Abstract
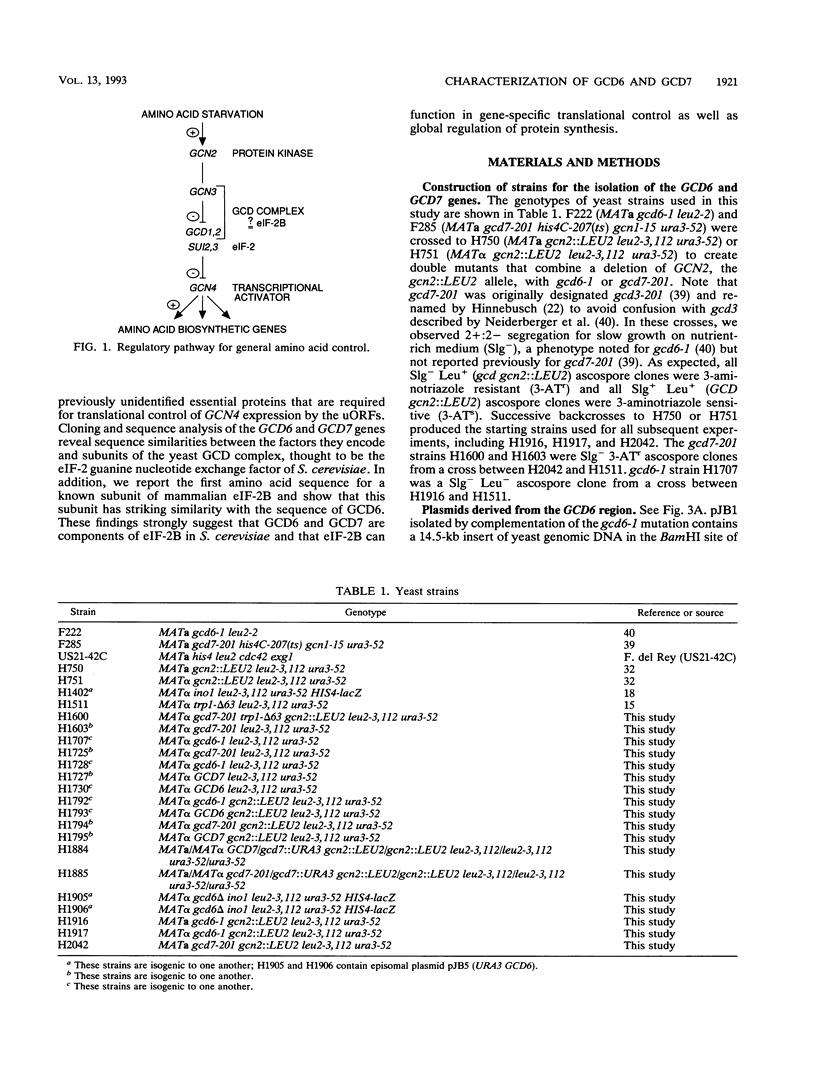
Starvation of the yeast Saccharomyces cerevisiae for an amino acid signals increased translation of GCN4, a transcriptional activator of amino acid biosynthetic genes. We have isolated and characterized the GCD6 and GCD7 genes and shown that their products are required to repress GCN4 translation under nonstarvation conditions. We find that both GCD6 and GCD7 show sequence similarities to components of a high-molecular-weight complex (the GCD complex) that appears to be the yeast equivalent of translation initiation factor 2B (eIF-2B), which catalyzes GDP-GTP exchange on eIF-2. Furthermore, we show that GCD6 is 30% identical to the largest subunit of eIF-2B isolated from rabbit reticulocytes. Deletion of either GCD6 or GCD7 is lethal, and nonlethal mutations in these genes increase GCN4 translation in the same fashion described for defects in known subunits of eIF-2 or the GCD complex; derepression of GCN4 is dependent on short open reading frames in the GCN4 mRNA leader and occurs independently of eIF-2 alpha phosphorylation by protein kinase GCN2, which is normally required to stimulate GCN4 translation. Together, our results provide evidence that GCD6 and GCD7 are subunits of eIF-2B in S. cerevisiae and further implicate this GDP-GTP exchange factor in gene-specific translational control.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaraju G. R., Hansen L. J., Jagus R. Increase in eukaryotic initiation factor 2B activity following fertilization reflects changes in redox potential. J Biol Chem. 1991 Dec 25;266(36):24451–24459. [PubMed] [Google Scholar]
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Chen J. J., Throop M. S., Gehrke L., Kuo I., Pal J. K., Brodsky M., London I. M. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7729–7733. doi: 10.1073/pnas.88.17.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991 Jun;11(6):3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa J., Vazquez de Aldana C. R., San Segundo P., del Rey F. Genetic mapping of 1,3-beta-glucanase-encoding genes in Saccharomyces cerevisiae. Curr Genet. 1992 Oct;22(4):283–288. doi: 10.1007/BF00317922. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992 Feb 7;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dholakia J. N., Wahba A. J. Phosphorylation of the guanine nucleotide exchange factor from rabbit reticulocytes regulates its activity in polypeptide chain initiation. Proc Natl Acad Sci U S A. 1988 Jan;85(1):51–54. doi: 10.1073/pnas.85.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Flotow H., Graves P. R., Wang A. Q., Fiol C. J., Roeske R. W., Roach P. J. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990 Aug 25;265(24):14264–14269. [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Rubino M. S., Starn T. K. Regulation of protein synthesis in rabbit reticulocyte lysate. Glucose 6-phosphate is required to maintain the activity of eukaryotic initiation factor (eIF)-2B by a mechanism that is independent of the phosphorylation of eIF-2 alpha. J Biol Chem. 1988 Sep 5;263(25):12486–12492. [PubMed] [Google Scholar]
- Hannig E. M., Hinnebusch A. G. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol Cell Biol. 1988 Nov;8(11):4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hinnebusch A. G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Struhl K. Molecular characterization of GCD1, a yeast gene required for general control of amino acid biosynthesis and cell-cycle initiation. Nucleic Acids Res. 1988 Oct 11;16(19):9253–9265. doi: 10.1093/nar/16.19.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Involvement of an initiation factor and protein phosphorylation in translational control of GCN4 mRNA. Trends Biochem Sci. 1990 Apr;15(4):148–152. doi: 10.1016/0968-0004(90)90215-w. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Novel mechanisms of translational control in Saccharomyces cerevisiae. Trends Genet. 1988 Jun;4(6):169–174. doi: 10.1016/0168-9525(88)90023-6. [DOI] [PubMed] [Google Scholar]
- Hwang Y. W., Miller D. L. A study of the kinetic mechanism of elongation factor Ts. J Biol Chem. 1985 Sep 25;260(21):11498–11502. [PubMed] [Google Scholar]
