Abstract
Natural resilin, the rubber-like protein that exists in specialized compartments of most arthropods, possesses excellent mechanical properties such as low stiffness, high resilience and effective energy storage. Recombinantly-engineered resilin-like polypeptides (RLPs) that possess the favorable attributes of native resilin would be attractive candidates for the modular design of biomaterials for engineering mechanically active tissues. Based on our previous success in creating a novel RLP-based hydrogel and demonstrating useful mechanical and cell-adhesive properties, we have produced a suite of new RLP-based constructs, each equipped with 12 repeats of the putative resilin consensus sequence and a single, distinct biologically active domain. This approach allows independent control over the concentrations of cell-binding, MMP-sensitive, and polysaccharide-sequestration domains in hydrogels comprising mixtures of the various RLPs. The high purity, molecular weight and correct compositions of each new polypeptide have been confirmed via high performance liquid chromatography (HPLC), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), and amino acid analysis. These RLP-based polypeptides exhibit largely random-coil conformation, both in solution and in the cross-linked hydrogels, as indicated by circular dichroic and infrared spectroscopic analyses. Hydrogels of various compositions, with a range of elastic moduli (1kPa to 25kPa) can be produced from these polypeptides, and the activity of the cell-binding and matrix metalloproteinase (MMP) sensitive domains was confirmed. Tris(hydroxymethyl phosphine) cross-linked RLP hydrogels were able to maintain their mechanical integrity as well as the viability of encapsulated primary human mesenchymal stem cells (MSCs). These results validate the promising properties of these RLP-based elastomeric biomaterials.
Introduction
Desired tissue function and regeneration rely on the dynamic interactions between cells and specific biophysical and biochemical cues,1-3 which facilitate critical cellular processes such as cell adhesion,4 migration,5 proliferation,6 differentiation7 and apoptosis.8 The ability to independently tailor specific cell-matrix interactions via tuning multiple properties of a scaffold, for example elastic micromechanical-transduction, integrin-mediated cell adhesion, matrix metalloproteinase-facilitated scaffold degradation, and sequestration of growth factors and cytokines, are critical parameters in regulating cell responses.9 Biomaterials derived from natural extracellular matrix (ECM) provide appealing biological activities and inherent biocompatibility; however, difficulties in selecting the mechanical properties, degradation rates, and nanostructures of these scaffolds narrow their potential application.10 In contrast, synthetic polymer-based materials have been widely employed,11-13 are relatively inexpensive, easily available and chemically versatile, although their lack of biological activity has been a limitation.14-17 The modular nature and sequence specificity of biosynthetically derived polypeptides provides opportunities to tune a range of materials properties and has fuelled the prominence of these materials in a variety of applications.18-22
An enormous variety of peptide modules have been easily incorporated into protein-based polymeric biomaterials, allowing the fine-tuning of the structural, mechanical, biological, and biodegradable properties for target cell niches.22-26 Recombinant elastin-like polypeptides, for example, have been widely employed over multiple decades as an elastomeric substrate for cell culture.25-35 More recently, polypeptides based on resilin have been employed with similar application in mind. Resilin is normally found in the specialized regions of cuticles of arthropods where fast, repetitive locomotion, and efficient energy storage are required.36 Early studies have revealed that natural resilin exhibits physicochemical properties distinct from the elastins, with high hydrophilicity and a low isoelectric point, while retaining useful mechanical features such as low stiffness, high resilience, large strain, reversible extensibility, long fatigue time, and excellent high-frequency-responsiveness.36-41 A variety of recombinant resilin-like polypeptides have been synthesized and in essentially all cases, the recombinant constructs mimic the mechanical properties of natural resilin,19, 42-51 suggesting their application in mechanically demanding tissue engineering applications.
The human vocal folds (VFs) are one of the most mechanically active tissues in the human body.52 They can sustain strain up to 30% at high frequencies of 100 to 1000 Hz and can reversibly recoil.53 Numerous deleterious environmental factors and pathological conditions can disrupt the natural pliability of this delicate tissue, resulting in a wide spectrum of voice disorders.54 While surgical techniques can be used to treat these conditions, they inevitably cause scarring, which inhibits voice production and compromises voice quality.54-57 Thus, RLP-based matrices could offer compelling alternatives in the treatment of vocal fold disorders.
Given the mechanical similarities between RLP hydrogels and vocal fold tissue (low modulus, high toughness, superior resilience, and high frequency responsiveness), we have sought to combine resilin’s excellent mechanical features with necessary biological functions to create a dynamic niche for engineering vocal fold tissues. In 2009, we first reported the design, synthesis, purification and characterization of a modular resilin-like polypeptide (RLP), containing 12 repetitive consensus resilin-like domains and additional sequences that confer biological activities such as cell adhesion, sensitivity to matrix metalloproteinases, and heparin binding.58 This RLP-based hydrogel was able to support the adhesion and proliferation of NIH 3T3 fibroblasts.58 A more thorough investigation of the mechanical properties of the RLP-based hydrogels was conducted via dynamic oscillatory rheology, tensile testing and torsional wave apparatus (TWA).59 Facile tuning of the protein concentrations and cross-linking ratios yielded materials with robust mechanical properties and high resilience, and Young’s moduli and high-frequency responses similar to those of vocal fold tissues.59 However, the original design did not permit variation of the relative concentrations of each biological domain.
We report here the production of multiple RLP-based constructs (RLP-X), each equipped with 12 repeats of the same 15 amino acid resilin consensus sequence, followed by a single and distinct biological domain (cell binding (RGD), RDG, MMP-sensitive, and heparin binding domains). Our results demonstrate that these constructs are easily expressed from E. coli expression hosts at high purity, maintain conformational similarity to other RLPs, are competent for MMP-based degradation and cell adhesion, and permit independent manipulation of the biological and mechanical properties of RLP-based hydrogels.
Materials and Methods
Materials
The plasmid DNA encoding RLP (flanked by Bam HI and Hind III) in pQE80 was purchased from Genscript Corporation (Piscataway, NJ). All customized double-stranded DNA oligos for generating other RLP constructs were purchased from Integrated DNA Technologies, Inc (Coralville, IA). Chemically competent cells of E. coli strain M15[pREP4] and Ni-NTA agarose resin were purchased from Qiagen (Valencia, CA). The trifunctional cross-linker tris(hydroxymethyl phosphine) (THP) was purchased from Strem Chemicals (Newburyport, MA). The recombinant human matrix metalloproteinase-1 (MMP-1) catalytic domain was purchased from Enzo Life Sciences (Farmingdale, NY). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO) and were used as received unless otherwise noted. Water was deionized and filtered through a NANOpure Diamond water purification system (Dubuque, IO).
