Abstract
Rhodopsin-containing marine microbes such as those in the class Flavobacteriia play a pivotal role in the biogeochemical cycle of the euphotic zone (Fuhrman JA, Schwalbach MS, Stingl U. 2008. Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol. 6:488–494). Deciphering the genome information of flavobacteria and accessing the diversity and ecological impact of microbial rhodopsins are important in understanding and preserving the global ecosystems. The genome sequence of the orange-pigmented marine flavobacterium Nonlabens dokdonensis (basonym: Donghaeana dokdonensis) DSW-6 was determined. As a marine photoheterotroph, DSW-6 has written in its genome physiological features that allow survival in the oligotrophic environments. The sequence analysis also uncovered a gene encoding an unexpected type of microbial rhodopsin containing a unique motif in addition to a proteorhodopsin gene and a number of photolyase or cryptochrome genes. Homologs of the novel rhodopsin gene were found in other flavobacteria, alphaproteobacteria, a species of Cytophagia, a deinococcus, and even a eukaryote diatom. They all contain the characteristic NQ motif and form a phylogenetically distinct group. Expression analysis of this rhodopsin gene in DSW-6 indicated that it is induced at high NaCl concentrations, as well as in the presence of light and the absence of nutrients. Genomic and metagenomic surveys demonstrate the diversity of the NQ rhodopsins in nature and the prevalent occurrence of the encoding genes among microbial communities inhabiting hypersaline niches, suggesting its involvement in sodium metabolism and the sodium-adapted lifestyle.
Keywords: heterotrophic picoplankton, Bacteroidetes, bacteriorhodopsin, xanthorhodopsin, sodium pump, metagenome
Introduction
Marine ecosystems, in which half of global primary production occurs, are home to oligotrophs that are responsible for the biogeochemical cycle and add an important axis to the Earth’s energy balance (Falkowski et al. 1998; Whitman et al. 1998; Azam and Malfatti 2007). Flavobacteria, which belong to the phylum Bacteroidetes, previously called the Cytophaga–Flavobacterium–Bacteroides (CFB) group, occupy an important position in the photometabolic system in marine environments. A comprehensive community analysis indicates that the phyla Proteobacteria (63%), Bacteroidetes (13%), Cyanobacteria (7.9%), Firmicutes (7.5%), and Actinobacteria (4.6%) are among the dominant members of the oceanic picoplankton (Venter et al. 2004). At the class level, Alpha- and Gammaproteobacteria and Flavobacteriia are believed to be major carriers of microbial rhodopsins called proteorhodopsins (PRs) (Giovannoni et al. 2005; Rusch et al. 2007). Among metagenome fragments recruited from the Global Ocean Sampling (GOS) expedition, two assembled flavobacterial genomes harboring the PR gene turned out to represent the dominant taxa in the Northwest Atlantic (Rusch et al. 2007; Woyke et al. 2009).
Solar energy is captured and converted into chemical energy by phototrophs that depend either on the chlorophyll-harboring photosynthetic reaction center or on the photoactive retinal-binding rhodopsin. Contrary to the multicomponent photosynthetic reaction centers, which are restricted to six bacterial phyla, single-molecule microbial rhodopsins show wide taxonomic distribution, possibly through horizontal gene transfer between domains and phyla (Bryant and Frigaard 2006; Sharma et al. 2006; Bryant et al. 2007). Despite the great diversity of microbial rhodopsins, these proteins share structural features such as seven-membrane-spanning helices (Fuhrman et al. 2008). The structure and function of archaeal bacteriorhodopsins (BRs) with the retinal chromophore have been studied most intensively to date. BRs move protons across the membrane out of the cell using light energy to generate an electrochemical proton gradient, which in turn can be used for ATP production (Lanyi 2004). Metagenomic approaches enabled the discovery of PRs, the first rhodopsin of bacterial origin from the uncultured marine gammaproteobacterial SAR86 group (Beja et al. 2000, 2001). PRs share high sequence similarity with BRs, and light-driven chemiosmotic proton translocation was observed after heterologous expression in Escherichia coli (Beja et al. 2000, 2001). Recently, in a PR-containing marine flavobacterial suspension, light-driven proton transport activity sufficient for ATP generation was demonstrated (Yoshizawa et al. 2012).
Among other well-known rhodopsins that generate proton-motive force, xanthorhodopsins (XRs), which have been discovered first in Salinibacter ruber, are unusual in that they require two chromophores, the carotenoids salinixanthin and retinal, to broaden the spectral range for light harvesting (Balashov et al. 2005). Actinorhodopsins (ActRs), which were recently found in actinobacteria from a hypersaline lagoon, an estuary, and a freshwater lake, are abundant in microbial communities in freshwater ecosystems (Sharma et al. 2009). Halorhodopsins (HRs), which are light-driven inward chloride pumps, exist in halophiles. These rhodopsins may participate in the regulation of ionic content and the osmotic state (Mongodin et al. 2005). Although most rhodopsins function as ion transporters, sensory rhodopsins (SRs) mediate phototaxis or signal transduction, like photoreceptors do (Fuhrman et al. 2008). The amino acid sequences of the proton-pumping rhodopsins necessary for retinal binding are highly conserved and differ from those of HRs, and SRs even lack the functional residues in the retinal-binding pocket.
In this study, we determined and analyzed the complete genome sequence of an orange-pigmented marine flavobacterium, Nonlabens (Donghaeana) dokdonensis DSW-6 (Yoon et al. 2006; Yi and Chun 2012). The genome information provides a glimpse to the survival strategy of DSW-6 as a photoheterotroph in the oligotrophic ocean. Importantly, in addition to a typical PR, we found a new type of rhodopsin whose retinal-binding sequences are distinct from those of well-studied rhodopsins. To better understand the characteristics of this new type of rhodopsin, its gene expression level in DSW-6 was monitored under various light intensities, nutrient concentrations, and NaCl concentrations. Similarity searches against completely sequenced genomes and expressed sequence tags were performed, revealing a number of homologs present in the classes Flavobacteriia, Alphaproteobacteria, Cytophagia, and Deinococci, and even a eukaryotic diatom. Finally, the frequency of this rhodopsin family in diverse aquatic ecosystems was investigated by searching through public databases of environmental sequence data.
Materials and Methods
Strain and Culture Conditions
Donghaeana dokdonensis DSW-6, recently reclassified as Nonlabens dokdonensis comb. nov. (Yi and Chun 2012), was isolated from the surface seawater collected between the two main islands of Dokdo, Republic of Korea (Yoon et al. 2006). This nonmotile strain grows under strictly aerobic conditions and exhibits optimal growth in the presence of 2% NaCl at 25°C (Yoon et al. 2006). Cells were grown on Marine Agar 2216 (Difco, USA) or Artificial Sea Water (ASW) prepared from sea salts (Sigma-Aldrich, USA) enhanced by 2.5% w/v peptone and 0.5% w/v yeast extract. The strain produces orange-colored carotenoid pigments.
Genome Sequencing and Annotation
A hybrid approach of Roche/454 pyrosequencing and Sanger sequencing followed by manual gap filling was applied to decipher the N. dokdonensis DSW-6 genome. Shotgun pyrosequence reads of approximately 30-fold genome coverage were generated from GS FLX (NICEM, Korea) and were assembled into 98 contigs using gsAssembler. A total of 2,035 paired-end Sanger sequence reads (GenoTech Co., Korea) from a 35-kb genomic library were incorporated to yield two scaffolds. Genomic regions containing nonribosomal peptide synthetase genes or IS elements could not be properly assembled because of their highly repeated sequence patterns. To disentangle these overcollapsed contigs, additional Sanger sequences were provided by random shotgun sequencing of 35-kb fosmid clones spanning each gap. All the remaining small gaps were closed by sequencing polymerase chain reaction (PCR)-amplified genomic fragments. The PHRED/PHRAP package (Ewing and Green 1998) was used for Sanger read base calling and partial mini-assembly, and all sequence editing procedures were conducted using CONSED (Gordon et al. 1998). The final assembly led to a single chromosome without plasmids. The sequence was validated and errors were rectified by comparing the final assembly with independent sequence data to further increase the accuracy of the assembly and to avoid sequence errors in the homopolymeric nucleotides. Trimmed high-quality 102-bp shotgun reads at 806-fold genome coverage were produced using the Illumina/Solexa GA II system and were mapped to the assembled sequence with CLC Genomics Workbench (CLC bio, Inc., Denmark).
