Abstract
The design of sensors to monitor the activity state of specific protein kinases is challenging due to the complexity of eukaryotic kinomes. Here we describe a peptide-based photaffinity probe that specifically labels the active conformation of the Abl tyrosine kinase.
Keywords: activity-based protein profiling, enzymes, photoaffinity labeling, protein kinase, phosphopeptides
Protein kinases are key players in intracellular signaling pathways that control all aspects of cell behavior. Because protein kinases often promote cell survival and growth, aberrant kinase activity frequently contributes to cancer, and kinase inhibitors have become important anti-cancer drugs.[1] Accordingly, much effort has been expended towards the design of protein or peptide-based reporters to probe the activity of specific protein kinases in complex systems (cell lysates, intact cells, tissues or tumors).[2] However, these approaches require that the substrate must be exquisitely specific for the kinase of interest, a major challenge given that there are over 500 human protein kinases that could potentially provide “off-target” activity.[3] An alternative to the use of substrates to profile enzyme activity in crude systems is to use affinity probes that bind to the enzyme in an activity-dependent manner. Because such activity-based probes (ABPs) covalently label their targets, binding to multiple enzymes is actually advantageous, as it can facilitate profiling of entire classes of molecules. ABPs have been developed and applied most extensively for hydrolytic enzymes, including lipid esterases, proteases, and phosphatases,[4] but have been lacking for protein kinases. We describe here a photoaffinity probe based on a consensus peptide substrate that specifically labels the active conformation of the oncogenic protein tyrosine kinase Abl. In some contexts this probe behaves like an ABP, reporting on the activation state of the kinase. Our approach should allow for the general design of conformation-specific probes for many protein kinases.
A nearly universal feature of kinase activation involves conformational reorganization of a region adjacent to the catalytic cleft called the activation loop.[5] The activation loop constitutes a portion of the protein substrate binding site, suggesting that probes based on a peptide substrate are likely to be specific for the active conformation. To test this hypothesis, we generated a probe targeting the non-receptor tyrosine kinase Abl (Scheme 1), an established oncology target deregulated in chronic myelogenous leukemia through a gene rearrangement leading to a constitutively active fusion protein (BCR-Abl). We based our probe on a consensus substrate (ABLtide, Figure 1) generated through peptide library screening.[6] ABPs typically consist of a reactive group that targets the enzyme linked to a reporter, either a fluorescent tag or affinity handle.[7] To allow covalent targeting of the kinase, we incorporated a photocrosslinking phenyl azide group (4-azidophenylalaine, Z) into the peptide sequence. We chose to insert the Z residue at the P+2 position (two residues downstream of the phosphorylation site), as X-ray crystal structures of Abl-peptide complexes[8] suggested that substitution at that position would not affect peptide binding. To facilitate detection, we incorporated a biotin moiety at the C-terminus to generate the probe (ABL-Z, Figure 1).
Scheme 1.
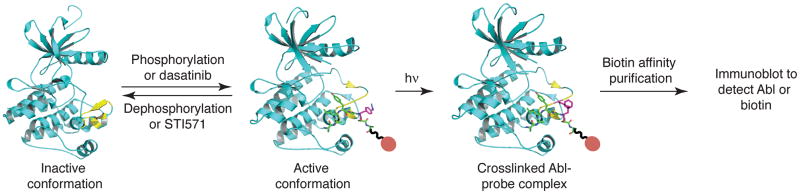
Labeling and detection of the Abl tyrosine kinase. The active conformation of Abl with an intact peptide substrate binding site can be induced either by phosphorylation of the kinase or by binding of specific inhibitors such as dasatinib. Conversely, dephosphorylation of Abl or binding of the inhibitor STI571 induces an inactive conformation that is incapable of binding peptide substrates. A biotin-conjugated peptide photoaffinity label binds to the substrate-binding cleft of Abl in its active conformation. Irradiation of the complex triggers photocrosslinking, and the extent of probe incorporation correlates with the activity state of the kinase.
Figure 1.