Cloning of RLP Constructs
pQE80-RLP plasmid DNA (Genscript Corporation) was first digested at 37 °C for an hour with either Kpn I or Sal I prior to treatment with calf intestinal phosphatase to generate vectors for building other RLP constructs. Customized double-stranded DNA oligo inserts encoding RGD (the Arg-Gly-Asp cell-binding sequence), RDG (Arg-Asp-Gly negative control), MMP (matrix metalloproteinase) and HBD (heparin binding domain) domains (see below) were then ligated into the dephosphorylated pQE80-RLP vectors purified via agarose gel extraction by treatment with T4 DNA ligase for one hour. The final plasmids, with sequences confirmed via DNA sequencing (University of Delaware Sequencing and Genotyping Center), were designated as pQE80-RLP-RGD, pQE80-RLP-RDG, pQE80-RLP-MMP, and pQE80-RLP-HBD, respectively. Each DNA plasmid was then transformed into the E. coli M15 [pREP4] strain to generate the expression host employed in protein production.
Expression and Purification of RLPs
The RLP-X constructs were produced as described for our other RLPs in previous reports.58, 59 A detailed description of procedures can be found in the Supporting Information. Approximately 20-30mg of each RLP construct per liter of cell culture was obtained after dialysis and lyophilization.
General Characterization of RLPs
The successful expression of the polypeptides RLP, RLP-RGD, RLP-RDG, and RLP-MMP was indicated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of purified samples, with visualization via Coomassie blue staining. The purity of each RLP construct was also confirmed via high performance liquid chromatography (HPLC). Each polypeptide was purified via preparative HPLC on a Waters Symmetry C18 column; under a linear gradient from 95:5 to 5:95 of water/acetonitrile (containing 0.1% of trifluoroacetic acid) at 5mL/min over 65 min. The molecular weights of all RLP constructs were characterized via matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry, which was conducted by the Yale Keck Facility (Yale University, New Haven, CT). Amino acid analysis was performed by the Molecular Structure Facility at the University of California, Davis (Davis, CA) using a Hitachi L-800 sodium citrate-based amino acid analyzer (Tokyo, Japan) to determine the composition of each RLP polypeptide.
Characterization of Conformation of RLPs
Circular dichroic spectra (CD) of RLP solutions were recorded on a Jasco J-810 spectropolarimeter (Jason Inc, Easton, MD). FTIR-ATR spectra of RLP solution and hydrogels were collected using a Nexus 670 FTIR spectrometer (Thermo Nicolet, Madison, Wisconsin). The detailed description of methods and analysis can be found in the Supporting Information.
Hydrogel Formation and Oscillatory Rheology Experiments
Bulk oscillatory rheology experiments of RLP-based hydrogels were conducted at 37 °C on a stress-controlled rheometer (AR-G2), TA Instruments, (New Castle, DE) with a 20 mm diameter cone-on-plate geometry, 1 degree cone angle and at a 25 μm gap distance. Dynamic oscillatory time, frequency and strain sweeps were performed. 8mg RLP or mixed RLP compositions were dissolved in degassed, pH 7.4 PBS at a final concentration of 200mg/mL. Stock solutions of both the RLP and the cross-linker THP (100mg/mL) were chilled on ice before mixing in order to slow the reaction speed, preventing cross-linking during handling. 2.6μL THP stock solution was added to the 37.4μL RLP or mixed RLP stock solutions at a final solution volume of 40μL to yield 20wt% gels with a 1:1 cross-linking ratio that is indicated as the molar ratio of lysine residues to reactive hydroxymethylphosphine (HMP) groups. To ensure homogeneous mixing, the mixture was vortexed gently for less than 5 seconds after the addition of the THP, and then followed by careful pipetting onto the bottom plate of the rheometer for in situ rheological characterization. Strain sweeps were performed on samples from 0.01% to a maximum strain of 1000% to determine the limit of the linear viscoelastic regime (LVE). Rheological properties were examined by frequency sweep experiments (ω = 0.1 - 100 rad/s) at fixed strain amplitude of 1%. Experiments were repeated on 3 to 4 samples and representative data presented.
Enzymatic Degradation of Soluble RLP and RLP-MMP
The degradation of soluble RLP and RLP-MMP polypeptides in PBS was characterized in triplicate. 200 μL solutions of protein polymers at 1 mg/mL concentration were incubated with 100nM MMP-1 at 37°C in PBS pH 7.4. Samples (30 μL each) were collected at specific time points (0, 1, 3, 6, 12, 24, and 48 hours) and immediately mixed with 6x SDS sample loading buffer (6 μL). MMP-1 was inactivated by incubating with 1% (w/v) SDS, and 5mM β-mercaptoethanol and heating at 100°C for 10 mins before storing the samples at −20°C. Proteolytic degradation of protein polymers over time was monitored by electrophoresis analysis on a 15% SDS-PAGE gel, staining of the degradation fragments with Coomassie Brilliant Blue, and densitometry analysis via ImageJ (NIH software, Bethesda, MD).
2D Cell Culture and Attachment of hMSCs
Human bone marrow-derived mesencymal stem cells (hMSCs) (Lonza, Walkersville, MD, passage 3-6) were sub-cultured at a seeding density of 5000-6000 cells/cm2 on T-150 flasks (Corning, New York, NY) at 37°C with 5% CO2 in MSC maintenance media (Lonza, MD). Medium was refreshed every 3 days. Upon reaching ~80% confluence, cells were trypsinized, counted, centrifuged, and resuspended in the same maintenance medium at a desired cell density. RLP hydrogels were prepared by adding 50μL of solution into one chamber of an eight-well plate (Electron Microscope Science). The cross-linking reaction was allowed to occur for 30 mins at room temperature followed by sterilization under UV light (254 nm) for 20 mins. The RLP hydrogels were subsequently washed twice with 300uL Dulbecco’s phosphate buffered saline (DPBS) for 15 mins at room temperature before seeding cells on the hydrogel surface at a density of 104 cells per well. Cell attachment and morphology were assessed by Live/Dead assay and F-Actin/Vinculin staining. For Live/Dead assay, RLP hydrogels were stained with propidium iodide (1:2000 in DPBS, Invitrogen, Carlsbad, CA) and Syto-13 (1:1000 in DPBS, Invitrogen, Carlsbad, CA) for 5-10 mins at room temperature and subsequently imaged with multiphoton confocal microscopy (Zeiss 510 NLO, Thornwood, NY). For F-Actin/Vinculin staining, a commercially available actin cytoskeleton and focal adhesion staining kit (Millipore, Bedford, MA) was utilized to assess cell morphology and attachment. RLP hydrogels were rinsed in DPBS, fixed in 4% paraformaldehyde (Electron Microscope Science, Hatfield, PA) for 15 min, and permeabilized in 0.1% Triton X-100 (Fisher, Wilmington, DE) on ice for 5 min. Hydrogels were subsequently blocked with 3% heat inactivated bovine serum albumin (BSA; Jackson ImmunoResearch, West Grove, PA) in DPBS for 30 min, followed by a 1 hour incubation with mouse anti-vinculin monoclonal primary antibody (1:100 in 1% BSA; Millipore, Bedford, MA). RLP hydrogels were subsequently co-stained with AlexaFluor-488 goat anti-mouse IgG (Invitrogen; 1:100 in 1% BSA) and TRITC-conjugated phalloidin (Millipore; 1:200 in 1% BSA) for another hour. Finally, RLP hydrogels were counterstained with Draq-5 (1:1000 in DPBS; Axxora LLC, San Diego, CA) for 5 min before being visualized via multiphoton confocal microscopy (Zeiss 510 NLO, Thornwood, NY).
