j.tig.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo CL, et al. Global microsatellite content distinguishes humans, primates, animals, and plants. Mol Biol Evol. 2009;26:2809–2819. doi: 10.1093/molbev/msp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Reyes A, Pesole G, Saccone C. Lineage-specific evolutionary rate in mammalian mtDNA. Mol Biol Evol. 2000;17:1022–1031. doi: 10.1093/oxfordjournals.molbev.a026383. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Schlötterer C. Microsatellites. Evolution and applications. New York: Oxford University Press; 1999. [Google Scholar]
- Green CL, Franck P Oldroyd BP. Characterization of microsatellite loci for Trigona carbonaria, a stingless bee endemic to Australia. Mol Ecol Notes. 2001;1:89–92. [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. New York: Cambridge University Press; 2005. [Google Scholar]
- Grover A, Sharma PC. Is spatial occurrence of microsatellites in the genome a determinant of their function and dynamics contributing to genome evolution? Curr Sci. 2011;100:859–869. [Google Scholar]
- Hölldobler B, Wilson EO. The ants. Cambridge (MA): Harvard University Press; 1990. [Google Scholar]
- Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé R, et al. Estimating the density of honeybee colonies across their natural range to fill the gap in pollinator decline censuses. Conserv Biol. 2010;24:583–593. doi: 10.1111/j.1523-1739.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- Janes DE, et al. Reptiles and mammals have differentially retained long conserved noncoding sequences from the amniote ancestor. Genome Biol Evol. 2011;3:102–113. doi: 10.1093/gbe/evq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Mazzi D, Ritchie MG, Hoikkala A. Sexual and postmating reproductive isolation between allopatric Drosophila montana populations suggest speciation potential. BMC Evol Biol. 2011;11:68. doi: 10.1186/1471-2148-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Suzuki T, Tsuchida K. Application of microsatellite primers for the social wasp Polistes to another social wasp genus, Parapolybia, to estimate genetic relationships among nestmates. Entomol Sci. 2007;10:1–5. [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- Keightley PD. Deleterious mutations and the evolution of sex. Science. 2000;290:331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, et al. What is a microsatellite: a computational and experimental definition based upon repeat mutational behavior at A/T and GT/AC repeats. Genome Biol Evol. 2010;2:620–635. doi: 10.1093/gbe/evq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrest A, et al. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Mol Biol. 2009;16:159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge (UK): Cambridge University Press; 1983. [Google Scholar]
- Kimura M, Ohta T. On the rate of molecular evolution. J Mol Evol. 1971;1:1–17. doi: 10.1007/BF01659390. [DOI] [PubMed] [Google Scholar]
- Laayouni H, Hasson E, Santos M, Fontdevila A. The evolutionary history of Drosophila buzzatii. XXXV. Inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol Biol Evol. 2003;20:931–944. doi: 10.1093/molbev/msg099. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Ho SYW, Love D, Bromham L. Mutation rate is linked to diversification in birds. Proc Natl Acad Sci U S A. 2010;107:20423–20428. doi: 10.1073/pnas.1007888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche J, Bousquet J. Evolution of the mitochondrial rps3 intron in perennial and annual angiosperms and homology to nad5 introns 1. Mol Biol Evol. 1999;16:441–452. doi: 10.1093/oxfordjournals.molbev.a026126. [DOI] [PubMed] [Google Scholar]
- Lattorff HMG, Moritz RF. Recombination rate and AT-content show opposite correlations in mammalian and other animal genomes. Evol Biol. 2008;35:146–149. [Google Scholar]
