Abstract
While there is general agreement that most forms of common disease develop as a consequence of a combination of factors, including genetic, environmental, and behavioral contributors, the actual mechanistic basis of how these factors initiate or promote diabetes, cancer, neurodegenerative, and cardiovascular diseases in some individuals while not in others with seemingly identical risk factor profiles, is not clearly understood. In this respect, consideration of the potential role for mitochondrial genetics, damage, and function in influencing common disease susceptibility seems merited, given that the prehistoric challenges were the original factors that molded cellular function, and these were based upon the mitochondrial-nuclear relationships which were established during evolutionary history. These interactions were likely refined during prehistoric environmental selection events that today, are largely absent. Contemporary risk factors such as diet, sedentary lifestyle and increased longevity that influence our susceptibility to a variety of chronic diseases were not part of the dynamics that defined the processes of mitochondrial – nuclear interaction, and thus, cell function. Consequently, the prehistoric challenges that contributed to cell functionality and evolution should be considered when interpreting and designing experimental data and strategies. Although several molecular epidemiologic studies have generally supported this notion, studies that probe beyond these associations are required. Such investigation will mark the initial steps for mechanistically addressing the provocative concept that contemporary human disease susceptibility is the result of prehistoric selection events for mitochondrial-nuclear function that increased the probability for survival and reproductive success during evolution.
Keywords: Mitochondria, Bioenergetics, Oxidants, Disease susceptibility
The genetic basis for common diseases associated with metabolism is generally appreciated as being complex. Overall, disease susceptibility is considered to be an interactive consequence of multiple modifiable (behavioral, lifestyle) and non-modifiable (genetic) factors, however, the actual mechanistic basis of this interaction is not clearly understood. The “common disease – common variants” hypothesis argues that common forms of disease involve multiple genes, each with a different effect that interact with other genes or environmental factors that modify their actions, contributing to individual disease susceptibility [1]. While this concept is important in that it introduces the notion of gene groups that can influence disease susceptibility and also provides a plausible explanation for differences in individual disease susceptibility, it is not yet evident whether it satisfactorily encompasses basic questions about risk for common diseases, such as ethnic disparity and why risk increases with age.
In this respect, mitochondrial genetics and biology could act in a complementary fashion with nuclear genetics to more lucidly explain individual disease susceptibility. Douglas Wallace proposed that mitochondrial DNA (mtDNA) missense mutations accumulated in humans as they migrated from Africa, and that these mutations changed aspects of mitochondrial bioenergetics that contributed to the establishment of human populations at more northern latitudes [2;3]. As described in this review, it is proposed that the prehistoric factors driving these selective changes in the mtDNA were the adaptations of cellular bioenergetics to increase fecundity under conditions of changing environmental challenges. Because the environment of “modern” humans has dramatically changed from our prehistoric ancestors, these prehistoric mtDNA mutations influence contemporary disease susceptibility. The following discussion will attempt to summarize the tenets of the “Mitochondrial paradigm for disease” and current evidence consistent and inconsistent with this paradigm.
MITOCHONDRIAL ORIGINS AND FUNCTIONS
While there is still controversy regarding the specific mechanics and nature of endosymbiotic origins of the eukaryotic cells, it is generally agreed that mitochondria are ancient bacterial endosymbionts, which were originally endocytosed by a larger, anaerobic nucleated cell (proto-eukaryote), that gave the resulting proto-eukaryotic organism a selective advantage for survival. Interestingly, while the basis of this advantage has historically been thought to be aerobic metabolism, this is now being challenged in that it may have originally been related to hydrogen or carbon compound based metabolism [4–9]. Nevertheless, in the presence of photosynthetic organisms, the atmosphere of the Precambrian world became increasingly oxygen laden, and aerobic mitochondria evolved. Hence, the observed compartmentalization of anaerobic (glycolytic) versus aerobic (mitochondrial) metabolism seen in cells today represents historical evidence of their past origins as endosymbionts. Considered in this light, it is possible that the ancestors of the mitochondrion and nucleus were likely to be on more equal terms in regulating cell function than commonly contemplated today.
By virtue of their origins as aerobic bacteria, mitochondria have their own DNA, RNA, and protein synthesis systems. Each cell contains hundreds to thousands of mitochondria and each mitochondrion contains multiple copies of a double stranded, closed circular, maternally inherited mtDNA. Interestingly, while it has been previously speculated that the reasons for this maternal inheritance were due to the location (sperm midpiece) and low numbers of paternal mtDNA copies versus maternal, recent evidence also suggests that the paternal mtDNA is degraded by fertilization triggered autophagy [10;11]. In humans, the mtDNA is 16,569 base pairs in size [12]. The mtDNA in our proto-eukaryotic ancestors was significantly larger in genetic complement, but transfer of mtDNA encoded genes to the nucleus has occurred over the 1.5 – 2 billion years since the origin of the eukaryotic cell, and today the mammalian mtDNA encodes 13 polypeptides, two rRNAs (12S and 16S) and 22 tRNAs that are essential for OXPHOS and proper cell function. Consequently, a unique feature of the molecular machinery controlling cellular bioenergetics is that electron transport and oxidative phosphorylation proteins are encoded by both nuclear and mitochondrial genomes. Whereas the nucleus encodes ~1500 mitochondrial proteins and the mtDNA only 13, the mtDNA retained key structural subunits required for the catalytic activity for NADH-ubiquinone oxidoreductase (commonly referred to as NADH dehydrogenase), ubiquinone-cytochrome c oxidoreductase (also referred to as cytochrome c reductase or the bc1 complex), cytochrome c oxidase, and ATP synthase. Accordingly, mutations in these mtDNA encoded subunits could alter features in mitochondrial metabolism or economy (bioenergetic function) [13]. Pathogenic mtDNA mutations have been associated with numerous types of diseases, including neurodegeneration, neuromuscular disease, deafness, blindness, diabetes, and cardiovascular disease [14]. However, the focus of this mini-review is upon the potential influence of mtDNA sequence variation that is not considered to be pathogenic, but polymorphic, upon mitochondrial function and disease susceptibility.