3D Encapsulation and Viability of hMSCs
The sub-culture conditions for the hMSCs in these experiments were identical to those described above for the 2D cell culture experiments. An aliquot of hMSC (P3-5) suspension was then added to UV-sterilized (20 mins) mixture of RLP and RLP-RGD (with THP) or a mixture of RLP-RGD and RLP-MMP (with THP) at room temperature at a 1:1 molar ratio of lysine residues and HMP functional groups at a final protein concentration of 200 mg/mL. The final density of the encapsulated cells was approximately 1 million per mL. After gently pipetting for 1-2 min at room temperature, 50μl of each RLP-cell mixture was then deposited onto an 8-well chamber and incubated at 37 °C for 30 min in a 5% CO2 incubator to allow for complete gelation, followed by adding 300μl media into the chamber. Media was changed every 15 mins at least 2 times to remove potential residual THP cross-linker. The constructs were cultured up to 7 days and the medium was changed every other day. To assess cell viability, cell-gel constructs were rinsed with DPBS, and stained with propidium iodide (Molecular Probes, 1:2000 in DPBS) and SYTO 13 (Molecular Probes, 1:1000 in DPBS) for 5-10 min at room temperature. The stained cells at day 1, 3 and 7 were visualized using multiphoton confocal microscopy (Zeiss 510 NLO, Thornwood, NY).
Results and Discussion
Vocal folds are the only tissue in humans that vibrates regularly at frequencies of 100-1000 Hz at amplitudes of about 1mm.57 Due to the stringent biomechanical properties of the human vocal fold, there have not been many investigations on creating artificial ECM scaffolds to treat vocal folds pathologies and disorders. Given our previous studies indicating the similarity of the mechanical properties (at both low and high frequencies) of RLP hydrogels and vocal fold tissues,58, 59 coupled with the ability of our RLP hydrogels to support the adhesion and proliferation of NIH 3T3 fibroblasts,58 we have sought to expand the versatility of these recombinant RLPs as a scaffold for engineering vocal fold therapeutics. Reported here is the production of multiple RLP-based constructs, in which each RLP construct contains the same repetitive resilin consensus motif but contains a different biologically active module (to impart cell adhesion, MMP-sensitivity, or protein sequestration) (Figure 1). It is thus possible, via simple mixing of the various RLP constructs, to independently modulate the concentrations of each biological domain in materials without altering the mechanical properties of the material.
Figure 1.
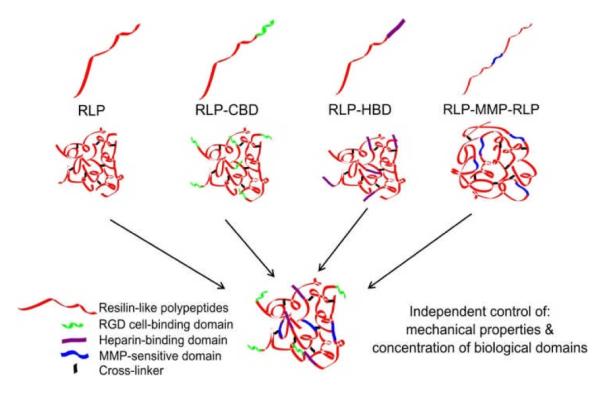
Schematic of the various RLP constructs.
Materials Design
The amino acid sequences of the RLPs produced in this study are listed in Table 1. Guided from our initial design of RLP12,58, 59 12 repeats of the pro-resilin putative consensus sequence (GGRPSDSYGAPGGGN) were employed; this sequence is derived from the first exon of the Drosophila melanogaster gene CG15920 and confers mechanical properties to resulting hydrogels that are similar to native resilin.43 We substituted tyrosine (Y) with phenylalanine (F) and methioine (M) to expand future options for cross-linking these materials through the incorporation of non-natural amino acids. Unlike the previous sequence where the cross-linking positions were randomly distributed along the chain, in this new design, five lysine-containing GGKGGKGGKGG sequences were incorporated and placed evenly along the polypeptide chain, with 45 intervening amino acids, in order to mimic the distance between the cross-links of natural resilin (30-60 amino acids between cross-links). The short amino-acid dyads between the various modules result from the restriction enzyme digestion sites employed for the cloning strategies. The integrin-binding domain GRGDSPG, derived from fibronectin, was employed, as it has been shown to promote adhesion of multiple types of endothelial cells, smooth muscle cells, and fibroblasts through αvβ3 and α5β1 integrins.30-32, 60 GRDGSPG is included as a scrambled negative control sequence for cell adhesion. The MMP-sensitive domain GPQGIWGQG, derived from human α(I) collagen, is also included in these constructs owing to not only its successful use in cell-mediated remodelling of polymeric biomaterials, but also because of its general relevance as a collagenase substrate to mediate cell invasion, migration, proliferation, and new tissue growth.61-63
Table 1.
Summary of the sequence and bioactive domains of RLPs
| Protein Name |
Bioactive Domain Sequence | MW (Da) |
|---|---|---|
| RLPa | N/A | 23127 |
| RLP-RGD | T-GRGDSPG-G | 23911 |
| RLP-RDG | T-GRDGSPG-G | 23911 |
| RLP-MMP | D-GGKGGKGGKGG- GPQGIWGQG-V |
24007 |
MRGSHHHHHH-GS-RS-GGKGGKGGKGG-(GGRPSDSF/MGAPGGGN)3-LQ-GGKGGKGGKG-LQ-(GGRPSDSF/MGAPGGGN)3-VD-GGKGGKGGKGG-VD-(GGRPSDSF/MGAPGGGN)3-EL-GGKGGKGGKGG-EL-(GGRPSDSF/MGAPGGGN)3-GGKGGKGGKGG-GT-KL
Expression and Purification of RLPs
The genetic construction of RLP constructs is shown schematically in Figure S1. The sequences of the recombinant plasmids were confirmed via DNA sequencing (Figure S2) and the plasmids (designated as pQE80-RLP, pQE80-RLP-RGD, pQE80-RLP-RDG, and pQE80-RLP-MMP) were then each transformed into E. coli M15 [pREP4] competent cells to generate relevant expression hosts. All RLP-X constructs were successfully expressed via traditional IPTG induction and easily purified under native conditions via Ni-NTA metal chelating affinity chromatography with a yield between 20-30mg/L. The purity, correct expression, and polypeptide composition of each construct were analyzed via HPLC, SDS-PAGE, MALDI-TOF mass spectroscopy and amino acid analysis. SDS-PAGE analysis of all RLP constructs (shown in Figure 2) shows a single polypeptide band of high purity (>97%) for each RLP construct at a position corresponding to a molecular mass of approximately 25kDa. This observation is consistent not only with theoretical molecular weight of each RLP construct listed in Table 2 but also with the observed single peak observed in the HPLC trace of each RLP construct (Figure S3). MALDI-TOF mass spectrometry further confirmed the expected molecular mass of each RLP construct (Table S1 and Figure S4) and the compositions of the polypeptides were determined to be within 5% of the expected amino acid compositions via amino acid analysis (Table S2). All RLPs exhibited heat stability with incubation at temperatures of 100 °C for 30 min, consistent with the properties of other reported recombinant RLPs and natural resilin.36, 44 RLP constructs isolated by this protocol were used directly for conformational analysis and for subsequent testing of mechanical and biological properties.