Gene prediction of the finished DSW-6 genome sequence was conducted using Glimmer 3.0 (Delcher et al. 2007). Functional assignment of the predicted genes was achieved by searching for homologs in public protein databases, such as UniRef100, Swiss-Prot, the GenBank nonredundant protein database, KEGG, and SMART, using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al. 1997). The outputs were automatically parsed using AutoFACT (Koski et al. 2005), and then all the annotations were manually curated. tRNA scan (Lowe and Eddy 1997) was applied to search for tRNA genes in the genome, and rRNA genes were identified using BLAST. Metabolic pathways were examined using the KEGG database (Aoki-Kinoshita and Kanehisa 2007) and BioCyc, which was generated by the PathoLogic program of the Pathway Tools software (Karp et al. 2010). The sequence and annotation have been deposited in GenBank under the accession number CP001397. The genome information is also available from the Genome Encyclopedia of Microbes (GEM; http://www.gem.re.kr, last accessed January 9, 2013).
Phylogenetic Analysis of Microbial Rhodopsins
To infer the phylogenetic groups of microbial rhodopsins, exemplary representatives of each class of ActRs, BRs, HRs, SRs, PRs, and XRs were retrieved from the UniProt or GenBank databases. The two rhodopsins of N. dokdonensis DSW-6 and nine NQ-type rhodopsins identified from complete genomes and expressed sequence tags were included in the analysis. They are from Gillisia limnaea R-8282 (Van Trappen et al. 2004), Krokinobacter (reclassified into Dokdonia; Yoon et al. 2012) sp. 4H-3-7-5 (Klippel et al. 2011), Hymenobacter roseosalivarius DSM 11622 (Hirsch et al. 1998), Citromicrobium bathyomarinum JL354 (Jiao et al. 2010), Citromicrobium sp. JLT1363 (Zheng et al. 2011), Fulvimarina pelagi HTCC2506 (Kang et al. 2010), Truepera radiovictrix DSM 17093 (Albuquerque et al. 2005), and Chaetoceros neogracile KOPRI AnM0002. A multiple sequence alignment was obtained using the MUSCLE algorithm (Edgar 2004), and ambiguously aligned regions were adjusted with the Gblocks program (Talavera and Castresana 2007). To construct phylogenetic trees, we used the MEGA5 package (Tamura et al. 2011) for maximum parsimony (MP) and neighbor-joining (NJ), and maximum likelihood (ML) methods, and MrBayes v3.1.2 (Zhang et al. 2012) for Bayesian inference. Phylogenetic trees were constructed by the NJ method based on the Jukes & Cantor distance model, followed by a 1,000-replicate bootstrap analysis for statistical support. For the application of ML and Bayesian methods, we ran ProtTest v2.4 (Abascal et al. 2005) to determine the appropriate model of amino acid replacement. Each computation was accompanied with 500,000-generation runs of four chains. Finally, Bayesian posterior probability from the set of nonparametric bootstrap replicate samples trees was supported by SumTrees program in DendroPy package (Sukumaran and Holder 2010). The 0.25 fraction of initial number of trees in each file was excluded from the analysis. The image of trees obtained by MrBayes was illustrated by using the Dendroscope 3 program (Huson and Scornavacca 2012).
NaCl-Dependent Expression of the NQ Rhodopsin Gene
For the study of NQ rhodopsin gene expression, DSW-6 was grown in 30 ml of ASW enriched with an additional carbon source. ASW prepared with sea salt (Sigma-Aldrich, USA) was passed through a 0.2-µm-pore-size filter, enriched by adding 0.15 g of peptone (Bacto Peptone, Difco) and 0.03 g yeast extract (Bacto Yeast Extract, Difco) and then autoclaved. The final dissolved organic carbon concentration was approximately 2,200 ppm, as measured using a TOC-V total organic carbon analyzer (Shimadzu, Japan). Triplicate cultures were incubated at 25°C under continuous light (135.05 µmol m−2 s−1 photons). When the culture’s optical density at 600 nm reached approximately 0.4, 3 ml of the culture was centrifuged, and the pellet was stored immediately in 500 µl of RNAlater (Ambion, USA) to yield the untreated control (UC) samples. For the remaining cultures, 4 M NaCl or the carbon-enriched ASW was added into each cell culture to yield NaCl concentrations of 2.5% (0.43 M), 5.0% (0.86 M), 7.5% (1.28 M), 10% (1.71 M), and 12.5% (2.14 M). The culture volume of each sample was adjusted to 50 ml, causing a dilution of approximately 2-fold. To make the 4 M NaCl stock, NaCl was dissolved in the ASW enriched with peptone and yeast extract to maintain the nutrient concentration of the culture medium. When the OD600 nm of the 2.5% NaCl culture reached approximately 0.4 again, induction (ID) samples were collected in 500 µl of RNAlater by the same method. Samples of 3 ml were collected for the 2.5% NaCl ID cultures, 5 ml samples were collected for the 5.0% NaCl ID cultures, and 6 ml samples were collected for the 7.5%, 10%, and 12.5% NaCl ID cultures. All of the RNAlater-treated samples were stored at 4°C until RNA extraction. Total RNA was extracted using the RNeasy kit (Qiagen, USA). The remaining chromosomal DNA was eliminated using a Turbo DNA-free Kit (Ambion, USA). No amplification was observed after 40 cycles of PCR when using the RNA samples as templates without the reverse transcription step, confirming complete DNA removal. cDNAs were obtained from reverse transcription polymerase chain reaction (M-MLV cDNA Synthesis Kit, Enzynomics, Korea).
Real-time PCRs were carried out on a CFX Connect Real-Time PCR Detection System (Bio-rad, USA) with the iQ SYBR Green Supermix (Bio-rad). Each cDNA sample was amplified with specific primers (qNR_F 5’-GAG AAT TAT GTA GGT GCT ACA GAC G, qNR_R 5’ GTG CCA AAT TAC CAA GTA ATA CAC CA, q16S_F 5’- AGG ACT TAA CCT GAC ACC TCA C, q16S_R 5’- GGG TTG TAA ACT ACT TTT GTA CAG, qPR_F 5’-CTA AAA TGG CCA CAG ATG ATT ATG TAG, and qPR_R 5’-TTG CAT CAC CAA TGT TGT AAA CTA CG) and quantified in triplicate. The PCR conditions were an initial denaturation step at 95°C for 5 min, followed by 40 cycles of amplification at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Finally, an additional step to establish the melting curve, in which the temperature was decreased from 95 to 65°C (0.05°C s−1), was performed. Threshold cycle values (Ct) for each measurement were determined. The relative quantification of gene expression was calculated by using the comparative critical threshold 2−ΔΔCT method, in which the amount of the RNA of interest is adjusted to an internal reference RNA (Livak and Schmittgen 2001). The 16S rRNA gene was used as the internal control in this study. The following equations were used:
 |
Metagenomic Data Analysis of the NQ Rhodopsins
To identify the new type of rhodopsin genes from the metagenomic data sets and to assess their abundance in diverse aquatic environments, approximately 58 Gb of unassembled reads from metagenome sequencing projects were downloaded from the CAMERA (Sun et al. 2011) and IMG/M (Markowitz et al. 2012) databases in October 2011. BLASTX searches of individual reads (identity ≥ 30%, E value ≤ 1e − 5) were performed against an in-house database consisting of prokaryotic rhodopsins selected for phylogenetic reconstruction retrieved from the GenBank and UniProt databases. Identity of the NQ-type rhodopsins among the recruited reads was assured by carrying out BLASTP searches of the translation products against the GenBank nonredundant protein database and selecting ones that matches the NQ-type rhodopsins in the database as one of the top three best hits. The metagenomic reads confirmed as NQ-type rhodopsins were aligned with 10 full-length NQ-type rhodopsins (see earlier), and their phylogenetic positions were assigned by reconstructing trees using the NJ method in the MEGA5 package (Tamura et al. 2011) with 1,000 iterations. If a read is not localized within the NQ-rhodopsin group or shows low phylogenetic correlations (bootstrap value ≤ 70), the corresponding read was discarded.
