Abstract
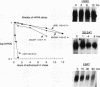
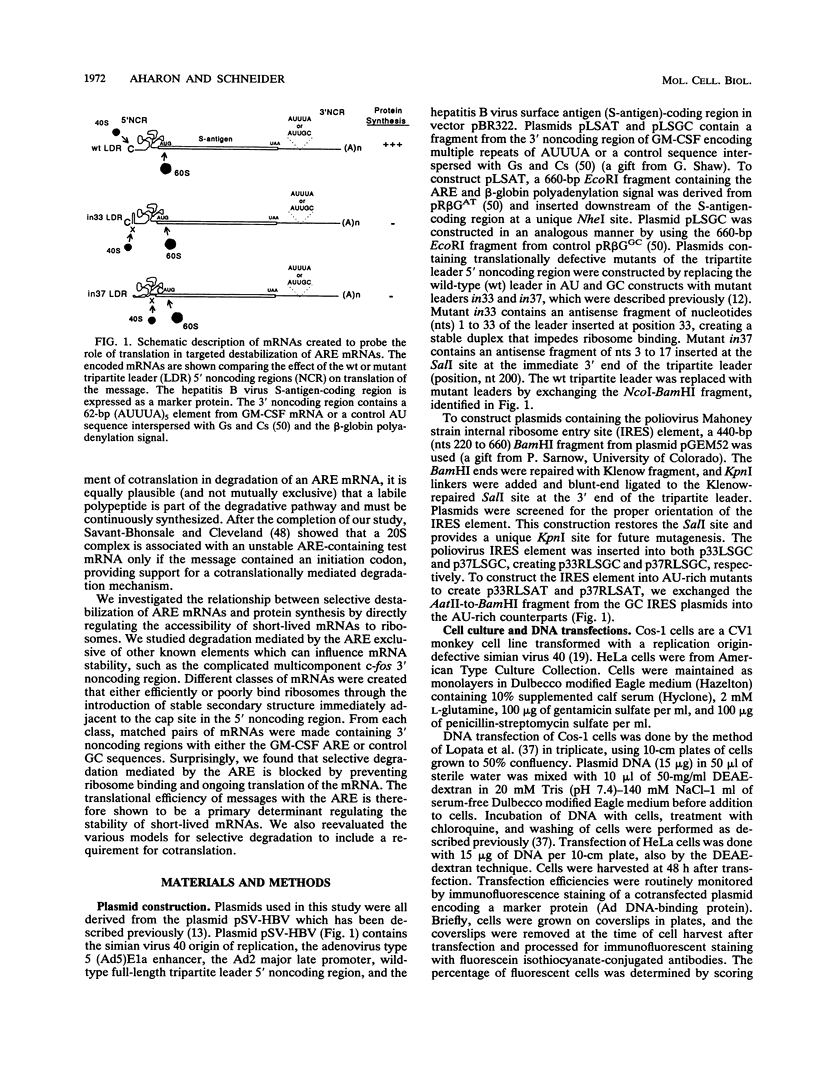
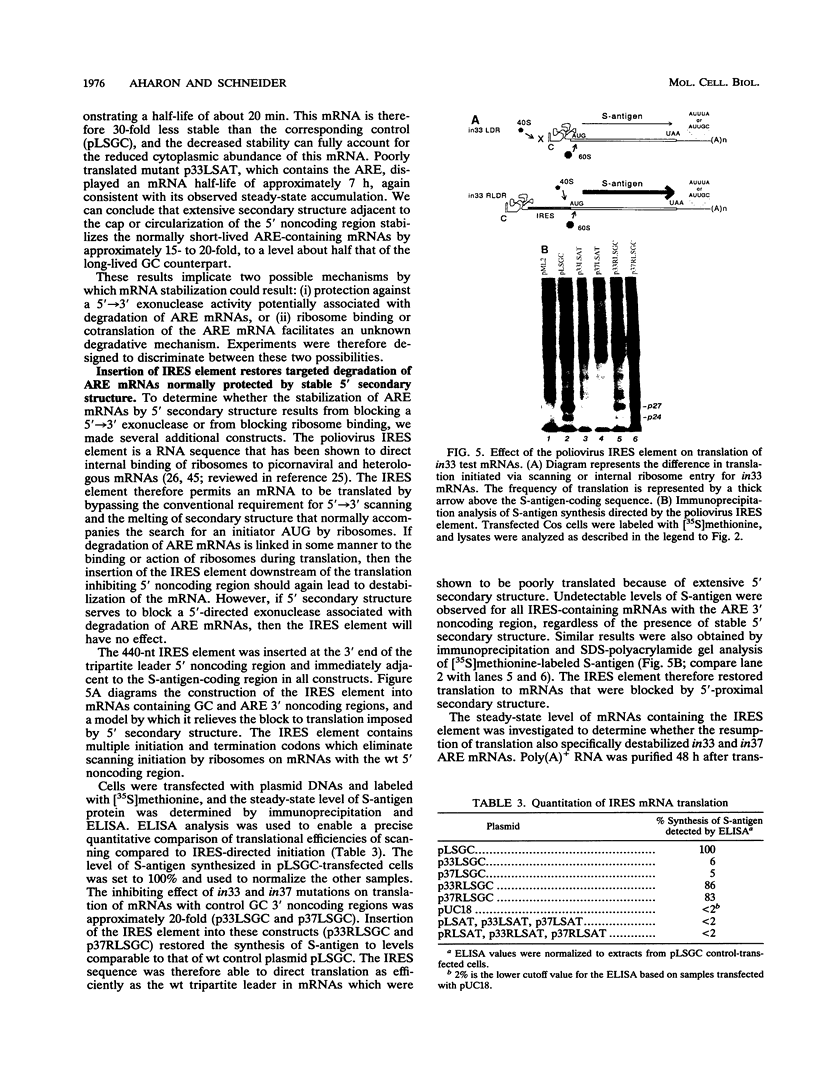
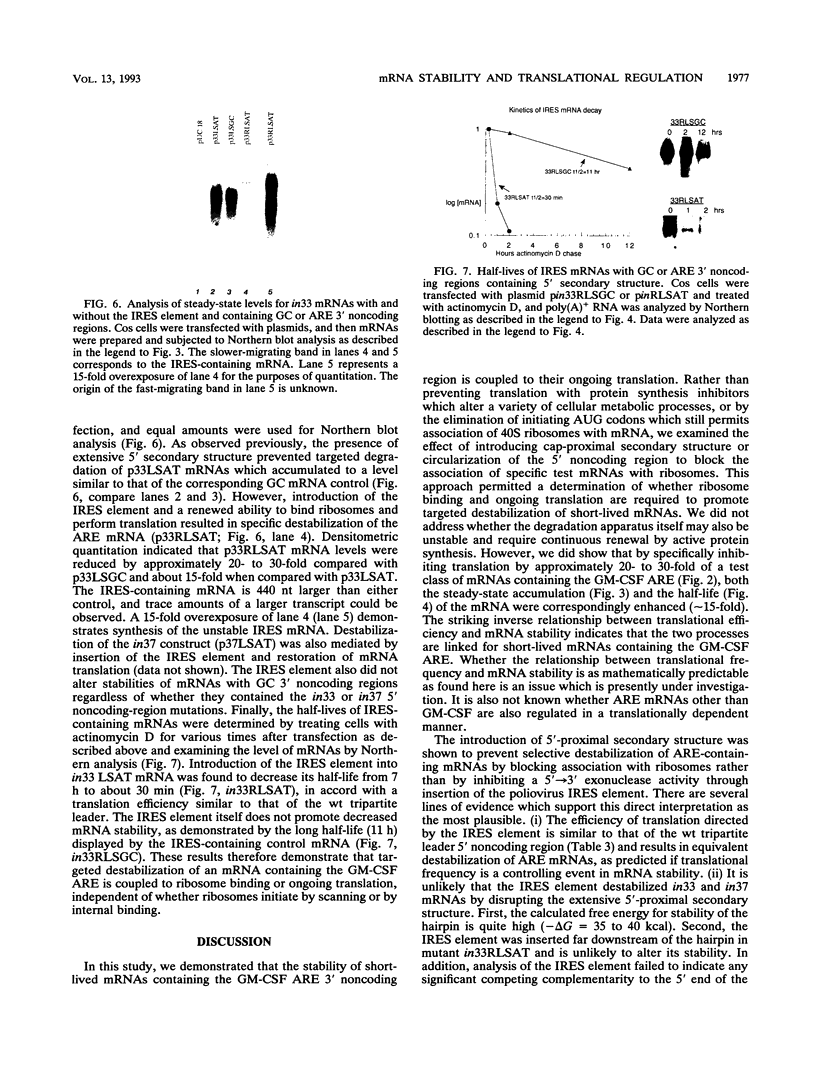
The 3' noncoding region element (AUUUA)n specifically targets many short-lived mRNAs for degradation. Although the mechanism by which this sequence functions is not yet understood, a potential link between facilitated mRNA turnover and translation has been implied by the stabilization of cellular mRNAs in the presence of protein synthesis inhibitors. We therefore directly investigated the role of translation on mRNA stability. We demonstrate that mRNAs which are poorly translated through the introduction of stable secondary structure in the 5' noncoding region are not efficiently targeted for selective destabilization by the (AUUUA)n element. These results suggest that AUUUA-mediated degradation involves either a 5'-->3' exonuclease or is coupled to ongoing translation of the mRNA. To distinguish between these two possibilities, we inserted the poliovirus internal ribosome entry site, which promotes internal ribosome initiation, downstream of the 5' secondary structure. Translation directed by internal ribosome binding was found to fully restore targeted destabilization of AUUUA-containing mRNAs despite the presence of 5' secondary structure. This study therefore demonstrates that selective degradation mediated by the (AUUUA)n element is coupled to ribosome binding or ongoing translation of the mRNA and does not involve 5'-to-3' exonuclease activity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988 May;8(5):2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Voloch Z., Bastos R., Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976 Aug;8(4):495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Sharp P. A. Effect of the tripartite leader on synthesis of a non-viral protein in an adenovirus 5 recombinant. Nucleic Acids Res. 1985 Feb 11;13(3):841–857. doi: 10.1093/nar/13.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989 May;9(5):1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph P. J., Huang J. T., Schneider R. J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990 Jun;64(6):2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph P. J., Racaniello V., Villamarin A., Palladino F., Schneider R. J. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J Virol. 1988 Jun;62(6):2059–2066. