Abstract
We have previously shown that a fish oil-rich diet increased the chemopreventive efficacy of tamoxifen (Tam) against N-methyl-N-nitrosourea (MNU)-induced rat mammary carcinogenesis. Herein, we provide evidence that tamoxifen treatment modifies gene expression of mammary tumors depending upon the type of dietary fat fed to the animals. Rats initiated with MNU and treated with Tam were fed a diet rich in corn oil (CO) or fish oil (FO). After 8 weeks, cribriform tumors were collected and gene expression analysis was performed. Increased RNA expression of genes such as SerpinB10, Wisp2 and Apod in tumors from FO-treated rats is indicative of highly differentiated tumors. Decreased expression of H19 and Igf2 mRNA in Tam-treated groups, and Sncg mRNA in FO+Tam group may be related to tumor growth impairment and lower metastatic capacity. Change in the expression of genes associated with immunity in animals in the FO+Tam group may suggest a shift in the immune response. These data show that, although tamoxifen modulates the expression of genes leading to tumor growth impairment, further modulations of genes are influenced by FO. FO modulation of Tam changes in gene expression accounts for its enhancing chemopreventive effect against MNU-induced mammary carcinogenesis.
Keywords: fish oil, corn oil, tamoxifen, breast cancer, chemoprevention
INTRODUCTION
Breast cancer is the most frequently diagnosed type of cancer and is the second cause of death in women in the United States (1). Based on current rates, it is estimated that more than 200,000 women were diagnosed with breast cancer in 2010 (1). Adjuvant and neoadjuvant endocrine therapies have shown benefits in estrogen receptor-positive breast cancer treatments. Adjuvant therapies proposed for the treatment of this malignancy include estrogen receptor antagonists, aromatase inhibitors and ovarian suppression (2). Treatment with the non-steroidal antiestrogen tamoxifen (Tam) increases recurrence-free and overall survival of patients with localized disease, irrespective of the nodal status, menopausal status or age (3). Tamoxifen has also been shown to be an effective chemopreventive agent (4). However, depending on the dose and length of treatment, some of Tam's side effects include a significant increase in the risk of developing endometrial cancer (5). In order to improve the chemopreventive action of Tam, our laboratories have tested the combination of this drug with a fish oil-rich diet in a model of MNU-induced mammary carcinogenesis (6). Omega-3 fatty acids, which are abundantly present in fish oil, have been shown to increase the cytotoxic action of drugs such as doxorubicin and mitomycin-C in breast cancer cells (7, 8). In addition, the consumption of an omega-3 fatty-acid rich diet protects against breast cancer in most preclinical studies (9). We have reported that the chemopreventive effect of Tam against MNU-induced mammary carcinogenesis was superior in FO fed rats compared to rats fed with CO (6). In order to understand the molecular mechanisms underlying the effects of FO alone and in combination with Tam on mammary carcinogenesis, we performed a transcriptomic analysis of tumors arising in rats fed a FO vs. CO rich diet in the presence and in the absence of concomitant Tam treatment.
MATERIAL AND METHODS
The mammary tumors used in this experiment were obtained from a previous chemoprevention study (6). Briefly, 21 day-old rats received a single i.p. injection of 50 mg MNU/kg body weight and were distributed into four different groups: Group 1 (CO group) received a diet with 20% of CO; Group 2 (CO+Tam group) received a diet with 20% of CO, and 100 μg/Kg s.c. 5 days/week of tamoxifen; Group 3 (FO group) received a diet with 17% of FO and 3% of CO; Group 4 (FO+Tam group) received 17% of FO and 3% of CO and tamoxifen. After week 8 of treatment, rats were euthanized by CO2 asphyxiation, and the palpable tumors were dissected and cut in half. One half was frozen in liquid nitrogen and kept at −80°C for molecular analysis and the other was fixed in 10% neutral buffered formalin, processed and paraffin embedded. Six micrometer sections were processed for routine H&E staining and tumor diagnosis. Only cribriform tumors were selected for RNA isolation, microarray and real time PCR analysis to exclude possible genomic expression heterogeneity among different tumor types.
Gene expression array
Total RNA was isolated from cribriform tumors using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA) following manufacture's protocol. After DNase I treatment, RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed by capillary electrophoresis with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Two hundred nanograms of total RNA from three cribriform tumors per group were processed for microarray hybridization using Quick Amp Labeling Kit-one color (Agilent Technologies, Palo Alto, CA) following the manufacturer's protocol. Labeled cRNAs were hybridized to microarray slides (Whole rat genome-4X44K oligo microarrays-G4112F, Agilent Technologies) at 65°C for 17 hours. Slides were then washed and scanned following Agilent's recommendations. The digitized images were decoded and submitted to quality control tests through Feature Extractor v9.5.3.1 using the default protocol GE1-v5_95_Feb07 (Agilent Technologies, Palo Alto, CA).