Relationships among the 10 NQ-type rhodopsins and the 55 metagenomic reads were further looked into through a clustering analysis, because many of the rhodopsin sequences from metagenome data sets are in small fragments and they often do not overlap to each other. We made a simple assumption that the identity values of a test sequence against the 10 reference sequences will be similar to those of its close relatives. Gap regions among the 10 references were determined by multiple sequence alignment based on the MUSCLE algorithm (Edgar 2004), and amino acid positions corresponding to the gaps were excluded from further analysis. Identity values between the test sequences and the references were calculated using the MEGA5 program with the pairwise deletion option. To estimate the similarities between the test sequences, the identity values were used as input characters for the UPGMA algorithm (Murtagh 1984) with root mean square deviation (Huang et al. 2005).
The prevalence of prokaryotic cells that possess the NQ-type rhodopsins in the metagenomic data sets was postulated by the ratio of the NQ-type rhodopsin reads to RecA, RpoB, and EF-Tu reads. These three proteins are commonly encoded by a single-copy gene in most prokaryotic genomes. BLASTX searches of the metagenomic reads from CAMERA and IMG/M against RecA, RpoB, and EF-Tu retrieved from the GenBank and UniProt databases followed by BLASTX of the resulting candidates against the COG database (Tatusov et al. 2003) identified the reads homologous to the three proteins. When estimating the number of each protein, the length of the protein should be taken into account; the longer a protein is, the higher the chance of that protein being sequenced from the metagenome pool. Thus, the number of reads assigned to each protein was divided by the average length of the protein. The normalized proportion (ra) of prokaryotes containing the new type of rhodopsins in a metagenomic data set was calculated as follows:
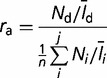 |
where Nd is the number of reads designated as NQ rhodopsins and Ni is the number of the i-th reference protein. ld and li are the lengths of the nucleotide sequences of each protein. n (=3) indicates the number of reference proteins.
Results
Genome Properties for Marine Heterotrophy
Isolated from the seawater sample taken at Dokdo in the East Sea of Korea, N. dokdonensis (basonym: Donghaeana dokdonensis) DSW-6 is a member of the class Flavobacteriia (Yoon et al. 2006). We determined the complete genome sequence of this marine flavobacterium, which harbors a single 3.9-Mb chromosome (supplementary fig. S1 and table S1, Supplementary Material online). Phylogenetic analysis of 16S rRNA genes suggested Gillisia, Gramella, Psychroflexus, and Zunongwangia as the sister genera of Nonlabens, whereas a tree based on broadly conserved proteins of the flavobacteria, whose genomes have been completely sequenced, indicated that Croceibacter, Gramella, Krokinobacter, Lacinutrix, and Zunongwangia are closely related to Nonlabens (supplementary fig. S2, Supplementary Material online).
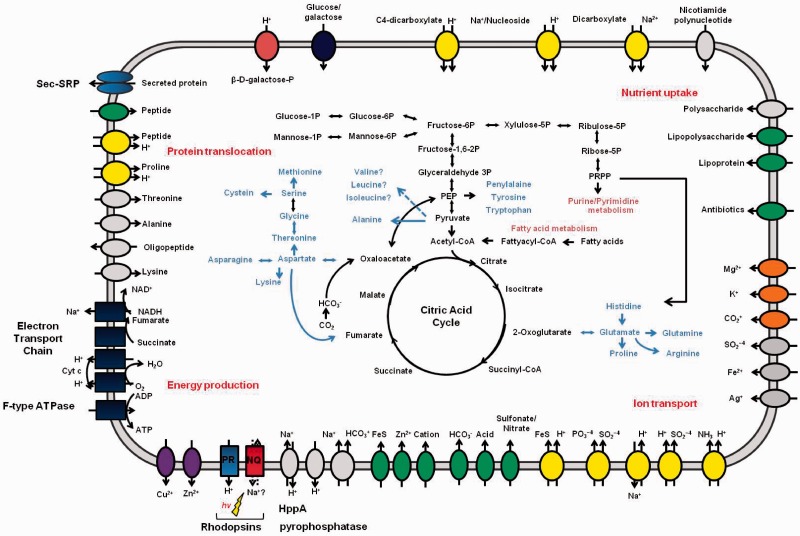
The genome sequence allowed us to reconstruct in silico the metabolic network of the bacterium. The primary metabolic network, including the amino acid biosynthetic pathways and transport systems, is summarized in figure 1 and supplementary table S2, Supplementary Material online. The genes encoding the enzymes for glycolysis and the pentose phosphate pathway were all present. These pathways provide the precursors of metabolites for nucleotide and fatty acid biosynthesis. All the enzymes in the tricarboxylic acid (TCA) cycle are also well conserved in this genome. Most amino acids are synthesized by DSW-6 itself or are derived from the digestion of proteins by secreted peptidases and then transported in across the membrane, where they serve as the main source of nitrogen. The urea cycle does not exist, but a pathway for the conversion of nitrite to ammonia to glutamine is conserved. Enzymes involved in the synthetic pathway for the three branched-chain amino acids leucine, isoleucine, and valine are missing, but genes encoding the enzymes responsible for the degradation of these amino acids exist in the genome.
Fig. 1.—
Overview of metabolism and transport in Nonlabens (Donghaeana) dokdonensis DSW-6. Transporters are grouped by their transport mechanisms: secondary transporters (yellow), ATP-driven transporters, ion channels (orange), P-type ATPase superfamily (purple), and unclassified transporters. Directions of substrate translocation are indicated by arrows.
Physiology for Survival in the Marine Environment
As a marine bacterium that inhabits surface seawater, DSW-6 has many attributes that may endow the bacterium with metabolic advantages that allow survival under oligotrophic conditions. Enzymes compensating for carbon limitation in central metabolism are well conserved in DSW-6. Anaplerotic enzymes replenish the pools of metabolic intermediates in the TCA cycle, which are used as precursors for the biosynthetic pathways, and maintain the oxidative carbon flux. Phosphoenolpyruvate (PEP) carboxylase (DDD_3148) and pyruvate carboxylase (DDD_1224) each create oxaloacetate from PEP or pyruvate by adding bicarbonate (Owen et al. 2002). Bicarbonate is imported into the cell by transporters encoded by a SulP-type Na+-dependent bicarbonate transporter (DDD_2127) or the Na+-dependent bicarbonate secondary transporter SbtA (DDD_0780). Carbonic anhydrase interconverts CO2 and bicarbonate to sustain a sufficient substrate level.