Figure 2.
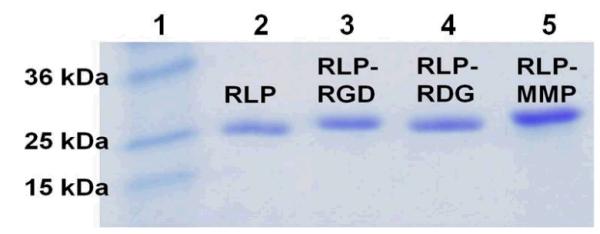
SDS-PAGE analysis of purified RLPs. Lane 1 is the protein standards; the identities of the samples in the other lanes are as marked in the figure.
Analysis of the Conformation of RLPs
CD spectroscopy was employed to probe the effects of specific amino acid substitutions in the putative resilin sequence and the addition of biological domains on the conformational properties of all RLP constructs. The CD spectra shown in Figure S5 were collected at a protein concentration of 10μM in PBS pH 7.4 at 37 °C in order to closely mimic physiological conditions. The spectra of all RLPs showed similar features, which are dominated by strong peaks near 198nm, indicating largely disordered random-coil structures. The positive peaks near 213nm and the minor negative peaks near 227nm indicate the contribution of type-II β-turns, which arise due to the presence of PG dyads in the sequence. These conformational features have also been observed for short resilin-mimetic peptides and resilin-like polypeptides, as well as natural resilin with similar sequences.40,46, 64 The amino acid substitution in the resilin sequence did not appreciably alter the conformational properties of RLPs, and the addition of the various biological domains did not cause the conformational behaviour of the various RLP constructs to differ significantly from one another, as is expected given the relatively small size of the biologically active amino acid sequences.
FTIR-ATR spectra were collected for uncross-linked RLP solutions and cross-linked RLP-based hydrogels of various compositions in D2O (at a concentration of RLP of 20wt%). Expanded amide-I regions of all the spectra were deconvoluted and spectra for the uncross-linked RLP and other hydrogel compositions, which are essentially identical to each other, are shown in Figure S6. The broad nature of the Amide-I band, centered at approximately 1650 cm−1, suggests contribution from multiple species. Deconvolution of the Amide-I regions yielded similar results for all samples, with a dominant peak at approximately 1645 cm−1 indicative of the random coil conformation with two minor peaks around 1672 cm−1 and 1618 cm−1, corresponding to those reported for β-turns and β-sheet structures respectively.58, 64, 65 Similarities in the contribution of disordered conformations in the uncross-linked (Figure S6, panel A) and cross-linked RLPs (Figure S6, panel B, C, D) are perfectly consistent with previous reports58 and indicate that the cross-linked RLP hydrogels exhibit significant chain flexibility and disorder. These secondary structures are reminiscent of conformers observed in other RLPs and other elastomeric proteins such as elastin, as well.46, 64, 66
Oscillatory Rheology
The vocal-fold mucosa, the major vibratory portion of the vocal fold, is directly involved in tissue repair, particularly in small-amplitude oscillations like phonation onset and offset.52 The viscoelastic mechanical properties of RLP-based hydrogels under shear are highly relevant as oscillation of the mucosa involves the propagation of a surface mucosal shear wave.53-55, 67 The mechanical properties of the RLP hydrogels were thus characterized via in situ dynamic oscillatory shear rheology using a cone-on-plate geometry; the hydrogels were cross-linked under mild aqueous conditions via the Mannich-type reaction of THP with primary amines of the lysine residues of the polypeptide.
We employed this Mannich-type cross-linking reaction because a similar cross-linking strategy employing [tris(hydroxymethyl phosphine)] propionic acid (THPP) has shown fast gelation and permitted direct cell encapsulation in the presence of fibroblasts without any deleterious effects.68 Protein solutions at various concentrations of RLP were pre-vortexed at room temperature with an appropriate amount of THP to ensure homogenous mixing, at an equimolar ratio of reactive lysines to HMP groups. Samples characterized using two cone-on-plates with different cone-degree angles resulted in similar gelation time, time to plateau, and final storage modulus, confirming the lack of slip during these dynamic oscillatory shear rheological experiments. Time sweep experiments were conducted in order to determine times of gelation; the data in Figure 3 showed the storage moduli (G’) at different protein concentrations for an RLP-only hydrogel. As shown in the data, all compositions were observed to gel rapidly after the temperature was increased to 37 °C, indicated by the considerable increase in viscosity and dynamic storage modulus (G’); all hydrogels reached a plateau modulus within 15 min with 50 to 100-fold differences between loss modulus (G’’), indicating the formation of elastic solid hydrogels. Increased protein concentrations decreased the time to obtain a plateau modulus and increased the final storage modulus, as expected, with G’ values ranging from 1000 Pa (5wt%) to 20 kPa (20wt%). This facile manipulation of shear moduli offers versatility in the choice of specific mechanical properties. Figure S7 shows frequency-sweep measurements of the various concentrations of the RLP hydrogels, illustrating that the G’ values remained constant during the entire time sweep (2h) even at the lowest protein concentration (5wt%), which demonstrates the anticipated stability and elastic-solid-like behaviour of these covalently cross-linked RLP hydrogels.
Figure 3.