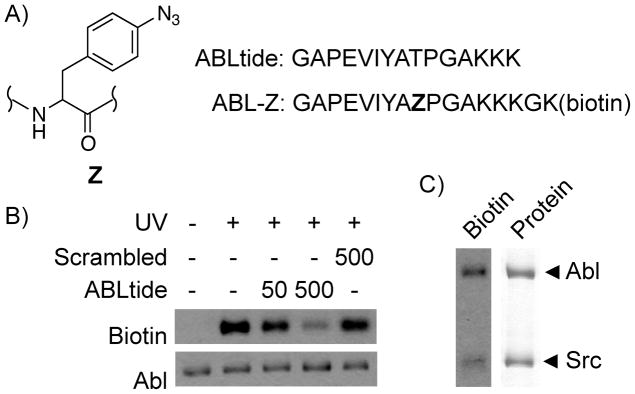
Peptide-based photoaffinity probe for Abl kinase. A, Probe and consensus peptide sequence. B, Labeling of purified Abl with 5 μM ABL-Z in the presence of indicated peptides (μM concentrations are shown). C, ABL-Z preferentially labels Abl in the presence of equimolar Src kinase. Left lane, biotin signal detected using streptavidin-HRP; right lane, Coomassie Blue stain showing total protein.
Incubation of purified recombinant Abl with ABL-Z followed by UV irradiation resulted in covalent incorporation of biotin into the kinase as judged by detection with streptavidin-horseradish peroxidase (HRP) following SDS-PAGE and transfer to a membrane (Figure 1). Labeling was UV-dependent, and could be competed with increasing concentrations of unlabeled peptide but not a scrambled peptide control. When labeling was carried out in the presence of equal amounts of another tyrosine kinase, Src, label was incorporated predominantly into Abl and to a lesser degree into the off-target kinase. These results suggest that ABL-Z crosslinks to Abl specifically through binding to the active site cleft. UV-dependent labeling of Abl also occurred in a crude cell lysate (Figure S1). In this context detection of label incorporation was accomplished by immunoblotting for the presence of Abl among proteins isolated using avidin resin. Biotin affinity isolation was necessary due to a high degree of labeling of other proteins, presumably due to excess label in free solution reacting through random collision. However, in crude lysates we could detect Abl with high sensitivity (<24 fmol kinase).
To establish whether ABL-Z could be used in a manner similar to an ABP, we first performed experiments with recombinant Abl in which we manipulated its activation state. Abl purified from mammalian cells exhibited a basal level of activity as detected by ABLtide phosphorylation (Figure 2). Abl activity is enhanced by phosphorylation at multiple sites within the kinase,[9] and lambda phosphatase (λPP) treatment reduces its activity. We also generated a triple point mutant (G2A/P242E/P249E, or GPP) that disrupts intramolecular interactions that restrain Abl kinase activity,[10] resulting in constitutively higher activity than wild-type (WT) Abl. These three forms of Abl were subjected to labeling with ABL-Z. The extent of labeling increased with increasing catalytic activity, indicating that in this context ABL-Z behaves as expected for an ABP (Figure 2).
Figure 2.
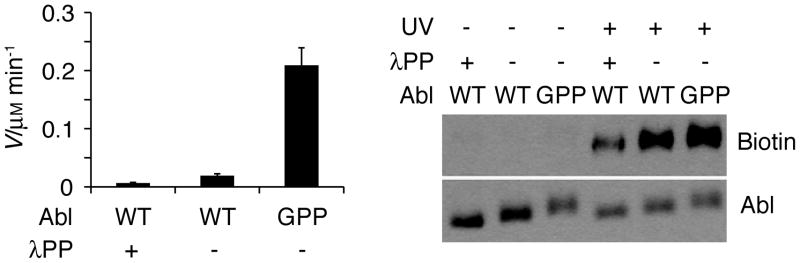
ABL-Z probe accessibility correlates with Abl kinase activity. Left panel, peptide kinase activity of either WT or constitutively active mutant (GPP) Abl is shown subsequent to mock treatment or dephosphorylation with λPP. Right panel, photoaffinity labeling of the same forms of Abl with ABL-Z.