The CFB group bacteria are known to be adapted to use high-molecular-weight organic matter, primarily polysaccharides and proteins (Kirchman 2002). In particular, during a mesocosm study of phytoplanktonic blooms, dominating flavobacteria were shown to play leading roles in the degradation of organic matter in micronutrient-rich environments (Riemann et al. 2000). Genome analysis of several marine flavobacteria revealed that these organisms are adapted to use polymeric organic matter that is occasionally available (Bauer et al. 2006; Gonzalez et al. 2008, 2011). Similar to the genomes of these strains, the DSW-6 genome encodes numerous degrading enzymes, including 14 glycosyl transferases, 20 glycosyl hydrolases, and 25 predicted peptidases. This genome also encodes 24 proteins with cell-surface-adhesion domains that may function in binding to organic particles for efficient breakdown, even though the bacterium is not motile. These proteins benefit the survival of DSW-6 in nutrient-poor oceans where blooms occur only occasionally.
DSW-6 shares features with other ocean-inhabiting bacteria that establish a Na+ gradient instead of a H+ gradient for generating motive force for the first step of their respiratory chain (Unemoto and Hayashi 1993). DSW-6 possesses Na+-translocating NADH:quinine reductase (NQR) subunits as parts of NADH dehydrogenase. In addition to the NADH dehydrogenase subunits, genes encoding the cytochrome c oxidase, cytochrome c, and ATP synthase are present, but genes for the bc1 complex do not exist (supplementary table S3, Supplementary Material online). Instead, the alternative complex III (Refojo et al. 2010) is present to feed electrons to cytochrome c. DSW-6 has numerous genes to cope with internal or environmental stresses, such as oxidative stress or osmotic shock (supplementary table S4, Supplementary Material online).
Many nonphotosynthetic marine bacteria use carotenoids to deal with the damage caused by solar radiation (Hader and Sinha 2005) or to adapt to the cold environment by modulating membrane fluidity (Jagannadham et al. 2000). DSW-6 is orange colored because of carotenoid pigments (Yoon et al. 2006), which are synthesized by clustered crt genes. The repertoire and organization of the carotenoid-biosynthetic genes are conserved between DSW-6, G. limnaea R-8282, and Krokinobacter sp. 4H-3-7-5 (supplementary fig. S3, Supplementary Material online). DSW-6 may express numerous photolyases/cryptochromes related to repairing UV-induced DNA damage and PAS/BLUF/PAC light-sensing domains located in membrane-bound sensor molecules (supplementary fig. S4 and table S5, Supplementary Material online). Carotenoids and these photoproteins are also related to the rhodopsin-based photosystem in the marine euphotic zone. β-Carotene, one of the carotenoid pigments, is a precursor of retinal that binds to rhodopsin, and phylogenetic analysis illustrates the coexistence of photolyase/cryptochrome and PRs among marine flavobacteria (Gonzalez et al. 2008).
Identification of a Unique Type of Microbial Rhodopsin
When mining through the DSW-6 genome, two rhodopsin genes, as well as the blh gene, whose product is responsible for the oxidative cleavage of β-carotene into two retinal molecules, were uncovered. Retinal binding is essential for rhodopsin function. E. coli BL21(DE3) expressing either of these rhodopsins and the purified protein were pinkish-red in color due to the binding of rhodopsin to trans-retinal, which was provided separately (data not shown).
Primary sequence analysis of the translation products suggested that one is a typical PR. Its sequence is highly similar to those of flavobacterial PRs. The functional residues in the ion transfer pathway are all conserved, as in the numerous other proton-pumping rhodopsins. Retinal is predicted to covalently bind to the ε-amino group of the Lys-233 residue in DSW-6 PR (Lys-216 for BR), yielding a protonated retinylidene Schiff base. When stimulated by light, Asp-87 (Asp-85 for BR) becomes a proton acceptor residue of the proton from the deprotonated retinal molecule. The proton release group of Arg-84 (Arg-82 for BR) pumps out the proton to the extracellular side of the membrane. Glu-97 (Asp-96 for BR) restores the original protonated form of the retinal molecule. Then, a proton from the cytosol reprotonates the donor residue, and the protonated acceptor residue gives a proton to the release group, and the proton-pumping cycle repeats (Hayashi et al. 2003).
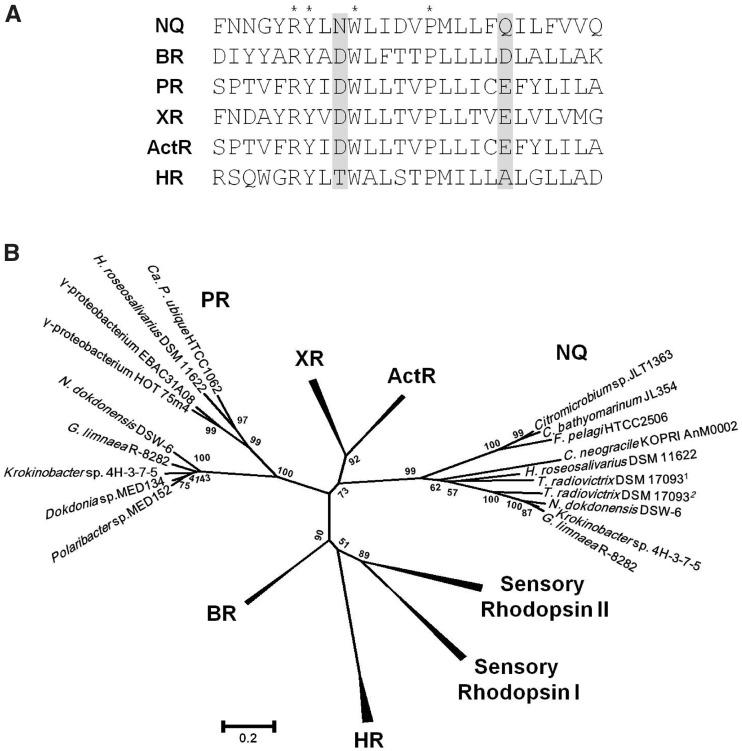
Interestingly, the other rhodopsin gene appears to encode a new type of microbial rhodopsin according to the amino acid sequence analysis. Although this rhodopsin has seven transmembrane domains, as do other rhodopsins, its sequence differs considerably. Notably, although carotenoid-binding Lys-255 (Lys-216 for BR) and proton-releasing Arg-109 (Arg-82 for BR) are conserved, the key active site residues in typical proton-pumping rhodopsins (Asp-85 and Asp-96 for BR) are replaced by Asn-112 and Gln-123 (fig. 2A), suggesting that this protein is functionally unique. We will refer to this Asn-(Xaa10)-Gln sequence as the NQ motif henceforth.
Fig. 2.—
Characteristic features of the new family of rhodopsins that contain the NQ motif. (A) Amino acid sequence alignment of the third transmembrane helix of the NQ rhodopsin of Nonlabens (Donghaeana) dokdonensis DSW-6 and representative microbial rhodopsins. Asterisks, invariant residues; shaded, active site residues of microbial rhodopsins. (B) Phylogenetic relationships among microbial rhodopsins. A tree inferred from 199 conserved amino acid positions was constructed using MEGA5. Bootstrap values for 1,000 replicates are shown next to the branches. NQ, NQ rhodopsin; BR, bacteriorhodopsin; PR, proteorhodopsin; XR, xanthorhodopsin; ActR, actinorhodopsin; HR, halorhodopsin. Accession numbers: Citromicrobium sp. JLT1363, ZP_08702831; Citromicrobium bathyomarinum JL354, ZP_06860850; Fulvimarina pelagi HTCC2506, ZP_01440547; Chaetoceros neogracile KOPRI AnM0002, EL620625; Truepera radiovictrix DSM 170931, YP_003705905; T. radiovictrix DSM 170932, YP_003706581; Gillisia limnaea R-8282, ZP_09669334; Krokinobacter (Dokdonia) sp. 4H-3-7-5, YP_004429763; Hymenobacter roseosalivarius DSM 11622, gene ID 2502504185 or locus tag HrosDRAFT_3745 in the IMG database.