Microarray data analyses
Preprocessing and differential expression analyses of microarray data was performed using LIMMA (Linear Methods for Microarray data Analysis) package (10) in the R/Bioconductor platform. Mean signal intensities were background-corrected using the normexp method, and quantile normalization was performed. Prior to differential expression analysis, a non-specific filter was applied, in which probes without a valid Entrez ID annotation were excluded. In addition, probes with signals close to the background noise level for all 4 groups under consideration were excluded. About 11,000 probes remained for further analysis. Empirical Bayes moderated t-statistics was used to assess significance among treatments (CO vs. CO+Tam; FO vs. FO+Tam and CO vs. FO comparisons). Genes with fold-changes of at least 3.0 at p < 0.01 were considered differentially expressed and used for bioinformatics analyses. Gene Ontology (GO) (11) enrichment analysis was performed using GeneSpring v.11.0 (Agilent Technologies, Palo Alto, CA) and biological processes with p value < 0.05 were considered enriched.
Validation by real time PCR
In order to validate the expression of selected genes found in the microarray analysis (Supplemental Table 1), real time PCR was performed on the same RNA samples used for microarray, as previously described by our group (12). Total RNA was reverse transcribed using the ABI High Capacity cDNA archive kit (Applied Biosystems, Foster City, CA). Standard curves were made using serial dilutions from pooled cDNA samples. Real time PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol and run on the ABI Prism 7300 Sequence Detection System. A TaqMan® assay was performed for RT1-Aw2 mRNA, following the manufacturer's instructions (Applied Biosystems). Quantification was performed for each gene and normalized to a housekeeping gene (18S ribosomal RNA). Normalized expression values were log2 transformed, analysis of variance (ANOVA), and Tukey's post hoc test were performed. Statistical analysis was performed in the JMP 7 software (SAS Institute, Cary NC). Differences were considered statistically significant with a P value less than 0.05.
RESULTS
The effects of our interventions on tumorigenesis and tumor multiplicity have been described in our previous report (6). We report herein the transcriptomic changes occurring in the tumors which developed after tamoxifen treatment and a concomitant CO or FO diet. The dietary influence on tamoxifen's effects was assessed by comparing the CO vs. CO+Tam and FO vs. FO+Tam groups. The comparison of CO vs. FO was performed to evaluate the effects of the diet per se. We performed 12 arrays in which, after quality control analysis, normalization and preprocessing of the microarray data, 11,079 out of 44,000 probes remained for our analysis. All the microarray data was deposited in the Gene Expression Omnibus (GEO) public repository under the series record GSE27465. We used enriched Gene Ontology analysis and analysis of the relation of each differentially expressed gene with cancer-related processes. After analysis, a set of genes of interest was chosen for further real time PCR validation.
Number of differentially expressed genes
Thirty two genes were differentially expressed in the comparison CO vs. CO+Tam (Supplemental Table 2), and 58 genes were differentially expressed between FO vs. FO+Tam (Supplemental Table 3). In the comparison CO vs. FO, 19 differentially expressed genes were found (Supplemental Table 4). An indirect analysis of the data was performed in order to assess the effects of tamoxifen independently of the diet. By studying the commonality between the effects of tamoxifen in a CO-rich diet (CO vs. CO+Tam comparison) and the effects of tamoxifen in FO-rich diet (FO vs. FO+Tam comparison), three genes were found to be differentially expressed by tamoxifen in either CO- or FO-rich diets.
Genes directly related to breast cancer profile
Differences in gene expression were found in the comparisons between FO vs. FO+Tam and CO vs. CO+Tam (H19, Igf2, Sncg, Thrsp, Wnt5b, Lcn2, Aqp1, Mme (CD10), Pdgfra, Lrrn1, Fmod, Chl1, Aspn, Spock2, Ptms and LOC298116). These genes play an active role in breast cancer progression, stage, prognosis, metastasis and disease-free survival. Regarding the effects of the diet per se in the expression profile of the tumor cells, genes that are typically expressed in tumors with a more differentiated phenotype were noticed in the FO-treated group than in the CO-treated group (SerpinB10, Apod and Wisp2).
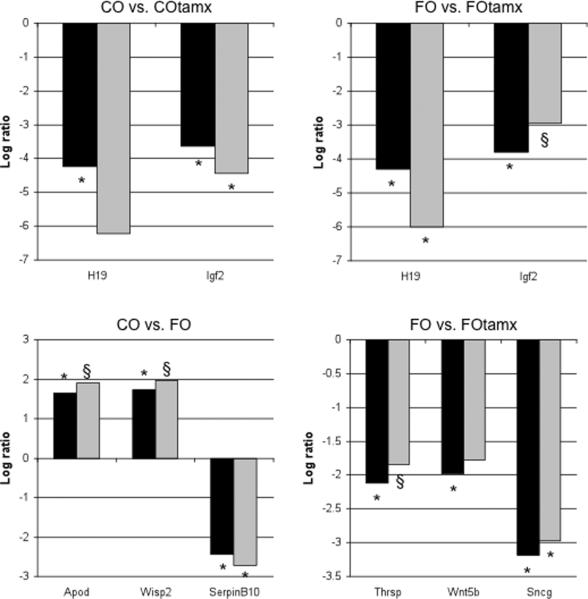
We have already reported that the administration of tamoxifen to CO- or FO-fed rats reduces mammary tumor incidence (6). In order to test the transcriptomic changes that were associated with these effects, eight of the differentially expressed genes directly related to the breast cancer profile were selected for validation by real time PCR: H19, Igf2, Serpinb10, Wisp2, Apod, Sncg, Thrsp and Wnt5b. H19 and Igf2 genes were selected in order to evaluate the effects of tamoxifen independently of the diet. Specifically, Thrsp, Sncg and Wnt5b mRNA were examined as genes responsible for tumor growth impairment, whereas SerpinB10, Wisp2 and Apod mRNAs as markers of differentiation in tumors.