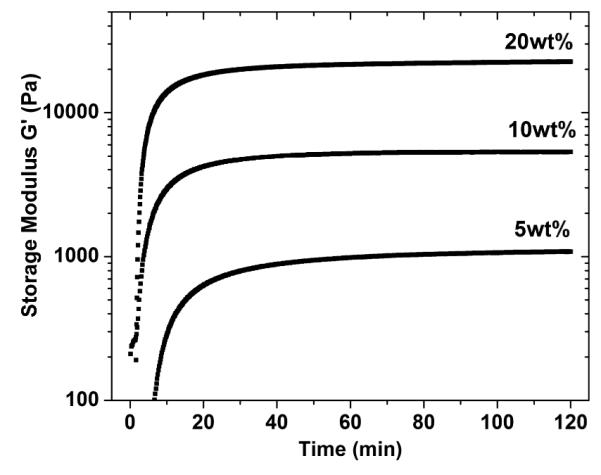
Dynamic oscillatory rheological characterization of hydrogels based on RLP alone at various protein concentrations. Time sweep of 5wt%, 10wt% and 20wt% RLP protein concentrations.
As shown in Figure 4, three different hydrogel scaffolds (comprising either RLP, 50% RLP with 50% RLP-RGD, or 50% RLP-RGD and 50% RLP-MMP) exhibited similar storage moduli at 10wt% and 20wt% polypeptide concentration (approximately 10±1.2 kPa or 20±3.5 kPa, respectively). These data illustrate that the concentration of biologically active domains in a hydrogel can be decoupled from the mechanical properties of the hydrogel simply by mixing different RLP constructs. Swelling ratio and water content measurements, shown in Figure S8, corroborate these observations. Both the swelling ratio (approximately 5) and water content (around 80%) are similar to each other despite the differences in hydrogel compositions and are also consistent with natural resilin, other RLP-based hydrogels and our previous reported RLP12 hydrogels.19, 43, 50, 58, 59, 69 The independent control of the content of biologically active domains over mechanical properties provides important opportunities for optimizing the compositions of artificial extracellular matrix.
Figure 4.
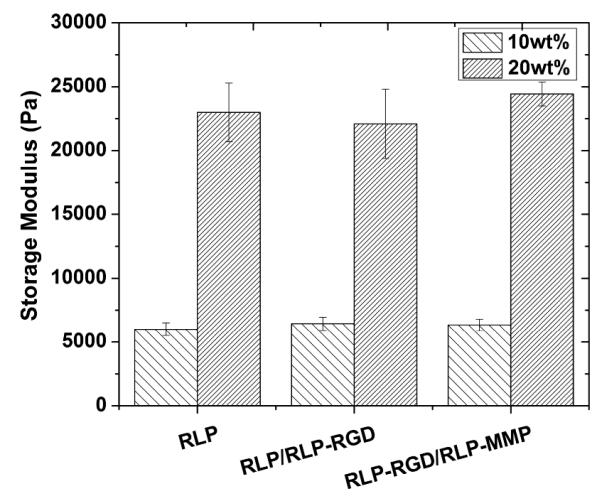
Oscillatory storage moduli of hydrogels formed from simple RLP-based or mixed RLP-based constructs at 10wt% and 20wt% final protein concentration at 1:1 (lysine : HMP) cross-linking ratio with error reported as the standard deviation of the average of 5 measurements.
Enzymatic Degradation of Soluble RLP and RLP-MMP
In order to mimic the naturally degradable ECM microenvironment, synthetic polymeric biomaterials have been equipped with short, enzyme-sensitive peptides and have demonstrated an increase in enzymatic sensitivity, resulting in the improvement of outgrowth, proliferation and migration of cells.1, 62, 63, 70 Biosynthetic strategies enable these short peptide sequences to be selectively localized in order to allow regional remodeling of the biomaterials in response to proteinases that are secreted by cells encapsulated within the scaffolds. The MMP-sensitive substrate was chosen because of its previous successful implementation in cell-mediated matrix turn-over of polymeric biomaterials, and because of its sensitivity to degradation by collagenases to mediate cell invasion, proliferation, migration, and tissue remodeling; indeed, collagens are main proteins in the vocal fold lamina propria. The degradation properties of these soluble RLP and RLP-MMP constructs were characterized in the presence of MMP-1 under physiological conditions. The high concentration of MMP-1 (100nM) used in this experiment was chosen because of its similarity to concentrations in previously reported MMP degradation experiments.61-63, 71 Gel electrophoresis was employed to monitor the degradation fragments of the RLP constructs within a range of time points and a representative RLP and RLP-MMP degradation profile is shown in Figure 5. As shown in the figure, the RLP construct lacking the MMP-sensitive domain remained intact during the entire incubation with MMP-1 (Figure 5A), however, the RLP-MMP construct equipped with the MMP-sensitive sequence degraded through the course of 48-hour reaction time (Figure 5B). ImageJ was used to perform densitometry analysis of the gels; the degradation profiles determined from these experiments are shown in Figure 5C. These results indicate that essentially 100% of the RLP remained intact while less than 10% of the RLP-MMP remained intact; the approximate half-life suggested from inspection of the densitometry analysis for the RLP-MMP is approximately 20 hours. Taken together, these results suggest the capacity to tune the degradation rate of a desired scaffold simply by mixing RLP-MMP with other RLPs. Experiments of degradation on hydrogels based on RLP-MMP compositions are underway.
Figure 5.
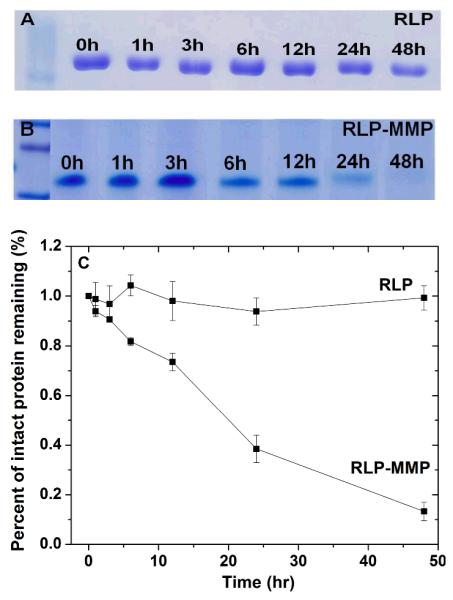
SDS-PAGE analysis of MMP degradation time course of (A) soluble RLP, (B) RLP-MMP, and (C) SDS-PAGE densitometry analysis of relative degradation rates of soluble RLP and RLP-MMP upon exposure to 100nM MMP-1 incubated at 37 °C in pH 7.4 PBS.