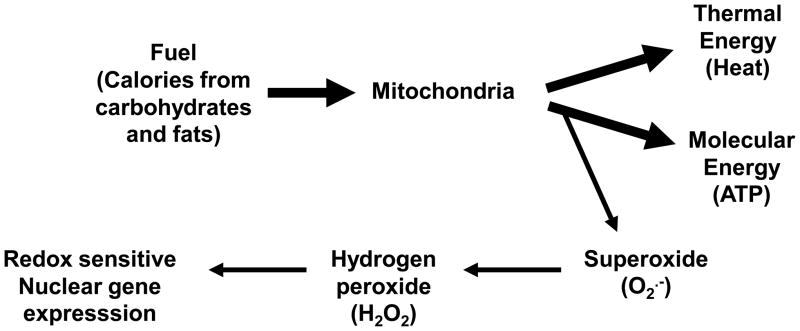
Historically, mitochondria have been characterized as “cellular powerplants”. In actuality, they are multifunctional organelles, and serve as the sites for electron transport, oxidative phosphorylation (OXPHOS), the citric acid cycle, β-oxidation, steroidogenesis, and many other important cellular functions including growth, oxidant generation and programmed cell death [13]. In some cells, the mitochondrion may serve as a source of regulated oxidant generation for cell signaling, whereas it will serve as an oxidative energetic source in others, or, a combination of these functions in still others. Mitochondria generate molecular energy (ATP), thermal energy (heat), and superoxide (O2·−) by utilizing electron flow generated from metabolism of cellular carbohydrates and fats (Figure 1). Nearly half of the energy released during electron flow is used to generate a transmembrane electrochemical gradient across the mitochondrial inner membrane; the potential energy in this gradient is used by ATP synthase to generate ATP. Energy lost during the formation of the transmembrane potential generates heat. Under conditions of high membrane potential, electrons can interact with oxygen to form O2·−, which is a fundamental building block for oxidant formation in the mitochondrion. Superoxide can be converted to highly diffusible hydrogen peroxide (H2O2) which functions as a redox signaling molecule, or in the presence of nitric oxide (·NO), reacts at diffusion limited rates to form the oxidant peroxynitrite (ONOO−)[15]. In the mitochondrion, ONOO− will likely react with CO2 (CO2 is generated in the citric acid cycle) to form the nitrating agent, nitrosperoxycarbonate (ONOOCO2−) [16;17].
Figure 1.
Schematic summarizing the basic mitochondrial functions. By utilizing electron flow generated from metabolism of carbohydrates and fats, mitochondria generate thermal energy (heat), molecular energy (ATP), and superoxide (O2·−). Energy lost during the transfer of electrons from electron transport generates heat, and the potential energy retained in the transmembrane electrochemical gradient across the mitochondrial inner membrane is used to generate ATP. Electrons also interact with oxygen to form superoxide (O2·−) that can be converted to highly diffusible hydrogen peroxide (H2O2) which functions as a redox signaling molecule that can influence redox sensitive signaling pathways and nuclear gene expression. Mitochondrial oxidant production is increased under conditions of high membrane potential.
MITOCHONDRIAL ECONOMY
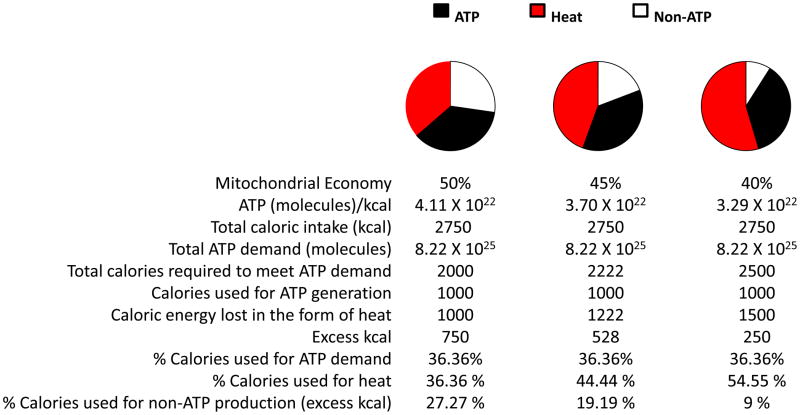
The ability of the mitochondrion to convert caloric energy into ATP influences non-ATP related processes (e.g. heat and oxidant production). Mitochondria that are highly economical in generating ATP with a minimal loss of caloric energy to heat are also more prone to produce O2·− under conditions of high membrane potentials and low energy demand compared to those less coupled or economical [13]. Consequently, while economical mitochondria generate ATP with less expenditure of energy (calories), they also have a greater propensity to generate oxidants under conditions of low energetic demand and high caloric intake (Figure 2). In addition to the basic mechanisms of energy generation associated with electron transport and the production of a transmembrane potential, several constantly changing factors such as local concentrations of O2, reactive species, mitochondrial antioxidants, cytokine concentrations, metabolic reducing equivalent availability, cellular energetic demand, uncoupling protein (UCP) activities, and overall organelle integrity influence mitochondrial economy [18].