The NQ Motif-Containing Rhodopsins Form a Distinct Phylogenetic Class
Homologous proteins were detected in broadly different taxa through similarity searches of public databases of microbial genomes. Genes encoding rhodopsins of the new type were present in the recently sequenced genomes of three CFB strains and three marine alphaproteobacterial strains (fig. 2B). Among these strains, two flavobacterial strains, G. limnaea R-8282 and Krokinobacter sp. 4H-3-7-5, and a cytophagal species, H. roseosalivarius, have both the PR gene and the new type of rhodopsin. The deinococcus T. radiovictrix, a radiation-resistant species recovered from sodium-rich hot spring runoff, carried two of these genes in addition to an HR gene and a rhodopsin gene of an unassigned family. All these rhodopsins contain the NQ motif instead of the DD motif, and the existence of genome sequences containing only the new type suggests that these organisms apparently do not require blh and idi for functioning (table 1). Surprisingly, a homologous sequence was found among the expressed sequence tags of the Antarctic marine planktonic diatom C. neogracile, suggesting that this class of rhodopsins is not restricted to prokaryotes. Comparative analyses based on NJ, ML, and Bayesian methods confirm, supported by ≥99% bootstrap values, that the NQ motif-containing rhodopsins form a distinct phylogenetic group (fig. 2B and supplementary fig. S5, Supplementary Material online).
Table 1.
Rhodopsin Genes and Retinal Biosynthetic Genes in Nonlabens (Donghaeana) dokdonensis DSW-6 and Other Strains with the NQ Motif in Rhodopsin Sequences
| Strain Name | NQ | PR | blh | crtE | crtB | crtI | crtY | ispA | idi | GenBank Accession |
|---|---|---|---|---|---|---|---|---|---|---|
| Nonlabens (Donghaeana) dokdonensis DSW-6 | + | + | + | + | + | + | + | + | + | CP001397 |
| Krokinobacter (Dokdonia) sp. 4H-3-7-5 | + | + | + | + | + | + | + | + | + | CP002528 |
| Gillisia limnaea R-8282 | + | + | + | + | + | + | + | + | + | AHKR00000000 |
| Hymenobacter roseosalivarius DSM 11622 | + | + | − | + | + | + | + | + | − | Taxon ID: 2502422321 (IMG) |
| Truepera radiovictrix DSM 17093 | ++ | − | − | + | + | + | − | + | − | CP002049 |
| Fulvimarina pelagi HTCC2506 | + | − | − | + | + | + | + | + | − | AATP00000000 |
| Citromicrobium bathyomarinum JL354 | + | − | − | + | + | + | + | + | − | ADAE00000000 |
| Citromicrobium sp. JLT1363 | + | − | − | + | + | + | + | + | − | AEUE00000000 |
Note.—++ indicates two copies of the same gene. NQ denotes the NQ rhodopsin gene; PR denotes the proteorhodopsin gene. CrtEBIY, Blh, Idi,and IspA are directly involved in beta-carotene and retinal biosynthesis.
The Expression of the NQ Rhodopsin Gene of DSW-6 Is Induced at High Concentrations of NaCl
Tracking gene expression patterns can be a reliable clue for deducing the function of a given gene, as cells change their gene expression in response to environmental perturbations. To infer the role of the DSW-6 rhodopsins, the gene expression levels under various nutritional and environmental conditions were measured (see supplementary materials and methods, Supplementary Material online). Quantitative real-time PCR (qRT-PCR) analysis indicated that both rhodopsin genes are highly induced when cells are incubated in the presence of light or in the absence of sufficient nutrients (supplementary fig. S6, Supplementary Material online). However, the absolute levels of the gene encoding the NQ motif-containing rhodopsin were much lower than those of the PR gene under these conditions.
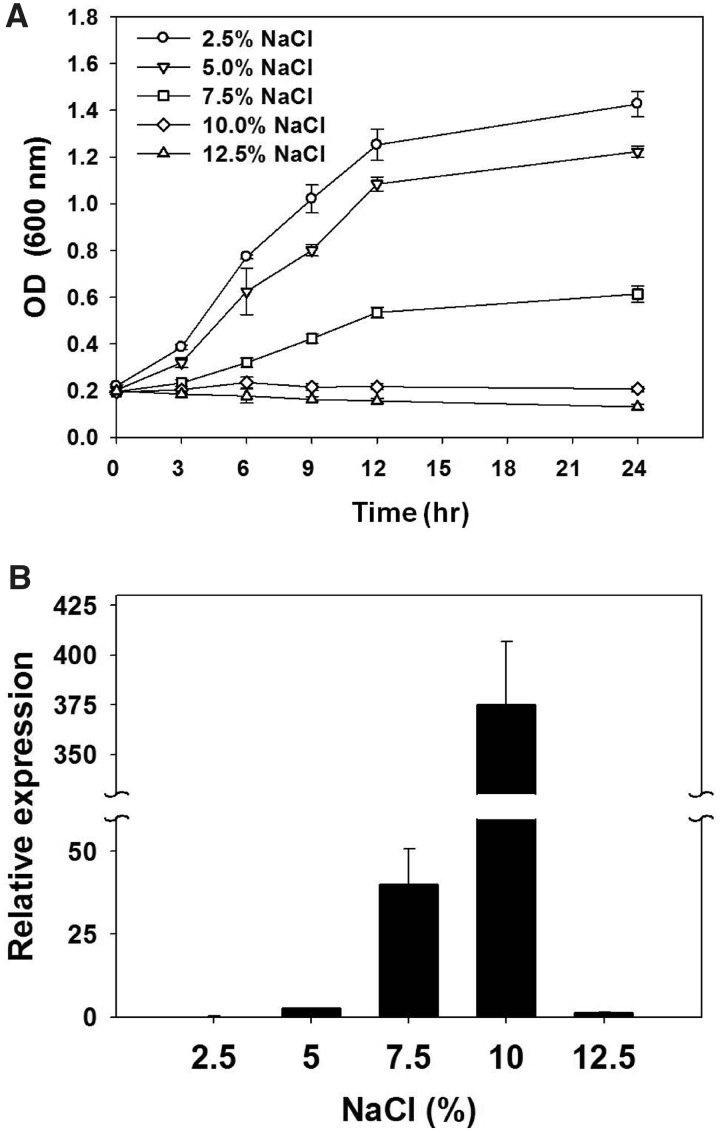
The presence of the unique NQ motif in the newly found class of rhodopsins and the presence of two rhodopsins containing this motif in a bacterium that thrives in a sodium-rich hot spring prompted us to analyze the expression of the NQ rhodopsin gene at various concentrations of NaCl to infer the function of the NQ rhodopsins. The artificial seawater (Huang et al. 2005) used for the culture medium contains 2.5% NaCl and represents the isosaline condition. DSW-6 cells were first grown in ASW enriched with additional carbon and nitrogen sources until the early log phase, and then the concentration of NaCl was set to 2.5%, 5.0%, 7.5%, 10%, or 12.5% (see Materials and Methods for details). Cells exposed to 2.5%, 5.0%, or 7.5% NaCl kept growing, whereas those exposed to 10% NaCl did not grow further after 6 h (fig. 3A). At 12.5% NaCl, the OD600 nm values decreased. After 3 h of exposure to each NaCl concentration, total RNA from each sample was extracted, and the relative expression level of the NQ rhodopsin gene was determined. No difference between the before and after treatment time points was observed for the 2.5% NaCl culture, as expected. As the NaCl concentration increased, gene expression was induced. The relative expression level was highest at 10% NaCl (of 375 ± 31.4). In 12.5% NaCl, the level of gene induction was lower than that in 5.0% NaCl (fig. 3B). In contrast, the expression levels of the PR gene were not affected by the NaCl concentration (data not shown).