Figure 1 shows the real time PCR results of the genes related to the breast cancer profile found to be differentially expressed in the microarray. H19 mRNA expression was decreased in the FO+Tam group. Moreover, Igf2 mRNA was decreased in CO+Tam group. The down-regulation of Igf2 and H19 transcript levels in the FO+Tam and CO+Tam groups was statistically significant in the microarray (P<0.05) but not in the real time PCR (0.05<P<0.15). FO treatment decreased SerpinB10 mRNA expression when compared to CO treatment. We also found a statistically significant increase in the expression of Apod and Wisp2 in the microarray data (P<0.05); although the real time PCR data shows the same trend, the difference was not statistically significant (P=0.058 and P=0.13, respectively; Fig. 1). Finally, FO+Tam treatment reduced Sncg mRNA expression both in the microarray data and in the real time PCR validation when compared to the FO treatment (Fig. 1, P<0.05). In the same comparison, the down-regulation of Wnt5b and Thrsp expression was statistically significant only in the microarray.
Figure 1.
Side-by-side log2 values of mRNA expression in microarray (Black bars) and real time PCR (grey bars) of genes directly related to the tumor profile. *p<0.05; § 0.05<p<0.20, with fold change > 3.0 (log2>1.58).
Genes related to immune response
GO enrichment analysis showed that fish oil treatment modified several genes related to immune response in comparison to CO-treated group (Table 1). Based on the GO classification and on previous reports, we chose two of the genes differentially expressed by FO to be validated by real time PCR (RT1-Aw2 and Irf7). Real time PCR data confirmed the increased mRNA expression for the gene Irf7 found in the microarray data in the FO vs CO group (fold change 4.99, P<0.05). The differential expression of RT1-Aw2 was not confirmed in the comparison CO vs. FO (data not shown).
Table 1.
Enrichment of GO terms (p<0.05) by differentially expressed genes between CO vs. FO treatments.
| GO:0006955 - Immune Response (p=3.66×10-4) | |||
| GO.0002376 - Immune System Process (p=0.003) | |||
|
| |||
| Probe name | Symbol | Gene name | |
| A_43_P15750 | Cfd | complement factor D (adipsin) | |
| A_44_P1039994 | Irf7 | interferon regulatory factor 7 | |
| A_44_P214846 | M×2 | myxovirus (influenza virus) resistance 2 | |
| A_44_P196401 | RT1-Aw2 | RT1 class Ib, locus Aw2 | |
| A_44_P274061 | RT1-CE3 | RT1 class I, CE3 | |
| A_44_P867246 | RT1-CE16 | RT1 class I, CE16 | |
| A_44_P422630 | Serping1 | serine (or cysteine) peptidase inhibitor, clade G, member 1 | |
|
| |||
| GO:0045087 - Innate Immune Response (p=0.01) | |||
|
| |||
| Probe name | Symbol | Gene name | |
| A_43_P15750 | Cfd | complement factor D (adipsin) | |
| A_44_P214846 | M×2 | myxovirus (influenza virus) resistance 2 | |
| A_44_P422630 | Serping1 | serine (or cysteine) peptidase inhibitor, clade G, member 1 | |
|
| |||
| GO:0019882 - Antigen Processing and Presentation (p=0.02) | |||
|
| |||
| Probe name | Symbol | Gene name | |
| A_44_P196401 | RT1-Aw2 | RT1 class Ib, locus Aw2 | |
| A_44_P274061 | RT1-CE3 | RT1 class I, CE3 | |
| A_44_P867246 | RT1-CE16 | RT1 class I, CE16 | |
|
| |||
| GO:0045861 - Negative Regulation of Proteolysis (p=0.03) | |||
| GO:0009895 - Negative Regulation of Catabolic Process (p=0.04) | |||
|
| |||
| Probe name | Symbol | Gene name | |
| A_44_P1004757 | Serpinb10 | serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 | |
| A_44_P422630 | Serping1 | serine (or cysteine) peptidase inhibitor, clade G, member 1 | |
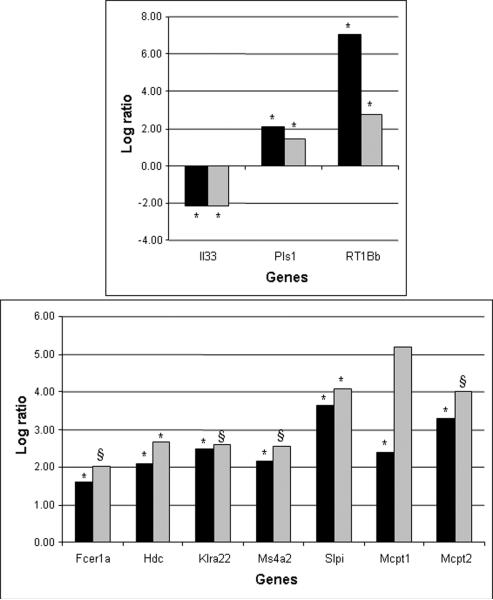
Similarly, enriched GO analysis showed that the transcripts differentially expressed in the FO+Tam group in comparison to FO group were related to immune response (Table 2). Considering the interaction and the function of these genes in the immune response, seven mRNA (Fcer1a, Hdc, Klra22, Mcpt1, Mcpt2, Ms4a2 and Slpi) were selected to be quantified by real time PCR (Figure 2). Hdc and Slpi mRNA levels were statistically significantly increased in the microarray data and validated by real time RT-PCR. mRNA levels of Fcer1a (P=0.20), Klra22 (P=0.17), Ms4a2 (P=0.07), Mcpt1 and Mcpt2 (p=0.13) were not statistically significant (Figure 2) when assessed by real time PCR. In the comparison between CO vs. CO+Tam, there were three differentially regulated genes related to immune response (RT1-Bb, Pls and Il33), and they were confirmed by real time PCR (Fig. 2).