As another means to assess the specificity of our probe for the active form of Abl, we used two clinical inhibitors known to bind specific conformations of the kinase. STI-571 (imatinib/Gleevec) induces an inactive conformation of Abl in which the active site cleft is occluded through an intramolecular pseudosubstrate interaction.[11] In contrast, the dual Src/Abl inhibitor dasatinib binds to the active conformation of Abl.[12] In these experiments WT or mutant Abl was expressed in mammalian cells and labeling was performed in crude cell lysates. As anticipated, STI-571 almost completely blocked labeling by ABL-Z, while dasatinib actually increased the extent of labeling (Figure 3). We confirmed that both compound inhibited Abl when purified from the same cell lysates (Figure S2). These results provide further evidence that ABL-Z specifically targets the active conformation of Abl.
Figure 3.
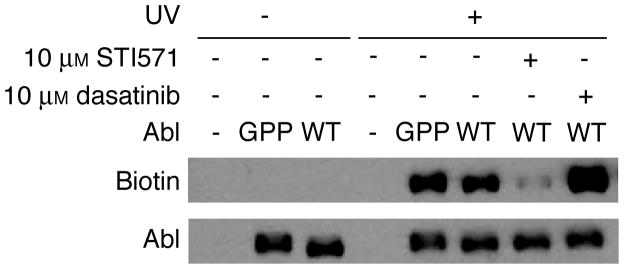
Labeling of Abl in the presence of inhibitors that bind to distinct conformations of the kinase. Crude lysates of HEK293T cells expressing WT or mutant Abl were treated with 5 μM ABL-Z in the presence of the indicated concentrations of inhibitor prior to irradiation and analysis of probe crosslinking.
These results illustrate the use of peptide substrate-based probes to assess the activity state of protein kinases in crude systems. Protein kinase affinity labels have previously been generated by derivatizing ATP or ATP-competitive small molecule inhibitors.[13] In addition, affinity resins consisting of immobilized inhibitors have been used to profile kinase expression in cell lysates.[14] These approaches have been valuable for profiling kinase expression and for assessing inhibitor selectivity and potency in situ. However, because ATP or inhibitor affinity need not correlate with the activity state of a kinase, these probes are not necessarily suitable for activity-based profiling. In contrast, because kinase activation can involve rearrangement of the peptide binding site, peptide substrate-based probes should frequently bind in an activity-dependent manner.
While the current probe was designed to target Abl, alterations in the peptide sequence should enable tuning the reactivity of such probes to target essentially any kinase of interest. It is unlikely that a short peptide sequence will be absolutely specific for a single kinase. However, cross-reactivity with other kinases does not limit use of such probes, since specific targets of interest can be detected by immunoblotting or by mass spectrometry following affinity isolation of bound proteins. In this way a limited number of probes can potentially be used to detect activity of a large number of kinases. While detection of endogenous kinase using the current probe is hampered by low affinity for Abl leading to low crosslinking efficiency (<1.3%), increasing peptide affinity through screening of peptide libraries[15] or the use of bivalent inhibitors[16] should allow detection of endogenous kinases in an activity-dependent manner. In addition, appending cell penetrating peptide sequences would allow their use in intact cells. In this way peptide substrate based probes should allow one to establish the repertoire of active kinases present in a cell line or tumor, and may thereby suggest new therapeutic targets.
Experimental Section
Experimental details are provided in the Supporting Information.