Attachment of hMSCs on RLP-based Hydrogel Surfaces
Adult human mesenchymal stem cells (hMSCs) can expand as undifferentiated cells in culture for more than 50 passages, indicating their proliferative capacity, and they also maintain their ability to selectively undergo osteogenic, chondrogenic, and adipogenic differentiation processes.72-76 Because primary cells from vocal fold tissue are not readily available and would not be a clinically viable selection, human bone-marrow derived MSCs were employed in cell studies; these cells are appropriate for future use in vocal fold cell-gel therapies. The ability of the cross-linked RGD-containing RLP-based hydrogels to support the attachment of hMSCs was investigated via seeding cells on the surface of 20wt% hydrogels with a 50% RLP and 50% RLP-RGD composition. Cells cultured on the surface of this hydrogel exhibited elongated morphology within 24 hours. The cells stained green via Live/Dead staining (with essentially no cells stained red), indicating their excellent viability (data not shown). Detailed cell morphology and the degree of cell spreading were assessed via F-Actin/vinculin co-staining, shown in Figure 6. After 24 hours, hMSCs were well-spread on 50% RLP and 50% RLP-RGD-based hydrogel and displayed an elongated cell morphology. Cell nuclei, stained by Draq 5 (blue, Figure 6A), indicated the relative location of hMSCs. Intense green spots (Figure 6B) are representative of focal adhesion complexes along the long axis of cell body. F-actin stress fibers (red, Figure 6C) along the edge of the cell body were also clearly visualized. In contrast, hMSCs seeded on the surface of a 20wt% RLP-only cross-linked hydrogel displayed a rounded morphology due to the lack of RGD integrin-binding motifs (Figure S9). Together, these data confirm that the RGD domain in the RLP-RGD hydrogel was able to support the attachment and spreading of hMSCs. Further comparisons of cell behavior on hydrogels of various RGD-content as well as on RDG-containing hydrogels are underway.
Figure 6.
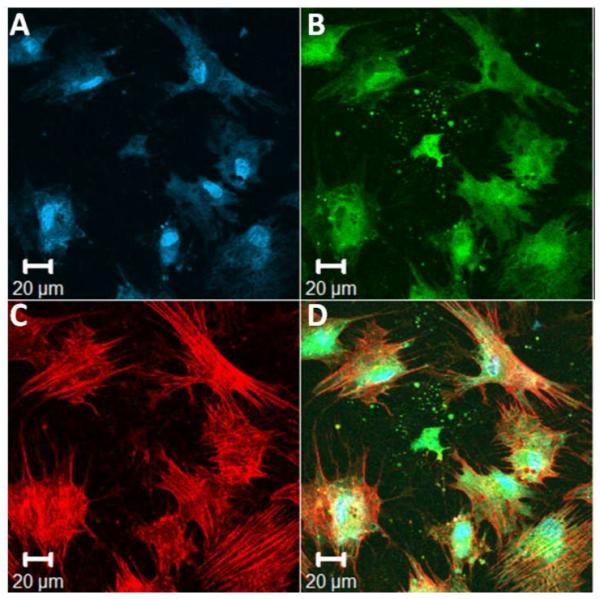
Representative images of hMSCs stained, after 24 hours, for visualization of nuclei, vinculin, and actin cytoskeleton, on the surface of a 20wt% RLP hydrogel with a composition of 50% RLP and 50% RLP-RGD (and with a 1:1 (lysine : HMP) cross-linking ratio). (A) Cell nuclei counterstained by Draq5 (blue); (B) focal adhesion sites visualized (green) by treatment with anti-vinculin and a FITC-labeled secondary antibody; (C) F-actin filaments visualized (red) by treatment with TRITC-phalloidin; and (D) the merged image of the triply stained hydrogels (Draq5, vinculin and TRITC-phalloidin).
3D Encapsulation of hMSCs in Cross-linked RLP-based Hydrogels
Initial cell encapsulation was investigated in the 50% RLP and 50% RLP-RGD hydrogels (the composition that supported cell adhesion in the 2D cell culture experiments) in order to confirm the cytocompatibility of the RLPs with bone-marrow derived hMSCs in the presence of the cross-linker THP, as well as to validate the mechanical integrity of cell-gel constructs over various periods of time in cell culture. The hMSC pellet was resuspended at room temperature with RLP solutions (and THP at a 1:1 molar ratio of lysine residues to HMP functional groups). The mixtures quickly formed hydrogels at 37 °C (within several minutes after incubation), consistent with the oscillatory rheology experiments. The cell-gel constructs were then cultured at 37 °C up to 7 days and cell viability/cytotoxicity was characterized via live/dead assay. The fluorescence images at day 1, 3 and 7 (visualized in situ at room temperature via multiphoton confocal microscopy) are shown in Figure 7. The data demonstrated the even distribution of hMSCs throughout the three dimensional gel as indicated by the similarities in each XY panel and Z direction at various times of cell culture. Although a small number of cells fluoresced red in these 3D cell culture experiments, indicating cell death, the majority of cells at either day 1, 3 or 7 fluoresced green, suggesting their viability in the presence of the cross-linker THP. These cell-gel constructs were also able to maintain their mechanical integrity and cell viability for an extended time of at least 21 days (data not shown). Given the fact that these hydrogels lacked the MMP-sensitive domain, it is consistent that hMSCs were not able to spread in this hydrogel composition. The potential for hMSCs to develop focal adhesions and elongated morphologies in 3D culture could be improved via tuning of the RGD density and introducing MMP-sensitive domains; such optimization is underway.
Figure 7.
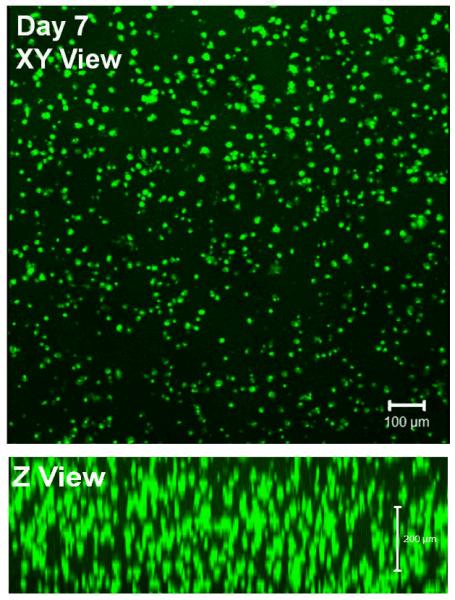
Day 7 Live/dead assay of 3D encapsulated hMSCs in 20wt% 1:1 (lysine : HMP) cross-linking ratio hydrogel based on 50% RLP and 50% RLP-RGD in gel composition.
Conclusions
These modular RLP constructs permit facile and independent tuning of the concentration of biologically active domains and mechanical properties of polypeptide hydrogels, and offer substantial opportunities for fabricating a regenerative scaffold that can be systematically tailored. Bacterial expression of all RLP constructs provided sufficient yield of protein with high purity. Both CD and ATR-FTIR spectra indicated the maintenance of largely random coil conformations derived from the repetitive resilin-like motifs in the polypeptides. Rheological characterization demonstrated that hydrogels can be rapidly formed upon mixing with the cross-linker THP and that various compositions of RLP hydrogels yield comparable mechanical properties independent of the identity and concentration of the biologically active domains in the hydrogel. SDS-PAGE and densitometry analysis confirmed that an RLP equipped with MMP-sensitive domains is susceptible to MMP-1 enzymatic degradation within 48 hours, in contrast to the complete stability of the RLP construct lacking the MMP-sensitive domain. An RGD-containing RLP hydrogel substrate supported the adhesion and spreading of hMSCs. 3D encapsulation of primary hMSCs in mixed-RLP constructs was possible; the integrity of hydrogels and viability of encapsulated cells were both maintained over extended time periods. Optimization of RGD density and MMP-degradation in these 3D RLP-based hydrogel constructs, in order to tune the activities of hMSCs for injectable therapies, is in progress.