Figure 2.
Hypothetical presentation of how differences in mitochondrial economy influence caloric utilization. Mitochondria with different economies have different capacities for generating ATP/kilo-calorie (kcal) consumed (where 50%, 45%, or 40% of the energy from electron flow is utilized for generating ATP in the hypothetical examples), and therefore those with lower economies will consume more calories to meet energetic demands (with an increased release of caloric energy as heat). It is assumed that electron flow is equal between mitochondria having different economies. A related consequence is that under conditions of excessive caloric consumption (excess kcal) relative to energy demand, mitochondria with higher economies will generate greater amounts of mitochondrial oxidants (a component non-ATP production). ATP/kcal calculations are based upon 1 mole of ATP (6.023 × 1023 molecules) = 7.33 kcal. Hence 1 kcal = 8.22 × 1022 molecules of ATP assuming 100% economy. This value is used to estimate ATP/kcal for 50%, 45% and 40% (4.11 × 1022, 3.70 × 1022 and 3.29 × 1022, respectively) economies. Total caloric consumption is 2750 kcal. Total ATP demand of 8.22 × 1025 molecules, based upon a demand of 1000 kcal assuming 50% economy, hence decreased economy (45% and 40%) requires utilization of more calories to meet ATP demand. Caloric energy lost in the form of heat is determined by subtracting calories required to meet a demand of 8.22 × 1025 ATP under conditions of 100% economy (1000 kcal, or, calories used for ATP generation) from “Total calories required to meet ATP demand” for each economy. Percentages of calories used for heat generation, ATP and non-ATP production are determined by dividing kcal used to for heat, ATP generation, and excess kcal, respectively, by total caloric intake (2750 kcal). Pie charts illustrate percentages.
MTDNA MUTATION AND ADAPTATION DURING PREHISTORY
It has been proposed that environmental factors influenced prehistoric human radiation patterns and survival by selecting for aspects of mitochondrial function (ATP production and thermogenesis) [2;3;19]. As prehistoric humans moved northward out of Africa they encountered climatic and dietary changes that played a role in the selective pressures of survival and reproduction [20;21]. It has been noted that as prehistoric humans migrated northward they accumulated mtDNA missense mutations that have been proposed to have altered mitochondrial bioenergetics and thus, economy [2;19]. These mtDNA mutations decreased mitochondrial economy resulting in the greater generation of heat/calorie consumed [2;3;19]; this apparent decrease in efficiency was tolerated due to the changes in diet also encountered – as humans moved from Africa, their diet changed from a low protein, low fat vegetarian diet to a high protein, high fat diet consisting of animal fats [20;21]. Hence, the decreased ATP generation/calorie (due to increased heat production) was offset by increased caloric intake associated with animal fats. It is therefore suggested that mitochondrial economy (via mtDNA mutation) significantly influenced survival and thus reproductive potential in prehistoric humans, in response to changes in the environment and diet.
The concept that changes in mitochondrial function allowed adaptation is not exclusive to Homo sapiens – the 13-lined ground squirrel (Spermophilus lateralis) upregulates mtDNA encoded NADH dehydrogenase mRNA during hibernation [22] which occurs concomitant with decreased metabolic rate and core body temperature compared to non-hibernating levels [23]. MtDNA-encoded ubiquinone-cytochrome c oxidoreductase subunits in S. lateralis liver mitochondria have also been reported to altered during hibernation, which may decrease the capacity of that electron transport complex [24]. Similarly, it has been shown that the bar-headed goose (Anser indicus) has evolved more subsarcolemmal mitochondria bordering capillaries with increased densities and numbers of oxidative fibers that enable them to sustain flight at high altitudes (up to 9000 m or 30,000 ft) compared to low altitude birds [25]. This adaptation is associated with decreased maximal cytochrome c oxidase activity, and a higher affinity of the enzyme for reduced cytochrome c that appears to be associated with a mutation in the mtDNA encoded subunit III for cytochrome c oxidase that may alter the structure of the enzyme complex that accounts for increased economy and high-altitude adaptation [26]. Examination of the mtDNA encoded subunits across placental mammals is currently underway to determine the degree of “adaptive” evolution which may provide useful information regarding key mtDNA mutations and their role on evolution in terms of cellular function [27].