Supplementary Material
Acknowledgments
We thank Titus Boggon for helpful comments on the manuscript. This work was supported by National Institutes of Health grants R21 CA147993 to B.E.T. and R01 NS39475 to A.J.K. Y.D. was supported by a James Hudson Brown-Alexander Brown Coxe postdoctoral fellowship from Yale University School of Medicine.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a) Brognard J, Hunter T. Curr Opin Genet Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang J, Yang PL, Gray NS. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Chen CA, Yeh RH, Lawrence DS. J Am Chem Soc. 2002;124:3840–3841. doi: 10.1021/ja017530v. [DOI] [PubMed] [Google Scholar]; b) Shults MD, Imperiali B. J Am Chem Soc. 2003;125:14248–14249. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]; c) Wu D, Mand MR, Veach DR, Parker LL, Clarkson B, Kron SJ. Anal Biochem. 2008;375:18–26. doi: 10.1016/j.ab.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lukovic E, Vogel Taylor E, Imperiali B. Angew Chem Int Ed Engl. 2009;48:6828–6831. doi: 10.1002/anie.200902374. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Placzek EA, Plebanek MP, Lipchik AM, Kidd SR, Parker LL. Anal Biochem. 2010;397:73–78. doi: 10.1016/j.ab.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Wang Q, Zimmerman EI, Toutchkine A, Martin TD, Graves LM, Lawrence DS. ACS Chem Biol. 2010;5:887–895. doi: 10.1021/cb100099h. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Xu X, Liu X, Nie Z, Pan Y, Guo M, Yao S. Anal Chem. 2011;83:52–59. doi: 10.1021/ac102786c. [DOI] [PubMed] [Google Scholar]; h) Sato M, Ozawa T, Inukai K, Asano T, Umezawa Y. Nat Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]; i) Ting AY, Kain KH, Klemke RL, Tsien RY. Proc Natl Acad Sci USA. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Veldhuyzen WF, Nguyen Q, McMaster G, Lawrence DS. J Am Chem Soc. 2003;125:13358–13359. doi: 10.1021/ja037801x. [DOI] [PubMed] [Google Scholar]; m) Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. J Biol Chem. 2004;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]; n) Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]; o) Dai Z, Dulyaninova NG, Kumar S, Bresnick AR, Lawrence DS. Chem Biol. 2007;14:1254–1260. doi: 10.1016/j.chembiol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; p) Wang Q, Dai Z, Cahill SM, Blumenstein M, Lawrence DS. J Am Chem Soc. 2006;128:14016–14017. doi: 10.1021/ja065852z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.a) Liu Y, Patricelli MP, Cravatt BF. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jessani N, Liu Y, Humphrey M, Cravatt BF. Proc Natl Acad Sci USA. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Chem Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]; d) Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]; e) Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Proc Natl Acad Sci USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Proc Natl Acad Sci USA. 2004;101:7943–7948. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Kalesh KA, Tan LP, Lu K, Gao L, Wang J, Yao SQ. Chem Commun (Camb) 2010;46:589–591. doi: 10.1039/b919744c. [DOI] [PubMed] [Google Scholar]
- 5.Huse M, Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 6.Songyang Z, Carraway KL, 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 7.Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 8.Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Tanis KQ, Veach D, Duewel HS, Bornmann WG, Koleske AJ. Mol Cell Biol. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Brasher B, Van Etten RA. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 10.a) Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]; b) Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 11.a) Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]; b) Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 12.Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, Xie D, Zhang Y, Klei HE. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 13.a) Shreder KR, Wong MS, Nomanbhoy T, Leventhal PS, Fuller SR. Org Lett. 2004;6:3715–3718. doi: 10.1021/ol048656a. [DOI] [PubMed] [Google Scholar]; b) Yee MC, Fas SC, Stohlmeyer MM, Wandless TJ, Cimprich KA. J Biol Chem. 2005;280:29053–29059. doi: 10.1074/jbc.M504730200. [DOI] [PubMed] [Google Scholar]; c) Blair JA, Rauh D, Kung C, Yun CH, Fan QW, Rode H, Zhang C, Eck MJ, Weiss WA, Shokat KM. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]; d) Cohen MS, Hadjivassiliou H, Taunton J. Nat Chem Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]; f) Liu Y, Wu J, Weissig H, Betancort JM, Gai WZ, Leventhal PS, Patricelli MP, Samii B, Szardenings AK, Shreder KR, Kozarich JW. Bioorg Med Chem Lett. 2008;18:5955–5958. doi: 10.1016/j.bmcl.2008.08.045. [DOI] [PubMed] [Google Scholar]; g) Kalesh KA, Sim DS, Wang J, Liu K, Lin Q, Yao SQ. Chem Commun (Camb) 2010;46:1118–1120. doi: 10.1039/b919888a. [DOI] [PubMed] [Google Scholar]; h) Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee JD, Gray NS, Kozarich JW. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Shi H, Zhang CJ, Chen GY, Yao SQ. J Am Chem Soc. 2012;134:3001–3014. doi: 10.1021/ja208518u. [DOI] [PubMed] [Google Scholar]; j) Ranjitkar P, Brock AM, Maly DJ. Chem Biol. 2010;17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]; b) Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]; c) Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan PF, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 16.a) Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Nat Struct Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]; b) Hill ZB, Perera BG, Andrews SS, Maly DJ. ACS Chem Biol. 2012;7:487–495. doi: 10.1021/cb200387g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