Table 2.
Enrichment of GO terms (p<0.05) by differentially expressed genes between FO vs. FO+Tam treatments.
| GO:0002526 - Acute Inflammatory Response (p=1.38×10-4) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_43_P21634 | C4b | complement component 4B (Childo blood group) | |
| A_44_P419064 | Cfb | complement factor B | |
| A_44_P468468 | Lbp | lipopolysaccharide binding protein | |
| A_42_P661600 | Reg3a | regenerating islet-derived 3 alpha | |
| A_44_P271658 | Reg3b | regenerating islet-derived 3 beta | |
| A_44_P524718 | Serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | |
|
| |||
| GO:0006954 - Inflammatory Response (p=1.72×10-4) | |||
| GO:0006952 - Defense Response (p=0.004) | |||
| GO:0009611 - Response to Wounding (p=0.006) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_44_P1052607 | Aox1 | aldehyde oxidase 1 | |
| A_43_P21634 | C4b | complement component 4B (Childo blood group) | |
| A_44_P304323 | Cc15 | chemokine (C-C motif) ligand 5 | |
| A_44_P419064 | Cfb | complement factor B | |
| A_44_P468468 | Lbp | lipopolysaccharide binding protein | |
| A_43_P15496 | Ms4a2 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | |
| A_42_P661600 | Reg3a | regenerating islet-derived 3 alpha | |
| A_44_P271658 | Reg3b | regenerating islet-derived 3 beta | |
| A_44_P524718 | Serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | |
|
| |||
| GO:0009605 - Response to External Stimulus (p=0.002) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_44_P393531 | Adipoq | adiponectin, C1Q and collagen domain containing | |
| A_44_P1017367 | Alb | albumin | |
| A_44_P1052607 | Aox1 | aldehyde oxidase 1 | |
| A_43_P21634 | C4b | complement component 4B (Childo blood group) | |
| A_44_P304323 | Ccl5 | chemokine (C-C motif) ligand 5 | |
| A_44_P419064 | Cfb | complement factor B | |
| A_44_P428326 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | |
| A_44_P468468 | Lbp | lipopolysaccharide binding protein | |
| A_43_P15496 | Ms4a2 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | |
| A_42_P661600 | Reg3a | regenerating islet-derived 3 alpha | |
| A_44_P271658 | Reg3b | regenerating islet-derived 3 beta | |
| A_44_P524718 | Serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | |
|
| |||
| GO:0006953 - Acute-Phase Response (p=0.004) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_44_P468468 | Lbp | lipopolysaccharide binding protein | |
| A_42_P661600 | Reg3a | regenerating islet-derived 3 alpha | |
| A_44_P271658 | Reg3b | regenerating islet-derived 3 beta | |
| A_44_P524718 | Serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | |
|
| |||
| GO:0043306 - Positive Regulation of Mast Cell Degranulation (p=0.017) | |||
| GO:0033005 - PositiveRregulation of Mast Cell Activation (p=0.017) | |||
| GO:0043302 - Positive Regulation of Leukocyte Degranulation (p=0.017) | |||
| GO:0043304 - Regulation of Mast Cell Degranulation (p=0.027) | |||
| GO:0033003 - Regulation of Mast Cell Activation (p=0.027) | |||
| GO:0043300 - Regulation of Leukocyte Degranulation (p=0.04) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_44_P461402 | Fcer1a | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | |
| A_43_P15496 | Ms4a2 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | |
|
| |||
| GO:0048583 - Regulation of Response to Stimulus (p=0.024) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_43_P21634 | C4b | complement component 4B (Childo blood group) | |
| A_44_P419064 | Cfb | complement factor B | |
| A_43_P12786 | Fabp4 | fatty acid binding protein 4, adipocyte | |
| A_44_P461402 | Fcer1a | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | |
| A_43_P15496 | Ms4a2 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | |
| A_43_P20937 | Tmprss6 | transmembrane protease, serine 6 | |
|
| |||
| GO:0006950 - Response to Stress (p=0.027) | |||
|
|
|||
| Probe name | Symbol | Gene name | |
| A_44_P393531 | Adipoq | adiponectin, C1Q and collagen domain containing | |
| A_44_P1017367 | Alb | albumin | |
| A_44_P1052607 | Aox1 | aldehyde oxidase 1 | |
| A_43_P21634 | C4b | complement component 4B (Childo blood group) | |