CONTEMPORARY IMPLICATIONS OF MTDNA MUTATIONS SELECTED FOR DURING PREHISTORIC ADAPTATION
Western society has all too frequently become characterized by a sedentary lifestyle and excessive diet. In terms of mitochondrial bioenergetics, this has become problematic because a low energy demand and chronic high caloric intake were not factors under which mtDNA mutations became fixed in the human population during human prehistory. Mitochondria that harbor mtDNA mutations associated with greater economy will generate increased oxidants under conditions of excessive caloric intake and physical inactivity compared to those with mtDNA mutations linked to decreased mitochondrial economy, and therefore the former (greater mitochondrial economy) will be more prone to succumb to diseases connected to oxidative stress, whereas the latter (decreased mitochondrial economy) will be comparatively less susceptible, yet not completely immune. Chronic, excessive caloric intake and physical inactivity will still result in sustained mitochondrial oxidant generation over time even in mitochondria having decreased economy which will result in cell damage and dysfunction. Hence, disease susceptibility associated with oxidative stress will represent interplay between mtDNA genetic background, diet, exercise, and other environmental factors such as smoking or exposure to other environmental oxidants.
Evidence consistent with this provocative notion is still being gathered. In addition to the original studies that link mtDNA mutations with human disease, several studies have shown that specific mtDNA mutations and genetic backgrounds are associated with increased risk for diseases thought or known to have an environmental component in humans [28–37]. MtDNA genetic background can influence tumor growth and age-related deafness [32;38], and has been shown to co-segregate with longevity in humans [34;35]. While it is hypothesized that mtDNAs associated with increased economy will be associated with certain types of cancer and neurodegenerative diseases associated with somatic mutation or oxidative stress, it has been also suggested that mtDNAs associated with “cold adaptation” (e.g. less economical) will be associated with increased life span, but increased predilection for clinical illnesses potentially associated with energetics such as blindness and central nervous system (CNS) defects [19].
Cybrid culture studies have provided conflicting results supporting the concept that the mtDNA influences cellular bioenergetics [39–42]. However, these studies by necessity have had to utilize transformed or immortalized cell lines and therefore do not exactly represent in vivo circumstances. Studies utilizing conplastic strains of mice (mice having different nuclear and mitochondrial genetic backgrounds, generated by breeding two different strains to generate F1 females and backcrossing them and subsequent filial female generations onto the nuclear background of the paternal strain), suggest that mtDNA background does influence aspects of cognition, behavior, reproductive behavior, and susceptibility to autoimmune disease [43–46]. A potential issue with even this approach is that if the mtDNA alters organelle economy (bioenergetics) which can affect numerous nuclear genes, the mtDNA may also play a role in modulating nuclear gene expression and perhaps play a role in allelic segregation and assortment during meiosis. If this is the case, it would represent another historical clue regarding the evolution of the eukaryotic cell and endosymbiosis, and thus, provide the basis for an additional paradigm in that the mtDNA influences the selection of certain nuclear – mitochondrial gene combinations and mitochondrial retrograde signaling [47–49]. If true, this would have significant implications regarding the use of transgenics (or the creation of congenics) derived from different strains of mice.
MITOCHONDRIAL OXIDANT PRODUCTION
The concept that mitochondrial oxidant production is simply a consequence of energy production and serves as a primary, chronic source of cellular stress that causes disease development seems logical when considered from a contemporary viewpoint. However, from an evolutionary perspective and in reality, mitochondrial oxidants likely serve as signals for mitochondrial – nuclear interaction which evolved to increased cell survival under conditions of limited caloric availability. Whether under conditions of caloric scarcity or plentitude, a system with a facile feedback/signaling mechanism (redox signaling) linked to energy requirements and caloric availability would be advantageous. Consequently, mitochondrial oxidants likely served as stimuli for insulin production and signaling molecules associated with insulin signaling pathways in non-insulin producing tissues. In the presence of abundant calories and low energy demand, mitochondria increased mitochondrial oxidant production, triggering signaling pathways leading to storage of calories [50–54]. As food availability became low and/or energy demand increased, oxidant production and caloric storage would decrease. Interestingly, studies have shown that mitochondrial oxidants or the alteration of mitochondrial UCP levels affect insulin secretion and sensitivity [51;55–57]. Although studies have shown that mitochondrial oxidants can inhibit insulin production and sensitivity, they usually employ conditions of chronic hyperglycemia and therefore do not represent the original prehistoric conditions that were likely present during the establishment of mitochondrial – nuclear communication. Nevertheless, a connection does clearly seem to exist between mitochondrial oxidants, insulin secretion and insulin signaling [50;58–61].