Supplementary Material
Acknowledgments
We would like to acknowledge support by grants from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (P20-RR017716 (K.L.K.) and P20-RR015588 for instrument resources). We also acknowledge the National Institute on Deafness and Other Communication Disorders (NIDCD, RO1 DC008965 to X.J.). MALDI-TOF experiments were conducted at the Yale University Keck Facility and amino acid analyses at the UC Davis’ molecular structure facility. Wenqiong Tang is acknowledged for assistance with the collection and analysis of ATR-FTIR data.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Hubbell JA. Bio-Technology. 1995;13:565–576. doi: 10.1038/nbt0695-565. [DOI] [PubMed] [Google Scholar]
- 3.Place ES, Evans ND, Stevens MM. Nat. Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 4.Zamir E, Geiger B. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 5.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 6.Boudreau N, Sympson CJ, Werb Z, Bissell MJ. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibue T, Weinberg RA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mooney D, Hansen L, Vacanti J, Langer R, Farmer S, Ingber D. J. Cell. Physiol. 1992;151:497–505. doi: 10.1002/jcp.1041510308. [DOI] [PubMed] [Google Scholar]
- 9.Straley KS, Heilshorn SC. Soft Matter. 2009;5:114–124. [Google Scholar]
- 10.Amiel GE, Komura M, Shapira O, Yoo JJ, Yazdani S, Berry J, Kaushal S, Bischoff J, Atala A, Soker S. Tissue Eng. 2006;12:2355–2365. doi: 10.1089/ten.2006.12.2355. [DOI] [PubMed] [Google Scholar]
- 11.Place ES, George JH, Williams CK, Stevens MM. Chem. Soc. Rev. 2009;38:1139–1151. doi: 10.1039/b811392k. [DOI] [PubMed] [Google Scholar]
- 12.Lin CC, Anseth KS. Pharm. Res. 2009;26:631–643. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel PN, Gobin AS, West JL, Patrick CW. Tissue Eng. 2005;11:1498–1505. doi: 10.1089/ten.2005.11.1498. [DOI] [PubMed] [Google Scholar]
- 14.Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH. Biomaterials. 2005;26:1211–1218. doi: 10.1016/j.biomaterials.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Bryant SJ, Anseth KS. J. Biomed. Mater. Res. Part A. 2003;64A:70–79. doi: 10.1002/jbm.a.10319. [DOI] [PubMed] [Google Scholar]
- 16.Salinas CN, Cole BB, Kasko AM, Anseth KS. Tissue Eng. 2007;13:1025–1034. doi: 10.1089/ten.2006.0126. [DOI] [PubMed] [Google Scholar]
- 17.Babensee JE, Anderson JM, McIntire LV, Mikos AG. Adv. Drug Deliv. Rev. 1998;33:111–139. doi: 10.1016/s0169-409x(98)00023-4. [DOI] [PubMed] [Google Scholar]
- 18.Kiick KL. Polym. Rev. 2007;47:1–7. [Google Scholar]
- 19.Lv S, Dudek DM, Cao Y, Balamurali MM, Gosline J, Li HB. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 20.Li LQ, Charati MB, Kiick KL. Polym. Chem. 2010;1:1160–1170. doi: 10.1039/b9py00346k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagapudi K, Brinkman WT, Thomas BS, Wright ER, Conticello VP, Chaikof EL. Cell Trans. 2003;12:170–171. [Google Scholar]
- 22.Maskarinec SA, Tirrell DA. Curr. Opin. Biotechnol. 2005;16:422–426. doi: 10.1016/j.copbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Wright ER, Conticello VP. Adv. Drug Deliv. Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 24.Nagapudi K, Brinkman WT, Leisen J, Thomas BS, Wright ER, Haller C, Wu XY, Apkarian RP, Conticello VP, Chaikof EL. Macromolecules. 2005;38:345–354. [Google Scholar]
- 25.Daamen WF, Veerkamp JH, van Hest JCM, van Kuppevelt TH. Biomaterials. 2007;28:4378–4398. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 26.Simnick AJ, Lim DW, Chow D, Chilkoti A. Polym. Rev. 2007;47:121–154. [Google Scholar]
- 27.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2008;9:222–230. doi: 10.1021/bm7007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyle S, Aggeli A, Ingham E, McPherson MJ. Trends Biotechnol. 2009;27:423–433. doi: 10.1016/j.tibtech.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellingham CM, Lillie MA, Gosline JM, Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, Keeley FW. Biopolymers. 2003;70:445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 30.Heilshorn SC, DiZio KA, Welsh ER, Tirrell DA. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu JC, Heilshorn SC, Tirrell DA. Biomacromolecules. 2004;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 32.Heilshorn SC, Liu JC, Tirrell DA. Biomacromolecules. 2005;6:318–323. doi: 10.1021/bm049627q. [DOI] [PubMed] [Google Scholar]
- 33.Cirulis JT, Keeley FW, James DF. J Rheol. 2009;53:1215–1228. [Google Scholar]
- 34.Baldock C, Oberhauser AF, Ma LA, Lammie D, Siegler V, Mithieux SM, Tu YD, Chow JYH, Suleman F, Malfois M, Rogers S, Guo LA, Irving TC, Wess TJ, Weiss AS. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4322–4327. doi: 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo GC, Baldock C, Tuukkanen A, Roessle M, Dyksterhuis LB, Wise SG, Matthews J, Mithieux SM, Weiss AS. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2878–2883. doi: 10.1073/pnas.1111615108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis-Fogh T, Andersen SO. Advances in Insect Physiology. Vol. 2. Academic Press; London: 1964. pp. 1–65. [Google Scholar]
- 37.Weis-Fogh T. J. Mol. Biol. 1961;3:648–667. [Google Scholar]
- 38.Weis-Fogh T. J. Mol. Biol. 1961;3:520–531. [Google Scholar]
- 39.Andersen SO. Biochim. Biophys. Acta. 1963;69:249–262. doi: 10.1016/0006-3002(63)91258-7. [DOI] [PubMed] [Google Scholar]
- 40.Elliott GF, Huxley AF, Weis-Fogh T. J. Mol. Biol. 1965;13:791–795. [Google Scholar]