| A_44_P244851 | Car3 | carbonic anhydrase 3 | |
| A_44_P304323 | Cc15 | chemokine (C-C motif) ligand 5 | |
| A_44_P419064 | Cfb | complement factor B | |
| A_44_P461402 | Fcer1a | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | |
| A_44_P468468 | Lbp | lipopolysaccharide binding protein | |
| A_43_P15496 | Ms4a2 | membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | |
| A_42_P661600 | Reg3a | regenerating islet-derived 3 alpha | |
| A_44_P271658 | Reg3b | regenerating islet-derived 3 beta | |
| A_44_P524718 | Serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | |
Figure 2.
Side-by-side log2 values of mRNA expression in microarray (Black bars) and real time PCR (grey bars) of genes related to the immune response. *p<0.05; § 0.05<p<0.20, with fold change > 3.0 (log2>1.58).
DISCUSSION
In the present study, it was observed that FO diet alone and in combination with Tam treatment is responsible for changes in the transcriptomic profile of mammary tumors that may influence the behavior of this malignancy. The changes observed are mainly in transcripts which encode for proteins related to tumor differentiation and progression and immune response.
We studied the expression of genes related to tumor differentiation both in animals fed a FO and CO diet. The microarray data show that the FO treatment decreased SerpinB10 mRNA expression when compared to CO treatment. We also found a statistically significant increase in the expression of Apod and Wisp2 in the microarray data. Wisp2 expression gradually decreases as breast cancer progresses from a well-differentiated to a poorly differentiated stage (13). This is confirmed by experiments that show high expression of Wisp2 in less aggressive human breast cancer cells (MCF7 and ZR-75-1), minimal expression in moderately aggressive breast cancer cells (SKBR-3) and no expression in highly aggressive breast cancer cells (MDA-MB-231) (14). Based on these results, it is possible that an Wisp2 increased expression in the present study indicates that tumors in FO-treated group are more differentiated than those observed in the CO-treated group. Likewise, breast tumors require a certain degree of differentiation to produce the Apod protein (15). As shown herein, this mRNA is increased in tumors of animals treated with FO. In addition, it has been shown a correlation of low Apod levels and a shorter relapse-free survival and poor survival of patients with breast cancer (15). Furthermore, the decrease in mRNA for SerpinB10 in the FO group may predict either a more differentiated mammary tumor, a reduction in the cellular proliferation and/or an increase of apoptosis. Decrease of SerpinB10 mRNA levels was observed in NRP-152 and NRP 154 prostatic epithelial cell lines after induction of differentiation (16). An up-regulation of this gene was observed in HL-60 cancer cells compared to normal myeloid cells (17). The trend of dysregulation of such transcripts in our study suggests that mammary tumors developed in FO-treated rats are more differentiated than tumors developed in CO-fed animals.
Tumors in the Tam-treated rats had decreased expression of H19 and Igf2 mRNA independently of the type of dietary fat fed to the animals (either FO or CO). In vitro studies correlated increased H19 with a more malignant cell phenotype, as assessed by colony formation capacity in soft-agar and enhanced adhesiveness in type I collagen (18). Adriaenssens et al. (19) showed that the non-coding gene H19 promotes cell progression of breast cancer cells. Similarly, Igf2 is a well-established growth factor both in vitro and in vivo (20). Cells with disrupted Igf2 function, when injected in transgenic mice, showed reduction in tumor cell growth, reduced malignancy and a significant number of apoptotic bodies (21). By using mRNA in situ hybridization, Manni et al. (22) found that the expression of Igf2 mRNA is under positive endocrine regulation, since its levels decreased in regressing tumors following ovariectomy, and the normal expression levels were re-established after estradiol repletion. In line with the results presented herein, in a T61 human breast cancer xenograft model, treatment of cells with tamoxifen produced a ten-fold reduction in the baseline level of Igf2 mRNA (23).