Mitochondrial oxidants are generated during electron transport, when O2 picks up electrons from the ubiquinone site in ubiquinone – cytochrome c oxidoreductase and flavin mononucleotide group of NADH dehydrogenase to generate O2·− [62–66]. In the mitochondrion, O2·− can be converted to H2O2 by mitochondrial manganese superoxide dismutase (MnSOD or SOD2). It has been estimated that mitochondrial H2O2 generation represents up to 2% of the total mitochondrial oxygen consumption under basal conditions and fluxes in mitochondrial H2O2 can be influenced by pharmaceuticals, inhibitors of respiration, uncouplers, redox cycling molecules, and by either endogenous or exogenous environmental changes [63;67–71]. This H2O2 produced by the mitochondrion acts as a redox signaling molecule that can stimulate growth and survival pathways or intrinsic apoptotic pathways, at low or high H2O2 concentrations, respectively [72–76].
Mitochondria may regulate many genes involved in cellular defense and immunological response via the generation of ROS that act as intermediate second messengers for NFκB activation by tumor necrosis factor alpha (TNFα) and interleukin – 1 [77–80]. Inhibition of mitochondrial respiration blocks H2O2 – induced transactivation of the endothelial growth factor receptor and stimulation of downstream targets [81]. As a result, antioxidant expression and cytokines such as TNF-α influence the steady state levels of specific mitochondrial oxidants, and therefore play a role in the modulation of mitochondrial mediated redox signaling. Cytokines such as platelet derived growth factor (PDGF) and TNFα directly or secondarily alter cellular oxidant production via mitochondrial pathways [82–84]. As with mitochondrial economy, oxidant generation from the mitochondrion is influenced by the concentrations of reactive species within the organelle, electron transport efficiency, availability of metabolic reducing equivalents (NADH and FADH2), UCP levels and activities, and organelle integrity. For example, nitric oxide (·NO) reversibly binds cytochrome c oxidase and can regulate its activity, influencing ATP production and O2 consumption [85–87]. Whereas lower ·NO concentrations can influence oxygen O2 diffusion and modest O2·− generation, higher concentrations can lead to greater O2·− generation and related ONOO− formation triggering cytoxicity [88].
FUTURE IMPLICATIONS
Greater discernment of the factors that shaped original mitochondrial function, and how mitochondrial – nuclear interaction worked will lead to greater appreciation and understanding of the actual role of the mitochondrion in the cell. Perhaps because the mtDNA in humans encodes only 37 genes compared to the estimated ~23,000 in the nucleus, this has led to the general perception that the answer for a genetic basis for common disease susceptibility and development would more likely lie within the nuclear genome. However, the mitochondrion and nucleus were originally both unique genetic entities that entered into a symbiotic relationship, and hence, were probably once more balanced than we completely understand. Since changes in key subunits in electron transport may alter mitochondria economy, which includes oxidant production, one mtDNA mutation can ultimately have a multitude of effects on nuclear gene expression due to redox signaling mechanisms.
Interpretation and designing experiments directed at understanding how mitochondrial – nuclear functionality evolved will undoubtedly lead to greater understanding of disease pathology because they will provide greater understanding of the basis of mitochondrial – nuclear interaction, and therefore the etiology of disease. In order to successfully achieve these aims however it will require basic experimental studies to utilize “normal” or non-diseased models that examine their response to various stimuli commonly thought to be the initial harbingers for disease risk. A potential pitfall in many translational studies today is that they rely on experimental models that represent disease states, and therefore may not provide information on the original role(s) of the mitochondrion in the cell. Overall, a greater understanding of how the original relationships between the mitochondrion and the nucleus evolved to allow for the adaptation of cell function will facilitate discovery of the mechanisms of how common disease develops.
Acknowledgments
The author wishes to acknowledge Drs. Victor Darley-Usmar and Jack Lancaster for their helpful comments and discussions.
FUNDING:
This work was supported by National Institutes of Health (Heart, Lung, Blood) grants RO1HL94518 and RO1HL103859.