Fig. 3.—
The relative expression levels of NQ rhodopsin genes in Nonlabens (Donghaeana) dokdonensis DSW-6 at different NaCl concentrations. (A) Cell growth was determined by optical density values at 600 nm for cultures incubated in 2.5% NaCl (circles), 5.0% NaCl (downward triangles), 7.5% NaCl (square), 10% (upward triangles), and 12.5% NaCl (diamonds). After 3 h, cultures were collected for RNA extraction to analyze the gene expression. Error bars indicate the standard deviations for triplicate samples. (B) The NQ rhodopsin expression level was quantified using qRT-PCR and the comparative critical threshold (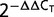 ) method (Livak and Schmittgen 2001). The 16S rRNA gene was used as an internal control, and the NQ rhodopsin transcript at 3 h was quantified relative to the NQ rhodopsin transcript in an UC (2.5% NaCl). The break in the y axis ranges from 60 to 330. Error bars indicate the standard deviations for triplicate reactions.
) method (Livak and Schmittgen 2001). The 16S rRNA gene was used as an internal control, and the NQ rhodopsin transcript at 3 h was quantified relative to the NQ rhodopsin transcript in an UC (2.5% NaCl). The break in the y axis ranges from 60 to 330. Error bars indicate the standard deviations for triplicate reactions.
NQ Rhodopsins Are Frequently Found in Hypersaline Environments
To gain information on the diversity and the prevalence of NQ rhodopsins in nature, we searched for this rhodopsin type in metagenomic sequences. A total of 55 reads orthologous to NQ rhodopsin genes in the microbial genomes described earlier were recruited from metagenomic data sets originating from various aquatic environments, including a hypersaline microbial mat, saltern and freshwater lakes, oceans, and even Antarctica (supplementary table S6, Supplementary Material online). Sixteen NQ motif-containing reads are shown in supplementary figure S7, Supplementary Material online.
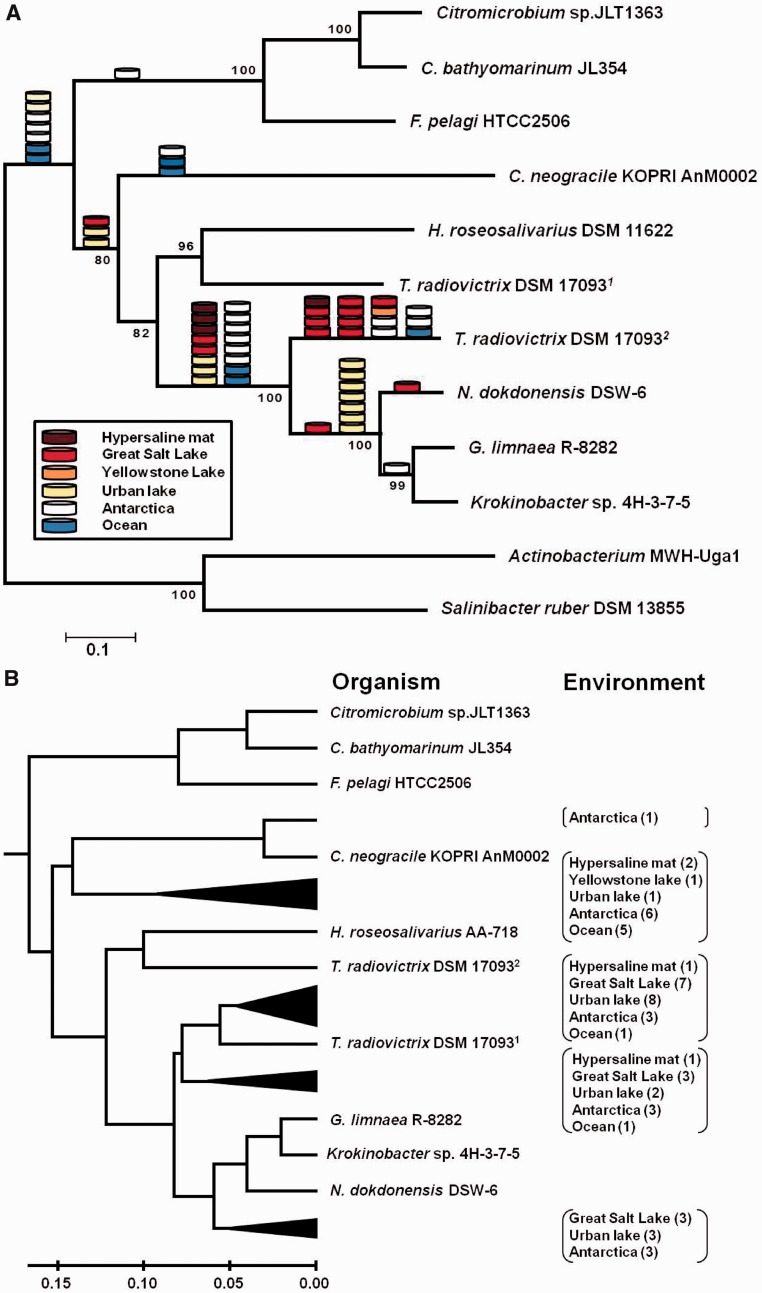
Phylogenetic relationships of the genome-derived NQ rhodopsins inferred through MP, NJ, ML, and Bayesian approaches indicated that overall branching patterns are almost identical among the trees (fig. 4A and supplementary fig. S8, Supplementary Material online). However, grouping of the branches of an NQ rhodopsin in T. radiovictrix DSM 17093 and that in H. roseosalivarius DSM 11622 in the MP, NJ, and Bayesian trees could not be supported in the ML tree (supplementary fig. S8A, Supplementary Material online). Positioning of the metagenomic reads on the NJ tree of the full-length NQ rhodopsins indicated that 40 of them (72.7%) can be anchored to the branches that comprised the ones present in N. dokdonensis, G. limnaea, and Krokinobacter sp., and YP_003706581 of T. radiovictrix (fig. 4A). Among the reads included in this subfamily are four out of four reads from a hypersaline microbial mat and 12 of 13 reads from the Great Salt Lake. YP_003706581 has the largest number (15) of reads that are closest. Results obtained from clustering of the metagenome reads and the full-length NQ rhodopsins using UPGMA showed that NQ rhodopsins from the metagenomic data sets can be divided into at least four groups (fig. 4B). Similar to the results from phylogenetic anchoring, 39 reads were clustered with those of the four species (YP_003706581 for T. radiovictrix). Out of the 13 reads from the Great Salt Lake, 10 reads seem to be closely related to YP_003706581. Among the reads from Antarctica, one clusters with the NQ rhodopsin of C. neogracile, whereas six others form another group and appear more distantly related.
Fig. 4.—
NQ rhodopsin-like sequences in the metagenomic data sets. (A) Phylogenetic positions of the NQ rhodopsin-like metagenomic reads. The cylinder symbol represents a metagenomic read originating from hypersaline microbial mat (brown), Greate Salt Lake (red), Yellowstone Lake (orange), urban lake (yellow), Antarctica (white), or oceans (blue). A phylogenetic tree of full-length NQ-rhodopsins was constructed based on the NJ distance analysis of 243 positions. (B) UPGMA clustering of the NQ rhodopsin-like metagenomic reads. The dendrogram was derived from UPGMA cluster analysis using identity values against completely sequenced genes. Metagenome projects from which NQ rhodopsin-like sequences were recruited: hypersaline mat, Guerrero Negro hypersaline microbial mat (CAM_PROJ_HypersalineMat); Great Salt Lake, Great Salt Lake (Gm00191 of GOLD ID in IMG database); Yellowstone Lake, Yellowstone Lake (CAM_PROJ_YLake); urban lake, LaBonte Lake (Gm00212 of GOLD ID in IMG database); Antarctica, Antarctica aquatic microbial metagenome (CAM_PROJ_AntarcticaAquatic); ocean, Global Ocean Sampling expedition (CAM_PROJ_GOS). Truepera radiovictrix DSM 170931, YP_003705905; T. radiovictrix DSM 170932, YP_003706581.