In addition to the changes that Tam caused irrespective of the diet, the combination of FO and Tam treatment altered the expression of genes that may lead to a better prognosis of mammary cancer. Importantly, the combination of FO and Tam affect the expression of genes that involved in tumor growth. Gamma Synuclein (Sncg) is highly associated with breast cancer and ovarian cancer progression. Sncg is undetectable in normal breast tissue and in most of the benign lesions, whereas this gene is expressed in breast cancer with a positive correlation with stage, poorer prognosis, metastasis, and negative correlation with disease-free survival and overall survival (24). Jiang et al. demonstrated that Sncg strongly stimulated the ligand-dependent transcriptional activity of estrogen receptor-α (ER-α) in breast cancer cells (25). They showed that overexpression of this protein stimulated the ligand-dependent cell proliferation, and suppression of endogenous Sncg expression significantly inhibited cell growth in response to estrogen. Over-expression of Sncg also increases motility and invasiveness of MDA-MB 435 cells, and metastatic potential in vivo (26). It is plausible that the decreased expression of Sncg mRNA by tamoxifen in presence of FO diet may be impairing tumor growth and/or reducing its metastatic potential, being the responsible for the enhanced chemopreventive efficacy of this combination regimen described previously (6).
Based on the gene expression pattern, some genes related to immune response are dysregulated by some of the treatments. In this context, real time PCR confirmed a 4.99-fold increase of Irf7 mRNA expression by FO treatment (P<0.05, data not shown). It is well established that Irf7 is a major regulator of IFN gene expression (27) and, in turn, IFN treatment can augment anti-tumor properties. Also, Irf7 increases antitumor activities of macrophages (28). It has been shown that BRCA1 is required for IFN-γ-mediated induction of Irf-7 and that BRCA1 sensitizes breast cancer cell lines to IFN-γ-mediated apoptosis (29). The supposition that a FO diet may improve the immune response against tumors is supported by improved immune response against tumors after n-3 PUFAs administration to animals, as reported in the literature (30, 31).
Surprisingly, FO+Tam strongly increased the expression of several mRNAs that may be related to the Th2 pattern of immune response. Generally, the augmentation of a Th2 response down-regulates the Th1 response (32). It is believed that a polarized immune response towards the Th2 pattern is related to a reduced cellular immunity against cancer (33–35). In fact, the increased transcript levels of Hdc and Slpi genes (P<0.05), and the trend of increase of Fcer1a, Klra22, Ms4a2 and Mcpt2 genes by the FO+Tam treatment suggests an increment of this pattern of response.
Fcer1a and Ms4a2 are subunits of Fc receptor located on mast cells (36, 37). The up-regulation of these genes triggers the IgE-mediated immune response. As a consequence, T-cell releases mediators that initiate the inflammatory response and allergic reaction (36, 37). Histamine is one of the mediators produced by mast cells and basophils, through the action of histidine decarboxylase (Hdc) (36, 38). Some studies show an increase in Hdc activity in breast cancer, in comparison to normal tissue. Moreover, histamine promotes migration of breast cancer cells, which may have an important role in invasion and metastasis and shifts pattern of immune response from Th1 to Th2 (38). The increase of Fcer1a, Ms4a2 and Hdc mRNAs in the FO+Tam group may in turn increase histamine release and the Th2 response pattern in the tumor microenvironment. Finally, Slpi mRNA is increased in FO+Tam group. This gene is commonly increased in gastric, ovarian and lung carcinomas; its serum level is positively correlated with tumor stage, and it may promote tumor formation and development through the prevention of the formation of endostatin, an anti-angiogenic factor (39–41). These data led us to hypothesize that mammary tumor cells in FO+Tam-treated group escaped from the immune response against tumors through a possible up-regulation of Th2 pattern of immune response in the tumor microenvironment. Further studies are needed to confirm or discard this hypothesis.
In conclusion, treatment of female rats with FO resulted in differential expression of several mRNAs that encode genes that promote more differentiated tumors and a more efficient immune response against tumors, when compared to tumors of CO fed counterparts. In addition, cribriform tumors of FO-fed animals that received Tam showed decreased mRNA for genes directly related to tumor growth and metastasis; thus, Tam treatment was more efficient in fish oil than in corn oil background. However, FO+Tam treatment increased transcripts of several molecules involved in the immune response.
Supplementary Material
ACKNOWLEDGEMENTS
L.T. Bidinotto was the recipient of a one-year fellowship PDEE (CAPES/Brazil -0675091). The support and advice of Dr. Luis Fernando Barbisan are greatly appreciated. We thank Daniel Hannon for his technical support. This project was supported by the Komen Foundation Grant (KG081632).