List of Abbreviations
- ATP
adenosine triphosphate
- CNS
central nervous system
- CO2
carbon dioxide
- H2O2
hydrogen peroxide
- MnSOD
manganese superoxide dismutase
- mtDNA
mitochondrial DNA
- NADH
nicotinamide adenine dinucleotide, reduced form
- ·NO
nitric oxide
- O2
oxygen
- O2·−
superoxide
- ONOO−
peroxynitrite
- OXPHOS
oxidative phosphorylation
- PDGF
platelet derived growth factor
- SOD2
manganese superoxide dismutase
- TNF-α
tumor necrosis factor alpha
Reference List
- 1.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: On the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Margulis L. The origin of plant and animal cells. Am Sci. 1971;59:230–235. [PubMed] [Google Scholar]
- 5.Margulis L, Dolan MF, Guerrero R. The chimeric eukaryote: origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci USA. 2000;97:6954–6959. doi: 10.1073/pnas.97.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embley TM, Martin W. A hydrogen-producing mitochondrion. Nature. 1998;396:517–519. doi: 10.1038/24994. [DOI] [PubMed] [Google Scholar]
- 7.Embley TM, van der GM, Horner DS, Dyal PL, Foster P. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci. 2003;358:191–201. doi: 10.1098/rstb.2002.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Embley TM, van der GM, Horner DS, Dyal PL, Bell S, Foster PG. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- 9.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 10.Al RS, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 11.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 12.Anderson S, Bankier AT, Barrell BG, de Bruin MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human miochonrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 13.Ballinger SW. Deregulation of mitochondrial function: a potential common theme for cardiovascular disease development. In: Miwa S, Beckman KB, Muller FL, editors. Aging Medicine: Oxidative Stress in Aging: From Model Systems to Human Diseases. Humana Press; Totowa, NJ: 2008. pp. 165–189. [Google Scholar]
- 14.Wallace D. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 15.Kissner R, Nauser T, Bugnon P, Lye PG, Loppenol WH. Formation and properties of peroxynitrite as studied by laser flash photolysis, high-pressure stopped-flow technique, and pulse radiolysis. Chem Res Toxicol. 1997;10:1285–1292. doi: 10.1021/tx970160x. [DOI] [PubMed] [Google Scholar]
- 16.Gow A, Duran D, Thom S, Ischiropoulos H. Carbon dioxide enhancement of peroxynitrite mediated protein tyrosine nitration. Arch Biochem Biophys. 1996;333:42–48. doi: 10.1006/abbi.1996.0362. [DOI] [PubMed] [Google Scholar]
- 17.Ischiropoulos H. Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 18.Krzywanski DM, Moellering DR, Fetterman JL, Dunham-Snary KJ, Sammy MJ, Ballinger SW. The mitochondrial paradigm for cardiovascular disease susceptibility and cellular function: a complementary concept to Mendelian genetics. Lab Invest. 2011;91:1122–1135. doi: 10.1038/labinvest.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milton K. Hunter-gatherer diets – a different perspective. Am J Clin Nutr. 2000;71:665–667. doi: 10.1093/ajcn/71.3.665. [DOI] [PubMed] [Google Scholar]
- 21.Cordain L, Brand-Miller J, Eaton SB, Mann N, Holt SHA, Speth JD. Plant-animal subsistence ratios and macronutrient energy estimations in worldwide hunter-gatherer diets. Am J Clin Nutr. 2000;71:682–692. doi: 10.1093/ajcn/71.3.682. [DOI] [PubMed] [Google Scholar]
- 22.Fahlman A, Storey JM, Storey KB. Gene up-regulation in heart during mammalian hibernation. Cryobiology. 2000;40:332–342. doi: 10.1006/cryo.2000.2254. [DOI] [PubMed] [Google Scholar]
- 23.Muleme HM, Walpole AC, Staples JF. Mitochondrial metabolism in hibernation: metabolic suppression, temperature effects, and substrate preferences. Physiol Biochem Zool. 2006;79:474–483. doi: 10.1086/501053. [DOI] [PubMed] [Google Scholar]
- 24.Shug AL, Ferguson S, Shrago E, Burlington RF. Changes in respiratory control and cytochromes in liver mitochondria during hibernation. Biochim Biophys Acta. 1971;226:309–312. doi: 10.1016/0005-2728(71)90097-1. [DOI] [PubMed] [Google Scholar]
- 25.Scott GR, Egginton S, Richards JG, Milsom WK. Evolution of muscle phenotype for extreme high altitude flight in the bar-headed goose. Proc Biol Sci. 2009;276:3645–3653. doi: 10.1098/rspb.2009.0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott GR, Schulte PM, Egginton S, Scott AL, Richards JG, Milsom WK. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 2011;28:351–363. doi: 10.1093/molbev/msq205. [DOI] [PubMed] [Google Scholar]
- 27.da Fonseca RR, Johnson WE, O’Brien SJ, Ramos MJ, Antunes A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics. 2008;9:119. doi: 10.1186/1471-2164-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally trasnmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 29.Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 30.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 31.Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup J. Human Genetics. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 32.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. American Journal of Medical Genetics. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 34.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. Faseb Journal. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 35.Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafe M, Franceschi C, De Benedictis G. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. European Journal of Human Genetics. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 36.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. American Journal of Human Genetics. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Doraiswamy PM, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neuroscience Letters. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nature Genetics. 2001;27:191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amo T, Yadava N, Oh R, Nicholls DG, Brand MD. Experimental assessment of bioenergetic differences caused by the common European mitochondrial DNA haplogroups H and T. Gene. 2008;411:69–76. doi: 10.1016/j.gene.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amo T, Brand MD. Were inefficient mitochondrial haplogroups selected during migrations of modern humans? A test using modular kinetic analysis of coupling in mitochondria from cybrid cell lines. Biochem J. 2007;404:345–351. doi: 10.1042/BJ20061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, de Cordoba SR, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nature Genetics. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Human Molecular Genetics. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 43.Roubertoux PL, Sluyter F, Carlier M, Marcet B, Maarouf-Veray F, Cherif C, Marican C, Arrechi P, Godin F, Jamon M, Verrier B, Cohen-Salmon C. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nature Genetics. 2003;35:65–69. doi: 10.1038/ng1230. [DOI] [PubMed] [Google Scholar]
- 44.Gimsa U, Kanitz E, Otten W, Ibrahim SM. Behavior and Stress Reactivity in Mouse Strains with Mitochondrial DNA Variations. Neuroimmunomodulation: from Fundamental Biology to Therapy. 2009;1153:131–138. doi: 10.1111/j.1749-6632.2008.03960.x. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Research. 2009;19:159–165. doi: 10.1101/gr.078865.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu XH, Wester-Rosenlof L, Gimsa U, Holzhueter SA, Marques A, Jonas L, Hagenow K, Kunz M, Nizze H, Tiedge M, Holmdahl R, Ibrahim SM. The mtDNA nt7778 G/T polymorphism affects autoimmune diseases and reproductive performance in the mouse. Human Molecular Genetics. 2009;18:4689–4698. doi: 10.1093/hmg/ddp432. [DOI] [PubMed] [Google Scholar]
- 47.Schieke SM, Finkel T. Mitochondrial signaling, TOR, and life span. Biol Chem. 2006;387:1357–1361. doi: 10.1515/BC.2006.170. [DOI] [PubMed] [Google Scholar]
- 48.Karu TI. Mitochondrial signaling in mammalian cells activated by red and near-IR radiation. Photochem Photobiol. 2008;84:1091–1099. doi: 10.1111/j.1751-1097.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- 49.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 50.Hoehn KL, Salmon AB, Hohnen-Behrens C, Turner N, Hoy AJ, Maghzal GJ, Stocker R, Van Remmen H, Kraegen EW, Cooney GJ, Richardson AR, James DE. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci USA. 2009;106:17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudina S, Sena S, Theobald H, Sheng XM, Wright JJ, Hu XX, Aziz S, Johnson JI, Bugger H, Zaha VG, Abel ED. Mitochondrial energetics in the heart in obesity-related diabetes - Direct evidence for increased uncoupled respiration and activation of uncoupling proteins. Diabetes. 2007;56:2457–2466. doi: 10.2337/db07-0481. [DOI] [PubMed] [Google Scholar]
- 52.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo Journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Civitarese AE, MacLean PS, Carling S, Kerr-Bayles L, McMillan RP, Pierce A, Becker TC, Moro C, Finlayson J, Lefort N, Newgard CB, Mandarino L, Cefalu W, Walder K, Collier GR, Hulver MW, Smith SR, Ravussin E. Regulation of Skeletal Muscle Oxidative Capacity and Insulin Signaling by the Mitochondria! Rhomboid Protease PARL. Cell Metabolism. 2010;11:412–426. doi: 10.1016/j.cmet.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiederkehr A, Wollheim CB. Minireview: Implication of mitochondria in insulin secretion and action. Endocrinology. 2006;147:2643–2649. doi: 10.1210/en.2006-0057. [DOI] [PubMed] [Google Scholar]
- 55.Gates AC, Bernal-Mizrachi C, Chinault SL, Feng C, Schneider JG, Coleman T, Malone JP, Townsend RR, Chakravarthy MV, Semenkovich CF. Respiratory uncoupling in skeletal muscle delays death and diminishes age-related disease. Cell Metabolism. 2007;6:497–505. doi: 10.1016/j.cmet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Mattiasson G, Sullivan PG. Comprehensive invited review - The emerging functions of UCP2 in health, disease, and therapeutics. Antioxidants & Redox Signaling. 2006;8:1–38. doi: 10.1089/ars.2006.8.1. [DOI] [PubMed] [Google Scholar]
- 57.Fisler JS, Warden CH. Uncoupling proteins, dietary fat and the metabolic syndrome. Nutrition & Metabolism. 2006:3. doi: 10.1186/1743-7075-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leloup C, Tourrel-Cuzin C, Magnan C, Karaca M, Castel J, Carneiro L, Colombani AL, Ktorza A, Casteilla L, Penicaud L. Mitochondrial Reactive Oxygen Species Are Obligatory Signals for Glucose-Induced Insulin Secretion. Diabetes. 2009;58:673–681. doi: 10.2337/db07-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pi JB, Bai YS, Zhang Q, Wong V, Floering LM, Daniel K, Reece JM, Deeney JT, Andersen ME, Corkey BE, Collins S. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56:1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 60.Dokken BB, Saengsirisuwan V, Kim JS, Teachey MK, Henriksen EJ. Oxidative stress-induced insulin resistance in rat skeletal muscle: role of glycogen synthase kinase-3. American Journal of Physiology-Endocrinology and Metabolism. 2008;294:E615–E621. doi: 10.1152/ajpendo.00578.2007. [DOI] [PubMed] [Google Scholar]