The occurrence of NQ rhodopsin-carrying prokaryotes in the metagenomic data sets was evaluated using the ratio of rhodopsin genes to conserved single-copy genes (supplementary table S7, Supplementary Material online). A hypersaline microbial mat sample from Guerrero Negro, Mexico, containing approximately 90 practical salinity units had an abundance of the NQ rhodopsin genes as high as 17.36%. In addition, they were found frequent in the samples from the Great Salt Lake (1.48%) and Labonte Lake (2.75%). Based on the abundance analysis of GOS data, however, 0.09% of the prokaryotes inhabiting the sea surface may possess this rhodopsin type. No prokaryotes possessing this gene were found in high salinity ponds from Chula Vista solar salterns, where haloarchaea dominate (Pasic et al. 2009).
Discussion
The genome analysis of DSW-6 demonstrates that this strain is highly adapted for living in the oligotrophic surface of the ocean. Similar to other ocean-inhabiting bacteria, DSW-6 is thought to use the Na+ gradient instead of the H+ gradient to generate the motive force for the respiratory chain (Unemoto and Hayashi 1993) and has many Na+-dependent transporter genes. Several stress-response gene products and pigments may protect DSW-6 from the harsh sunlight. In light of light utilization, genes for a PR and retinal biosynthesis in addition to those for light-sensing proteins exist in the DSW-6 genome. In the case of the PR-carrying marine flavobacterium Polaribacter sp. MED152, uptake of bicarbonate increased in the presence of light, suggesting that a PR-mediated proton gradient drives anaplerotic inorganic carbon fixation toward more efficient anabolism (Gonzalez et al. 2008). Similar to MED152, enzymes involved in the anaplerotic pathway are well conserved in DSW-6.
Because Beja et al. (2000) discovered PR in an uncultured marine bacterial clade, extensive research, including metagenomic and photochemical approaches, has been conducted to discover the reasons for the great success of this widespread rhodopsin family. Currently, these light-driven proton pumps are considered to power cell growth or extend the survival of marine oligotrophs. Light stimulates growth of the PR-containing marine flavobacterium Dokdonia sp. MED134 (Gomez-Consarnau et al. 2007). Vibrio sp. AND4, another PR-containing strain, exhibited longer survival during starvation than its corresponding PR deletion mutant (Gomez-Consarnau et al. 2010). When expressed in E. coli, a PR from the SAR-86 clade of the Gammaproteobacteria promotes proton-motive force that turns the flagellar motor during light illumination (Walter et al. 2007). Several lines of evidence suggest that the PR in DSW-6 should be a functional equivalent of other PRs and may play similar roles to contribute to the growth or survival of the bacterium in oligotrophic environments. First, it exhibits the key features of a typical PR, and all the functional residues in the proton transfer pathway are conserved. Second, the encoding gene is highly expressed in the presence of light or in the absence of sufficient nutrients. Most significantly, purified PR protein binds to trans-retinal and pumps out proton (Han S-I, Kwon S-K, Kim JF, and Jung K-H, unpublished data).
Microbial rhodopsins are functionally versatile and may transit nonproton ions across the plasma membrane. HRs existing in halophiles pump in chloride instead of proton in a light-dependent manner (Kolbe et al. 2000). The channelrhodopsins of green algae are light-gated cation channels (Kato et al. 2012). In this article, we propose the existence of a new type of rhodopsin with a function that is unique among proton pumps and is related to salinity; this proposal was based on the following: These proteins, which we dubbed the NQ rhodopsins, have seven transmembrane domains and the key amino acids required for carotenoid binding or proton releasing, but the proton-donor and proton-acceptor residues in the PRs (Asp-87 and Glu-97) or the BRs (Asp-85 and Asp-96) are substituted by Asn and Gln. In case of a PR, the replacement of Asp-87 with Asn or of Glu-97 with Gln leads to variants that can no longer pump protons (Dioumaev et al. 2003; Saeedi et al. 2012). In addition, the expression levels of the NQ rhodopsin gene in DSW-6 correlate with illumination, nutrient levels, and, most importantly, the NaCl concentration. It is noteworthy that these novel rhodopsins are widely distributed in geographic environments exposed to high salinity, according to homology searches against genomic and metagenomic data sets. The NQ rhodopsin gene in DSW-6 showed the highest expression at 10% NaCl, which is consistent with the observation from a metagenomic survey that NQ sequences are most abundant in a hypersaline microbial mat with the salinity of 9%.
Sodium and chloride are the two most predominant dissolved ions in seawater; which could be the substrate of the NQ rhodopsins? An environmental clue supports the possibility that the NQ rhodopsins act as sodium pumps. A strain of T. radiovictrix isolated from a high-sodium environment (Albuquerque et al. 2005) has two NQ rhodopsin genes in its genome. Moreover, the key functional residues of the chloride-pump HRs differ from those of the NQ rhodopsins (Kolbe et al. 2000). In particular, Asn-112 and Gln-123 of the NQ rhodopsin in DSW-6 are Thr and Ala in the HRs, respectively. Site-directed mutagenesis of Asp-85 in a proton-pump BR to Thr or Ser enables the protein to transport chloride inward as an HR does (Sasaki et al. 1995).
As sodium pumps, NQ rhodopsins may simply be involved in the regulation of the osmotic state. However, NQ rhodopsins can be more productive if they function in generating sodium-motive force. Some extremophilic archaea and marine bacteria use the sodium-motive force to power energy-transducing machinery, either exclusively or by coupling it with the proton-motive force (Albers et al. 2001). In the sodium-rich marine environment, employing the sodium-motive force instead of the proton-motive force can be advantageous for marine bacteria that require sodium for their growth (Kogure 1998). These bacteria generate sodium-motive force by possessing either a respiration-dependent primary Na+ pump that directly couples Na+ translocation to a chemical reaction or a Na+/H+ antiporter that converts the H+ gradient generated by primary H+ pumps into a Na+ gradient (Hase et al. 2001). The cell yields of Dokdonia sp. MED134 cultures exposed to light decrease in the presence of an inhibitor of Na+-translocating NQR (Kimura et al. 2011). Among the transcripts of MED134, Na+-translocating NQR and Na+ transporters were greatly up-regulated in cultures grown under light (Kimura et al. 2011). Therefore, the sodium ion gradient essentially functions in light-enhanced growth promotion in a PR-containing marine bacterium, suggesting that the existence of sodium-pumping rhodopsins is plausible.
Restricted distribution of the NQ rhodopsins to bacterial species in the classes Alphaproteobacteria, Cytophagia, Flavobacteriia, and Deinococci suggests the horizontal transfer of the encoding genes among bacteria inhabiting in the saline environments. Furthermore, identification of the same type from the expressed sequence tags of a diatom in the Coscinodiscophyceae raises an interesting possibility of interdomain gene transfer between bacteria and eukaryotes. Diatoms are ubiquitous eukaryotic phytoplankton in the ocean (Armbrust 2009; Amin et al. 2012), and the genus Chaetoceros is considered as one of the most abundant and widespread diatom groups (Nagasaki 2008). Metagenomic analysis indicates that, even from the data sets that were supposed to be mostly prokaryotic, a number of NQ rhodopsin-like sequence reads are most closely related to that of C. neogracile.