- 41.Andersen SO. In: Elastomeric Proteins. Editon edn T. Shewry PR, A.S., Bailey AJ, editors. Cambridge University Press; Cambridge: 2003. pp. 259–278. [Google Scholar]
- 42.Ardell DH, Andersen SO. Insect Biochem. Mol. Biol. 2001;31:965–970. doi: 10.1016/s0965-1748(01)00044-3. [DOI] [PubMed] [Google Scholar]
- 43.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DCC, Merritt DJ, Dixon NE. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 44.Lyons RE, Lesieur E, Kim M, Wong DCC, Huson MG, Nairn KM, Brownlee AG, Pearson RD, Elvin CM. Protein Eng. Des. Sel. 2007;20:25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- 45.Kim M, Elvin C, Brownlee A, Lyons R. Protein Expr. Purif. 2007;52:230–236. doi: 10.1016/j.pep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Bochicchio B, Pepe A, Tamburro AM. Chirality. 2008;20:985–994. doi: 10.1002/chir.20541. [DOI] [PubMed] [Google Scholar]
- 47.Nairn KM, Lyons RE, Mulder RJ, Mudie ST, Cookson DJ, Lesieur E, Kim M, Lau D, Scholes FH, Elvin CM. Biophys. J. 2008;95:3358–3365. doi: 10.1529/biophysj.107.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyons RE, Nairn KM, Huson MG, Kim M, Dumsday G, Elvin CM. Biomacromolecules. 2009;10:3009–3014. doi: 10.1021/bm900601h. [DOI] [PubMed] [Google Scholar]
- 49.Qin GK, Lapidot S, Numata K, Hu X, Meirovitch S, Dekel M, Podoler I, Shoseyov O, Kaplan DL. Biomacromolecules. 2009;10:3227–3234. doi: 10.1021/bm900735g. [DOI] [PubMed] [Google Scholar]
- 50.Qin GK, Rivkin A, Lapidot S, Hu X, Preis I, Arinus SB, Dgany O, Shoseyov O, Kaplan DL. Biomaterials. 2011;32:9231–9243. doi: 10.1016/j.biomaterials.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bracalello A, Santopietro V, Vassalli M, Marletta G, Del Gaudio R, Bochicchio B, Pepe A. Biomacromolecules. 2011;12:2957–2965. doi: 10.1021/bm2005388. [DOI] [PubMed] [Google Scholar]
- 52.Titze IR. J. Acoust. Soc. Am. 1989;85:901–906. doi: 10.1121/1.397562. [DOI] [PubMed] [Google Scholar]
- 53.Hammond TH, Gray SD, Butler J, Zhou RX, Hammond E. Otolaryngol. Head Neck Surg. 1998;119:314–322. doi: 10.1016/S0194-5998(98)70071-3. [DOI] [PubMed] [Google Scholar]
- 54.Kriesel KJ, Thibeault SL, Chan RW, Suzuki T, VanGroll PJ, Bless DM, Ford CN. Ann. Otol. Rhinol. Laryngol. 2002;111:884–889. doi: 10.1177/000348940211101004. [DOI] [PubMed] [Google Scholar]
- 55.Zeitels SM, Hillman RE, Mauri M, Desloge R, Doyle PB. Ann. Otol. Rhinol. Laryngol. 2002;111:21–40. doi: 10.1177/0003489402111s1203. [DOI] [PubMed] [Google Scholar]
- 56.Benninger MS, Alessi D, Archer S, Bastian R, Ford C, Koufman J, Sataloff RT, Spiegel JR, Woo P. Otolaryngol. Head Neck Surg. 1996;115:474–482. doi: 10.1177/019459989611500521. [DOI] [PubMed] [Google Scholar]
- 57.Kutty JK, Webb K. J. Biomater. Sci.-Polym. Ed. 2009;20:737–756. doi: 10.1163/156856209X426763. [DOI] [PubMed] [Google Scholar]
- 58.Charati MB, Ifkovits JL, Burdick JA, Linhardt JG, Kiick KL. Soft Matter. 2009;5:3412–3416. doi: 10.1039/b910980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li LQ, Teller S, Clifton RJ, Jia XQ, Kiick KL. Biomacromolecules. 2011;12:2302–2310. doi: 10.1021/bm200373p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 61.Nagase H, Fields GB. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 62.Lutolf MR, Weber FE, Schmoekel HG, Schense JC, Kohler T, Muller R, Hubbell JA. Nat. Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 63.Rizzi SC, Hubbell JA. Biomacromolecules. 2005;6:1226–1238. doi: 10.1021/bm049614c. [DOI] [PubMed] [Google Scholar]
- 64.Tamburro AM, Panariello S, Santopietro V, Bracalello A, Bochicchio B, Pepe A. ChemBioChem. 2009;11:83–93. doi: 10.1002/cbic.200900460. [DOI] [PubMed] [Google Scholar]
- 65.Grieshaber SE, Nie T, Yan CQ, Zhong S, Teller SS, Clifton RJ, Pochan DJ, Kiick KL, Jia XQ. Macromol. Chem. Phys. 2011;212:229–239. doi: 10.1002/macp.201000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamburro AM, Guantieri V, Pandolfo L, Scopa A. Biopolymers. 1990;29:855–870. doi: 10.1002/bip.360290419. [DOI] [PubMed] [Google Scholar]
- 67.Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Philos. Trans. R. Soc. Lond. Ser. B-Biol. Sci. 2002;357:121–132. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463–1470. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bailey K, Weis-Fogh T. Biochim. Biophys. Acta. 1961;48:452–459. doi: 10.1016/0006-3002(61)90043-9. [DOI] [PubMed] [Google Scholar]
- 70.Rizzi SC, Ehrbar M, Halstenberg S, Raeber GP, Schmoekel HG, Hagenmuller H, Muller R, Weber FE, Hubbell JA. Biomacromolecules. 2006;7:3019–3029. doi: 10.1021/bm060504a. [DOI] [PubMed] [Google Scholar]
- 71.Raeber GP, Lutolf MP, Hubbell JA. Acta Biomater. 2007;3:615–629. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 72.Burdick JA, Vunjak-Novakovic G. Tissue Eng. Part A. 2009;15:205–219. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noth U, Steinert AF, Tuan RS. Nat. Clin. Pract. Rheumatol. 2008;4:371–380. doi: 10.1038/ncprheum0816. [DOI] [PubMed] [Google Scholar]
- 74.Lee K, Chan CK, Patil N, Goodman SB. J. Biomed. Mater. Res. Part B. 2009;89B:252–263. doi: 10.1002/jbm.b.31199. [DOI] [PubMed] [Google Scholar]
- 75.Rada T, Reis RL, Gomes ME. Tissue Eng. Part B-Rev. 2009;15:113–125. doi: 10.1089/ten.teb.2008.0423. [DOI] [PubMed] [Google Scholar]
- 76.Sterodimas A, De Faria J, Correa WE, Pitanguy I. Ann. Plast. Surg. 2009;62:97–103. doi: 10.1097/SAP.0b013e3181788ec9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