- Leclercq S, Rivals E, Jarne P. DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome Biol Evol. 2010;2:325–335. doi: 10.1093/gbe/evq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Nei M. Persistence of common alleles in two related populations or species. Genetics. 1977;86:901–914. doi: 10.1093/genetics/86.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Lim S, Notley-McRob L, Lim M, Carter D. A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet Biol. 2004;41:1025–1036. doi: 10.1016/j.fgb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Mayer C. Phobos 3.3.11. 2006–2010. Available from: http://www.rub.de/spezzoo/cm/cm_phobos.htm (last accessed January 2011)
- Mayer C, Leese F, Tollrian R. Genome-wide analysis of tandem repeats in Daphnia pulex—a comparative approach. BMC Genomics. 2010;11:277. doi: 10.1186/1471-2164-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglécz E, et al. Microsatellite flanking region similarities among different loci within insect species. Insect Mol Biol. 2007;16:175–185. doi: 10.1111/j.1365-2583.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Molnar RI, Witte H, Dinkelacker I, Villate L, Sommer RJ. Tandem-repeat patterns and mutation rates in microsatellites of the nematode model organism Pristionchus pacificus. G3. 2012;2:1027–1034. doi: 10.1534/g3.112.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers AO, Harvey PH. Metabolic rate, generation time, and the rate of molecular evolution in birds. Mol Phylogenet Evol. 1994;3:344–350. doi: 10.1006/mpev.1994.1040. [DOI] [PubMed] [Google Scholar]
- Moore SS, Hale P, Byrne K. NCAM: a polymorphic microsatellite locus conserved across eutherian mammal species. Animal Genet. 1998;29:33–36. doi: 10.1046/j.1365-2052.1998.00234.x. [DOI] [PubMed] [Google Scholar]
- Moran C. Sex-linked effective population size in control populations, with particular reference to honeybees (Apis mellifera L.) Theor Appl Genet. 1984;67:317–322. doi: 10.1007/BF00272867. [DOI] [PubMed] [Google Scholar]
- Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the ants: diversification in the age of angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- Mueller JC, Pulido F, Kempenaers B. Identification of a gene associated with avian migratory behaviour. Proc Biol Sci. 2011;278:2848–2856. doi: 10.1098/rspb.2010.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. Strong variation of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol. 2008;25:120–130. doi: 10.1093/molbev/msm248. [DOI] [PubMed] [Google Scholar]
- Nolte V, Schlötterer C. African Drosophila melanogaster and D. simulans populations have similar levels of sequence variability, suggesting comparable effective population sizes. Genetics. 2008;178:405–412. doi: 10.1534/genetics.107.080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady P, Desalle R. Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae) Biol Lett. 2008;4:195–199. doi: 10.1098/rsbl.2007.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RE, Owen ARG. Effective population size in social Hymenoptera with worker-produced males. Heredity. 1989;63:59–65. doi: 10.1038/hdy.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannebakker B, Niehuis O, Hedley A, Gadau J, Shuker DM. The distribution of microsatellites in the Nasonia parasitoid wasp genome. Insect Mol Biol. 2010;19(Suppl 1):91–98. doi: 10.1111/j.1365-2583.2009.00915.x. [DOI] [PubMed] [Google Scholar]
- Paxton RJ, Zobel MU, Steiner J, Zillikens A. Microsatellite loci for Euglossa annectans (Hymenoptera: Apidae) and their variability in other orchid bees. Mol Ecol Resour. 2009;9:1221–1223. doi: 10.1111/j.1755-0998.2009.02612.x. [DOI] [PubMed] [Google Scholar]
- Peeters C, Liebig J. Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insectes Sociaux. 2000;47:325–332. [Google Scholar]
- Petit N, Barbadilla A. Selection efficiency and effective population size in Drosophila species. J Evol Biol. 2009;22:515–526. doi: 10.1111/j.1420-9101.2008.01672.x. [DOI] [PubMed] [Google Scholar]
- Powell JR, et al. Nonrecombining genes in a recombination environment: the Drosophila “dot” chromosome. Mol Biol Evol. 2011;28:825–833. doi: 10.1093/molbev/msq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmer CR, Møller AP, Ellegren H. A wide-range survey of cross-species microsatellite amplification in birds. Mol Ecol. 1996;5:365–378. doi: 10.1111/j.1365-294x.1996.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Rasnitsyn AP, Quicke DL. History of insects. Dordrecht (The Netherlands): Springer; 2002. [Google Scholar]