Taken together, our analysis reveals a novel family of rhodopsins whose genes are broadly present in microorganisms adapted to ecosystems that may experience hypersaline conditions and which seem to play an important role in sodium metabolism. This finding echoes the coexistence of the H+-pumping ATPases and the Na+-pumping ATPases (Morth et al. 2011).
Supplementary Material
Supplementary materials and methods, tables S1–S7, and figures S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Seung-Hwan Park, Kwang-Hwan Jung, Haeyoung Jeong, Jang-Cheon Cho, Dong-Woo Lee, Myung Hee Kim, and Kyung-Mo Kim, as well as GEM members, for helpful discussions and invaluable comments. This work was supported by the National Research Foundation of Korea (2011-0017670) and the 21C Frontier Microbial Genomics and Applications Center of the Ministry of Education, Science and Technology, Republic of Korea.
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Albers SV, Van de Vossenberg JL, Driessen AJ, Konings WN. Bioenergetics and solute uptake under extreme conditions. Extremophiles. 2001;5:285–294. doi: 10.1007/s007920100214. [DOI] [PubMed] [Google Scholar]
- Albuquerque L, et al. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol Lett. 2005;247:161–169. doi: 10.1016/j.femsle.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol. 2007;396:71–91. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- Armbrust EV. The life of diatoms in the world's oceans. Nature. 2009;459:185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Balashov SP, et al. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, et al. Whole genome analysis of the marine Bacteroidetes “Gramella forsetii” reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Béjà O, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Frigaard NU. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Bryant DA, et al. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science. 2007;317:523–526. doi: 10.1126/science.1143236. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioumaev AK, Wang JM, Bálint Z, Váró G, Lanyi JK. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry. 2003;42:6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Falkowski PG, Barber RT, Smetacek VV. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Gómez-Consarnau L, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- Gómez-Consarnau L, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci U S A. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, et al. Genomics of the proteorhodopsin-containing marine flavobacterium Dokdonia sp. strain MED134. Appl Environ Microbiol. 2011;77:8676–8686. doi: 10.1128/AEM.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hader DP, Sinha RP. Solar ultraviolet radiation-induced DNA damage in aquatic organisms: potential environmental impact. Mutat Res. 2005;571:221–233. doi: 10.1016/j.mrfmmm.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Häse CC, Fedorova ND, Galperin MY, Dibrov PA. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev. 2001;65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Tajkhorshid E, Schulten K. Molecular dynamics simulation of bacteriorhodopsin's photoisomerization using ab initio forces for the excited chromophore. Biophys J. 2003;85:1440–1449. doi: 10.1016/S0006-3495(03)74576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P, et al. Hymenobacter roseosalivarius gen. nov., sp. nov. from continental Antartica soils and sandstone: bacteria of the Cytophaga/Flavobacterium/Bacteroides line of phylogenetic descent. Syst Appl Microbiol. 1998;21:374–383. doi: 10.1016/s0723-2020(98)80047-7. [DOI] [PubMed] [Google Scholar]
- Huang HC, Nagaswamy U, Fox GE. The application of cluster analysis in the intercomparison of loop structures in RNA. RNA. 2005;11:412–423. doi: 10.1261/rna.7104605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Jagannadham MV, et al. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch Microbiol. 2000;173:418–424. doi: 10.1007/s002030000163. [DOI] [PubMed] [Google Scholar]
- Jiao N, Zhang R, Zheng Q. Coexistence of two different photosynthetic operons in Citromicrobium bathyomarinum JL354 as revealed by whole-genome sequencing. J Bacteriol. 2010;192:1169–1170. doi: 10.1128/JB.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I, et al. Genome sequence of Fulvimarina pelagi HTCC2506T, a Mn(II)-oxidizing alphaproteobacterium possessing an aerobic anoxygenic photosynthetic gene cluster and xanthorhodopsin. J Bacteriol. 2010;192:4798–4799. doi: 10.1128/JB.00761-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. Pathway tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Young CR, Martinez A, Delong EF. Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 2011;5:1641–1651. doi: 10.1038/ismej.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- Klippel B, et al. Complete genome sequences of Krokinobacter sp. strain 4H-3-7-5 and Lacinutrix sp. strain 5H-3-7-4, polysaccharide-degrading members of the family Flavobacteriaceae. J Bacteriol. 2011;193:4545–4546. doi: 10.1128/JB.05518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K. Bioenergetics of marine bacteria. Curr Opin Biotechnol. 1998;9:278–282. doi: 10.1016/s0958-1669(98)80059-1. [DOI] [PubMed] [Google Scholar]
- Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 2012;40:D123–D129. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, et al. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci U S A. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth JP, et al. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat Rev Mol Cell Biol. 2011;12:60–70. doi: 10.1038/nrm3031. [DOI] [PubMed] [Google Scholar]
- Murtagh F. Complexities of hierarchic clustering algorithms: state of the art. Comput Stats Q. 1984;1:101–113. [Google Scholar]
- Nagasaki K. Dinoflagellates, diatoms, and their viruses. J Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Pašić L, et al. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics. 2009;10:570. doi: 10.1186/1471-2164-10-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo PN, Sousa FL, Teixeira M, Pereira MM. The alternative complex III: a different architecture using known building modules. Biochim Biophys Acta. 2010;1797:1869–1876. doi: 10.1016/j.bbabio.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Riemann L, Steward GF, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66:578–587. doi: 10.1128/aem.66.2.578-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeedi P, Moosaabadi JM, Sebtahmadi SS, Behmanesh M, Mehrabadi JF. Site-directed mutagenesis in bacteriorhodopsin mutants and their characterization for bioelectrical and biotechnological equipment. Biotechnol Lett. 2012;34:455–462. doi: 10.1007/s10529-011-0731-4. [DOI] [PubMed] [Google Scholar]
- Sasaki J, et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Sharma AK, et al. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 2009;3:726–737. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Sun S, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemoto T, Hayashi M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J Bioenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- Van Trappen S, Vandecandelaere I, Mergaert J, Swings J. Gillisia limnaea gen. nov., sp. nov., a new member of the family Flavobacteriaceae isolated from a microbial mat in Lake Fryxell, Antarctica. Int J Syst Evol Microbiol. 2004;54:445–448. doi: 10.1099/ijs.0.02922-0. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci U S A. 2007;104:2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Chun J. Unification of the genera Nonlabens, Persicivirga, Sandarakinotalea and Stenothermobacter into a single emended genus, Nonlabens, and description of Nonlabens agnitus sp. nov. Syst Appl Microbiol. 2012;35:150–155. doi: 10.1016/j.syapm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Kang SJ, Lee CH, Oh TK. Donghaeana dokdonensis gen. nov., sp. nov., isolated from sea water. Int J Syst Evol Microbiol. 2006;56:187–191. doi: 10.1099/ijs.0.63847-0. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Kang SJ, Park S, Oh TK. Reclassification of the three species of the genus Krokinobacter into the genus Dokdonia as Dokdonia genika comb. nov., Dokdonia diaphoros comb. nov. and Dokdonia eikasta comb. nov., and emended description of the genus Dokdonia Yoon et al. 2005. Int J Syst Evol Microbiol. 2012;62: 1896–1901. doi: 10.1099/ijs.0.035253-0. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Kawanabe A, Ito H, Kandori H, Kogure K. Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ Microbiol. 2012;14:1240–1248. doi: 10.1111/j.1462-2920.2012.02702.x. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rannala B, Yang Z. Robustness of compound Dirichlet priors for Bayesian inference of branch lengths. Syst Biol. 2012;61:779–784. doi: 10.1093/sysbio/sys030. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhang R, Jiao N. Genome sequence of Citromicrobium strain JLT1363, isolated from the South China Sea. J Bacteriol. 2011;193:2074–2075. doi: 10.1128/JB.00121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.