- 61.Loh K, Deng HY, Fukushima A, Cai XC, Boivin B, Galic S, Bruce C, Shields BJ, Skiba B, Ooms LM, Stepto N, Wu B, Mitchell CA, Tonks NK, Watt MJ, Febbraio MA, Crack PJ, Andrikopoulos S, Tiganis T. Reactive Oxygen Species Enhance Insulin Sensitivity. Cell Metabolism. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Marcillat O, Giulivi C, Ernster L, Davies KJ. The oxidative inactivation of mitochondrial electron transport chain components and ATPase. J Biol Chem. 1990;265:16330–16336. [PubMed] [Google Scholar]
- 63.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lambert A, Brand M. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem J. 2004 doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- 66.Han D, Canali R, Rettori D, Kaplowitz N. Effect of glutathione depletion on sites and topology of superoxide and hydrogen peroxide production in mitochondria. Mol Pharmacol. 2003;64:1136–1144. doi: 10.1124/mol.64.5.1136. [DOI] [PubMed] [Google Scholar]
- 67.Forman HJ, Boveris A. In: Free Radicals In Biology. Prior WA, editor. Academic Press; Orlando: 1982. pp. 65–90. [Google Scholar]
- 68.Forman HJ, Boveris A. Superoxide radical and hydrogen peroxide in mitochondria. In: Prior WA, editor. Free Radicals In Biology. Academic Press; Orlando: 1982. pp. 65–90. [Google Scholar]
- 69.Boveris A, Turrens JF. Production of superoxide anion by the NADH dehydrogenase of mammalian mitochondria. In: Bannister J, Hill H, editors. Chemical and biochemical aspects of superoxide and superoxide dismutase. Elsevier; Amsterdam: 1980. pp. 84–91. [Google Scholar]
- 70.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 72.Baas AS, Berk BC. Differential activation of mitogen-activated protein kinases by H2O2 and O2- in vascular smooth muscle cells. Circ Res. 1995;77:29–36. doi: 10.1161/01.res.77.1.29. [DOI] [PubMed] [Google Scholar]
- 73.Chen K, Vita JA, Berk BC, Keaney JF., Jr c-Jun-N-terminal Kinase Activation by Hydrogen Peroxide in Endothelial Cells Involves Src-dependent Epidermal Growth Factor Receptor Transactivation. J Biol Chem. 2001;276:16045–16050. doi: 10.1074/jbc.M011766200. [DOI] [PubMed] [Google Scholar]
- 74.Guyton KZ, Liu Y, Gorospe M, Xu Q, Holbrook NJ. Activation of Mitogen-activated Protein Kinase by by H2O2. J Biol Chem. 1996;271:4138–4142. doi: 10.1074/jbc.271.8.4138. [DOI] [PubMed] [Google Scholar]
- 75.Kim SM, Byun JS, Jung YD, et al. The effects of oxygen radicals on the activity of nitric oxide synthase and guanylate cyclase. E Exp Mol Med. 1998;30:221–6. doi: 10.1038/emm.1998.32. [DOI] [PubMed] [Google Scholar]
- 76.Sundaresan M, Yu ZX, Fererans VJ, Irani K, Finkel T. Requirement for generation of H202 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 77.Schulze-Osthoff K, Los M, Baeuerle PA. Redox signaling by transcription factors NF-κB and AP-1 in lymphocytes. Biochem Pharmacol. 1995;50:735–741. doi: 10.1016/0006-2952(95)02011-z. [DOI] [PubMed] [Google Scholar]
- 78.Devary Y, Rosette C, DiDonato JA, Karin M. NF-kappa B activation by ultraviolet light not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 79.Mohan N, Meltz MM. Induction of nuclear factor κB after low-dose ionizing radiation involves a reactive oxygen intermediate signaling pathway. Rad Res. 1994;140:97–104. [PubMed] [Google Scholar]
- 80.Schulze-Osthoff K, Beyaert R, Vandervoorde V, Haegeman G, Fiers W. Depletion of the mitochondrial electron transport abrogates the cytotoxic and gene-inductive effects of TNF. EMBO J. 1993;12:3095–3104. doi: 10.1002/j.1460-2075.1993.tb05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen K, Thomas SR, Albano A, Murphy MP, Keaney JF. Mitochondrial function is required for hydrogen peroxide-induced growth factor receptor transactivation and downstream signaling. J Biol Chem. 2004 doi: 10.1074/jbc.M404859200. [DOI] [PubMed] [Google Scholar]
- 82.Manna SK. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappaB and activated protein-1. J Biol Chem. 1998;273:13245–54. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 83.Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992;267:5317–23. [PubMed] [Google Scholar]
- 84.Maehara K, Oh-Hashi K, Isobe KI. Early growth-responsive-1-dependent manganese superoxide dismutase gene transcription mediated by platelet-derived growth factor. FASEB J. 2001;15:2025–2026. doi: 10.1096/fj.00-0909fje. [DOI] [PubMed] [Google Scholar]
- 85.Clementi E, Brown G, Foxwell N, Moncada S. On the mechanism by which vascular endothelial cells regulate their oxygen consumption. Proc Natl Acad Sci USA. 1999;96:1559–1562. doi: 10.1073/pnas.96.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol. 2002;3:214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 87.Cassina A, Radi R. Differential inhibitory action of nitric oxide and peroxynitrite on mitochondrial electron transport. Arch Biochem Biophys. 1996;328:309–316. doi: 10.1006/abbi.1996.0178. [DOI] [PubMed] [Google Scholar]
- 88.Ramachandran A, Levonen AL, Brookes PS, Ceaser E, Shiva S, Barone MC, Darley-Usmar VM. Mitochondria, nitric oxide, and cardiovascular dysfunction. Free Radic Biol Med. 2002;33:1465–1474. doi: 10.1016/s0891-5849(02)01142-5. [DOI] [PubMed] [Google Scholar]