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985 Oct 25;13(20):7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3' UTR of certain maternal mRNAs. Genes Dev. 1990 Dec;4(12B):2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- Gillis P., Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991 Feb 15;266(5):3172–3177. [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Goldberg D. A. Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5794–5798. doi: 10.1073/pnas.77.10.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Huarte J., Belin D., Vassalli A., Strickland S., Vassalli J. D. Meiotic maturation of mouse oocytes triggers the translation and polyadenylation of dormant tissue-type plasminogen activator mRNA. Genes Dev. 1987 Dec;1(10):1201–1211. doi: 10.1101/gad.1.10.1201. [DOI] [PubMed] [Google Scholar]
- Hyman L. E., Wormington W. M. Translational inactivation of ribosomal protein mRNAs during Xenopus oocyte maturation. Genes Dev. 1988 May;2(5):598–605. doi: 10.1101/gad.2.5.598. [DOI] [PubMed] [Google Scholar]
- Jackson R. J. RNA translation. Picornaviruses break the rules. Nature. 1988 Jul 28;334(6180):292–293. doi: 10.1038/334292a0. [DOI] [PubMed] [Google Scholar]
- Jang S. K., Davies M. V., Kaufman R. J., Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5' nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989 Apr;63(4):1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J. Identification of the components necessary for adenovirus translational control and their utilization in cDNA expression vectors. Proc Natl Acad Sci U S A. 1985 Feb;82(3):689–693. doi: 10.1073/pnas.82.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeller D. M., Horowitz J. A., Casey J. L., Klausner R. D., Harford J. B. Translation and the stability of mRNAs encoding the transferrin receptor and c-fos. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7778–7782. doi: 10.1073/pnas.88.17.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986 May;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V., Marinx O., Shaw G., Deschamps J., Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989 Aug 25;245(4920):852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- Laird-Offringa I. A., Elfferich P., van der Eb A. J. Rapid c-myc mRNA degradation does not require (A + U)-rich sequences or complete translation of the mRNA. Nucleic Acids Res. 1991 May 11;19(9):2387–2394. doi: 10.1093/nar/19.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3655–3659. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenhaupt K., Lingrel J. B. A change in the stability of globin mRNA during the induction of murine erythroleukemia cells. Cell. 1978 Jun;14(2):337–344. doi: 10.1016/0092-8674(78)90119-8. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Marbaix G., Huez G., Burny A., Cleuter Y., Hubert E., Leclercq M., Chantrenne H., Soreq H., Nudel U., Littauer U. Z. Absence of polyadenylate segment in globin messenger RNA accelerates its degradation in Xenopus oocytes. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3065–3067. doi: 10.1073/pnas.72.8.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer V. E., Lee S. I., Steitz J. A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT alpha 1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Calame K. Differential stability of c-myc mRNAS in a cell-free system. Mol Cell Biol. 1988 Jul;8(7):2860–2868. doi: 10.1128/mcb.8.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988 Jul 28;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Raj N. B., Pitha P. M. Analysis of interferon mRNA in human fibroblast cells induced to produce interferon. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7426–7430. doi: 10.1073/pnas.78.12.7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal E. T., Ruderman J. V. Widespread changes in the translation and adenylation of maternal messenger RNAs following fertilization of Spisula oocytes. Dev Biol. 1987 May;121(1):237–246. doi: 10.1016/0012-1606(87)90155-2. [DOI] [PubMed] [Google Scholar]
- Savant-Bhonsale S., Cleveland D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992 Oct;6(10):1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Cole M. D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988 Dec 23;55(6):1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Swartwout S. G., Kinniburgh A. J. c-myc RNA degradation in growing and differentiating cells: possible alternate pathways. Mol Cell Biol. 1989 Jan;9(1):288–295. doi: 10.1128/mcb.9.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis N. G., Cleveland D. W. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992 Feb;12(2):791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. B., Challoner P. B., Neiman P. E., Groudine M. Expression of the c-myb proto-oncogene during cellular proliferation. 1986 Jan 30-Feb 5Nature. 319(6052):374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Yen T. J., Gay D. A., Pachter J. S., Cleveland D. W. Autoregulated changes in stability of polyribosome-bound beta-tubulin mRNAs are specified by the first 13 translated nucleotides. Mol Cell Biol. 1988 Mar;8(3):1224–1235. doi: 10.1128/mcb.8.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Dolph P. J., Schneider R. J. Secondary structure analysis of adenovirus tripartite leader. J Biol Chem. 1989 Jun 25;264(18):10679–10684. [PubMed] [Google Scholar]