REFERENCES
- 1.American Cancer Society . Cancer Facts and Figures. American Cancer Society; Atlanta: 2010. [Google Scholar]
- 2.Cleator SJ, Ahamed E, Coombes RC, Palmieri C. A 2009 update on the treatment of patients with hormone receptor-positive breast cancer. Clin Breast Cancer. 2009;9(Suppl 1):S6–S17. doi: 10.3816/CBC.2009.s.001. [DOI] [PubMed] [Google Scholar]
- 3.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 4.Vogel VG. Selective Estrogen Receptor Modulators and Aromatase Inhibitors for Breast Cancer Chemoprevention. Curr Drug Targets. 2010 Dec 15; doi: 10.2174/138945011798184164. Epub. [DOI] [PubMed] [Google Scholar]
- 5.White IN. The tamoxifen dilemma. Carcinogenesis. 1999;20:1153–1160. doi: 10.1093/carcin/20.7.1153. [DOI] [PubMed] [Google Scholar]
- 6.Manni A, Xu H, Washington S, Aliaga C, Cooper T, et al. The impact of fish oil on the chemopreventive efficacy of tamoxifen against development of N-methyl-N-nitrosourea induced rat mammary carcinogenesis. Cancer Prev Res (Phila) 2010;3:322–330. doi: 10.1158/1940-6207.CAPR-09-0173. [DOI] [PubMed] [Google Scholar]
- 7.Shao Y, Pardini L, Pardini RS. Dietary menhaden oil enhances mitomycin C antitumor activity toward human mammary carcinoma MX-1. Lipids. 1995;30:1035–1045. doi: 10.1007/BF02536289. [DOI] [PubMed] [Google Scholar]
- 8.Hardman WE, Avula CP, Fernandes G, Cameron IL. Three percent dietary fish oil concentrate increased efficacy of doxorubicin against MDA-MB 231 breast cancer xenografts. Clin Cancer Res. 2001;7:2041–2049. [PubMed] [Google Scholar]
- 9.Calviello G. Dietary omega-3 polyunsaturated fatty acids and cancer. Springer; Dordrecht, New York: 2010. [Google Scholar]
- 10.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopinathan L, Hannon DB, Smith RW, Iii, Peters JM, Vanden Heuvel JP. Regulation of peroxisome proliferator-activated receptors by e6-associated protein. PPAR Res. 2008;2008:746935. doi: 10.1155/2008/746935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee S, Dhar G, Haque I, Kambhampati S, Mehta S, et al. CCN5/WISP-2 expression in breast adenocarcinoma is associated with less frequent progression of the disease and suppresses the invasive phenotypes of tumor cells. Cancer Res. 2008;68:7606–7612. doi: 10.1158/0008-5472.CAN-08-1461. [DOI] [PubMed] [Google Scholar]
- 14.Dhar G, Banerjee S, Dhar K, Tawfik O, Mayo MS, et al. Gain of oncogenic function of p53 mutants induces invasive phenotypes in human breast cancer cells by silencing CCN5/WISP-2. Cancer Res. 2008;68:4580–4587. doi: 10.1158/0008-5472.CAN-08-0316. [DOI] [PubMed] [Google Scholar]
- 15.Diez-Itza I, Vizoso F, Merino AM, Sanchez LM, Tolivia J, et al. Expression and prognostic significance of apolipoprotein D in breast cancer. Am J Pathol. 1994;144:310–320. [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart LV, Song K, Hsing AY, Danielpour D. Regulation of trespin expression by modulators of cell growth, differentiation, and apoptosis in prostatic epithelial cells. ExpCell Res. 2003;284:303–315. doi: 10.1016/s0014-4827(02)00037-x. [DOI] [PubMed] [Google Scholar]
- 17.Ulger C, Toruner GA, Alkan M, Mohammed M, Damani S, et al. Comprehensive genome wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet. 2003;147:28–35. doi: 10.1016/s0165-4608(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 18.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 19.Adriaenssens E, Lottin S, Dugimont T, Fauquette W, Coll J, et al. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18:4460–4473. doi: 10.1038/sj.onc.1202819. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen AA, Cullen KJ. Paracrine/autocrine regulation of breast cancer by the insulin like growth factors. Breast Cancer Res Treat. 1998;47:219–233. doi: 10.1023/a:1005903000777. [DOI] [PubMed] [Google Scholar]
- 21.Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–418. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- 22.Manni A, Badger B, Wei L, Zaenglein A, Grove R, et al. Hormonal regulation of insulin like growth factor II and insulin-like growth factor binding protein expression by breast cancer cells in vivo: evidence for stromal epithelial interactions. Cancer Res. 1994;54:2934–2942. [PubMed] [Google Scholar]
- 23.Brunner N, Yee D, Kern FG, Spang-Thomsen M, Lippman ME, et al. Effect of endocrinetherapy on growth of T61 human breast cancer xenografts is directly correlated to a specific down-regulation of insulin-like growth factor II (IGF-II) Eur J Cancer. 1993;29A:562–569. doi: 10.1016/s0959-8049(05)80152-2. [DOI] [PubMed] [Google Scholar]