- Raychoudhury R, Grillenberger B, Gadau J. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial-Wolbachia sweep in North America. Heredity. 2010;104:318–326. doi: 10.1038/hdy.2009.160. [DOI] [PubMed] [Google Scholar]
- Reber Funk C, Schmid-Hempel R, Schmid-Hempel P. Microsatellite loci for Bombus spp. Mol Ecol Notes. 2006;6:83–86. [Google Scholar]
- Rico C, Rico I, Hewitt G. 470 million years of conservation of microsatellite loci among fish species. Proc Biol Sci. 1996;263:549–557. doi: 10.1098/rspb.1996.0083. [DOI] [PubMed] [Google Scholar]
- Ross CL, et al. Rapid divergence of microsatellite abundance among species of Drosophila. Mol Biol Evol. 2003;20:1143–1157. doi: 10.1093/molbev/msg137. [DOI] [PubMed] [Google Scholar]
- Sarich VM, Wilson AC. Generation time and genomic evolution in primates. Science. 1973;179:1144–1147. doi: 10.1126/science.179.4078.1144. [DOI] [PubMed] [Google Scholar]
- Schlötterer C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365–371. doi: 10.1007/s004120000089. [DOI] [PubMed] [Google Scholar]
- Schlötterer C. The evolution of molecular markers—just a matter of fashion? Nat Rev Genet. 2004;5:63–69. doi: 10.1038/nrg1249. [DOI] [PubMed] [Google Scholar]
- Smýkal P, Kalendar R, Ford R, Macas J, Griga M. Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity. 2009;103:157–167. doi: 10.1038/hdy.2009.45. [DOI] [PubMed] [Google Scholar]
- Stolle E, et al. Novel microsatellite DNA loci for Bombus terrestris (Linnaeus, 1758) Mol Ecol Resour. 2009;9:1345–1352. doi: 10.1111/j.1755-0998.2009.02610.x. [DOI] [PubMed] [Google Scholar]
- Stolle E, et al. A second generation genetic map of the bumblebee Bombus terrestris (Linnaeus, 1758) reveals slow genome and chromosome evolution in the Apidae. BMC Genomics. 2011;12:48. doi: 10.1186/1471-2164-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JX, Mullikin JC, Patterson N, Reich DE. Microsatellites are molecular clocks that support accurate inferences about history. Mol Biol Evol. 2009;26:1017–1027. doi: 10.1093/molbev/msp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber SW. The world of the harvester ants. College Station (TX): Texas A&M University Press; 1998. [Google Scholar]
- Taber SW. Fire ants. College Station (TX): Texas A&M University Press; 2000. [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tay WT, Behere GT, Batterham P, Heckel DG. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol Biol. 2010;10:144. doi: 10.1186/1471-2148-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. Vienna (Austria): R Foundation for Statistical Computing; 2011. R: a language and environment for statistical computing. [Google Scholar]
- Thomas JA, Welch JJ, Lanfear R, Bromham L. A generation time effect on the rate of molecular evolution in invertebrates. Mol Biol Evol. 2010;27:1173–1180. doi: 10.1093/molbev/msq009. [DOI] [PubMed] [Google Scholar]
- Thomou C, et al. Acquired somatic mutations in the microsatellite DNA, in children with bronchial asthma. Pediatr Pulmonol. 2009;44:1017–1024. doi: 10.1002/ppul.21097. [DOI] [PubMed] [Google Scholar]
- Tóth G, Gáspári Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiman D, et al. Conservation of a syntenic group of microsatellite loci between cattle and sheep. Mamm Genome. 1994;5:310–314. doi: 10.1007/BF00389547. [DOI] [PubMed] [Google Scholar]
- Wang Q, et al. Polymorphism of CAG repeats in androgen receptor of carnivores. Mol Biol Rep. 2012;39:2297–2303. doi: 10.1007/s11033-011-0979-8. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Bininda-Emonds OR, Bromham L. Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol Biol. 2008;8:53–64. doi: 10.1186/1471-2148-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity. 2007;98:189–197. doi: 10.1038/sj.hdy.6800950. [DOI] [PubMed] [Google Scholar]
- Woolfit M, Bromham L. Population size and molecular evolution on islands. Proc Biol Sci. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A. Effective population size in Hymenoptera with complementary sex determination. Heredity. 2004;93:627–630. doi: 10.1038/sj.hdy.6800588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.