- Janssen G. M., Maessen G. D., Amons R., Möller W. Phosphorylation of elongation factor 1 beta by an endogenous kinase affects its catalytic nucleotide exchange activity. J Biol Chem. 1988 Aug 15;263(23):11063–11066. [PubMed] [Google Scholar]
- Johnson D. I., Jacobs C. W., Pringle J. R., Robinson L. C., Carle G. F., Olson M. V. Mapping of the Saccharomyces cerevisiae CDC3, CDC25, and CDC42 genes to chromosome XII by chromosome blotting and tetrad analysis. Yeast. 1987 Dec;3(4):243–253. doi: 10.1002/yea.320030405. [DOI] [PubMed] [Google Scholar]
- Konieczny A., Safer B. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983 Mar 10;258(5):3402–3408. [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel E. A., Mulligan J. A., Sommercorn J., Krebs E. G. Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J Biol Chem. 1987 Jul 5;262(19):9136–9140. [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiori F., Meggio F., Marin O., Borin G., Calderan A., Ruzza P., Pinna L. A. Synthetic peptide substrates for casein kinase 2. Assessment of minimum structural requirements for phosphorylation. Biochim Biophys Acta. 1988 Oct 7;971(3):332–338. doi: 10.1016/0167-4889(88)90149-8. [DOI] [PubMed] [Google Scholar]
- Matts R. L., Levin D. H., London I. M. Effect of phosphorylation of the alpha-subunit of eukaryotic initiation factor 2 on the function of reversing factor in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1983 May;80(9):2559–2563. doi: 10.1073/pnas.80.9.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Myers P. L., Skvirsky R. C., Greenberg M. L., Greer H. Negative regulatory gene for general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Sep;6(9):3150–3155. doi: 10.1128/mcb.6.9.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger P., Aebi M., Hütter R. Identification and characterization of four new GCD genes in Saccharomyces cerevisiae. Curr Genet. 1986;10(9):657–664. doi: 10.1007/BF00410913. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hannig E. M., Hinnebusch A. G. Amino acid sequence similarity between GCN3 and GCD2, positive and negative translational regulators of GCN4: evidence for antagonism by competition. Genetics. 1989 Jul;122(3):551–559. doi: 10.1093/genetics/122.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hinnebusch A. G. gcd12 mutations are gcn3-dependent alleles of GCD2, a negative regulator of GCN4 in the general amino acid control of Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):543–550. doi: 10.1093/genetics/122.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain V. M. Initiation of protein synthesis in mammalian cells. Biochem J. 1986 May 1;235(3):625–637. doi: 10.1042/bj2350625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins K. K., Dailey G. M., Tjian R. In vitro analysis of the Antennapedia P2 promoter: identification of a new Drosophila transcription factor. Genes Dev. 1988 Dec;2(12A):1615–1626. doi: 10.1101/gad.2.12a.1615. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rowlands A. G., Montine K. S., Henshaw E. C., Panniers R. Physiological stresses inhibit guanine-nucleotide-exchange factor in Ehrlich cells. Eur J Biochem. 1988 Jul 15;175(1):93–99. doi: 10.1111/j.1432-1033.1988.tb14170.x. [DOI] [PubMed] [Google Scholar]
- Rowlands A. G., Panniers R., Henshaw E. C. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988 Apr 25;263(12):5526–5533. [PubMed] [Google Scholar]
- Shou C., Farnsworth C. L., Neel B. G., Feig L. A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992 Jul 23;358(6384):351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- Siekierka J., Manne V., Ochoa S. Mechanism of translational control by partial phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Proc Natl Acad Sci U S A. 1984 Jan;81(2):352–356. doi: 10.1073/pnas.81.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989 Jun 16;57(6):947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Ursic D., Culbertson M. R. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991 May;11(5):2629–2640. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]