- 24.Guo J, Shou C, Meng L, Jiang B, Dong B, et al. Neuronal protein synuclein gamma predicts poor clinical outcome in breast cancer. Int J Cancer. 2007;121:1296–1305. doi: 10.1002/ijc.22763. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Liu YE, Lu A, Gupta A, Goldberg ID, et al. Stimulation of estrogen receptor signaling by gamma synuclein. Cancer Res. 2003;63:3899–3903. [PubMed] [Google Scholar]
- 26.Jia T, Liu YE, Liu J, Shi YE. Stimulation of breast cancer invasion and metastasis by synuclein gamma. Cancer Res. 1999;59:742–747. [PubMed] [Google Scholar]
- 27.Ning S, Pagano JS, Barber GN. IRF7: Activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romieu-Mourez R, Solis M, Nardin A, Goubau D, Baron-Bodo V, et al. Distinct roles for IFN regulatory factor (IRF)-3 and IRF-7 in the activation of antitumor properties of human macrophages. Cancer Res. 2006;66:10576–10585. doi: 10.1158/0008-5472.CAN-06-1279. [DOI] [PubMed] [Google Scholar]
- 29.Andrews HN, Mullan PB, McWilliams S, Sebelova S, Quinn JE, et al. BRCA1 regulates the interferon gamma-mediated apoptotic response. J Biol Chem. 2002;277:26225–26232. doi: 10.1074/jbc.M201316200. [DOI] [PubMed] [Google Scholar]
- 30.Berquin IM, Edwards IJ, Chen YQ. Multi-targeted therapy of cancer by omega-3 fatty acids. Cancer Lett. 2008;269:363–377. doi: 10.1016/j.canlet.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calder PC, Yaqoob P, Thies F, Wallace FA, Miles EA. Fatty acids and lymphocyte functions. Br J Nutr. 2002;87(Suppl 1):S31–48. doi: 10.1079/bjn2001455. [DOI] [PubMed] [Google Scholar]
- 32.Dobrzanski MJ, Reome JB, Dutton RW. Type 1 and type 2 CD8+ effector T cell subpopulations promote long-term tumor immunity and protection to progressively growing tumor. J Immunol. 2000;164:916–925. doi: 10.4049/jimmunol.164.2.916. [DOI] [PubMed] [Google Scholar]
- 33.Sheu BC, Lin RH, Lien HC, Ho HN, Hsu SM, et al. Predominant Th2/Tc2 polarity of tumor-infiltrating lymphocytes in human cervical cancer. J Immunol. 2001;167:2972–2978. doi: 10.4049/jimmunol.167.5.2972. [DOI] [PubMed] [Google Scholar]
- 34.Dobrzanski MJ, Reome JB, Hylind JC, Rewers-Felkins KA. CD8-mediated type 1antitumor responses selectively modulate endogenous differentiated and nondifferentiated T cell localization, activation, and function in progressive breast cancer. J Immunol. 2006;177:8191–8201. doi: 10.4049/jimmunol.177.11.8191. [DOI] [PubMed] [Google Scholar]
- 35.Hegyesi H, Colombo L, Pallinger E, Toth S, Boer K, et al. Impact of systemic histamine deficiency on the crosstalk between mammary adenocarcinoma and T cells. J Pharmacol Sci. 2007;105:66–73. doi: 10.1254/jphs.fp0070636. [DOI] [PubMed] [Google Scholar]
- 36.Joshi N, Johnson LL, Wei WQ, Abnet CC, Dong ZW, et al. Gene expression differences in normal esophageal mucosa associated with regression and progression of mild and moderate squamous dysplasia in a high-risk Chinese population. Cancer Res. 2006;66:6851–6860. doi: 10.1158/0008-5472.CAN-06-0662. [DOI] [PubMed] [Google Scholar]
- 37.Kraft S, Kinet JP. New developments in FcepsilonRI regulation, function and inhibition. Nat Rev Immunol. 2007;7:365–378. doi: 10.1038/nri2072. [DOI] [PubMed] [Google Scholar]
- 38.Medina V, Croci M, Crescenti E, Mohamad N, Sanchez-Jimenez F, et al. The role of histamine in human mammary carcinogenesis: H3 and H4 receptors as potential therapeutic targets for breast cancer treatment. Cancer Biol Ther. 2008;7:28–35. doi: 10.4161/cbt.7.1.5123. [DOI] [PubMed] [Google Scholar]
- 39.Cheng WL, Wang CS, Huang YH, Liang Y, Lin PY, et al. Overexpression of a secretory leukocyte protease inhibitor in human gastric cancer. Int J Cancer. 2008;123:1787–1796. doi: 10.1002/ijc.23746. [DOI] [PubMed] [Google Scholar]
- 40.Devoogdt N, Rasool N, Hoskins E, Simpkins F, Tchabo N, et al. Overexpression of protease inhibitor-dead secretory leukocyte protease inhibitor causes more aggressive ovarian cancer in vitro and in vivo. Cancer Sci. 2009;100:434–440. doi: 10.1111/j.1349-7006.2009.01076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devoogdt N, Hassanzadeh Ghassabeh G, Zhang J, Brys L, De Baetselier P, et al. Secretory leukocyte protease inhibitor promotes the tumorigenic and metastatic potential of cancer cells. Proc Natl Acad Sci U S A. 2003;100:5778–5782. doi: 10.1073/pnas.1037154100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.