SUMMARY
Aberrant Skp2 signaling has been implicated as a driving event in tumorigenesis. Although the underlying molecular mechanisms remain elusive, cytoplasmic Skp2 correlates with more aggressive forms of breast and prostate cancers. Here, we report that Skp2 is acetylated by p300 at K68 and K71, which is a process that can be antagonized by the SIRT3 deacetylase. Inactivation of SIRT3 leads to elevated Skp2 acetylation, which leads to increased Skp2 stability through impairment of the Cdh1-mediated proteolysis pathway. As a result, Skp2 oncogenic function is increased, whereby cells expressing an acetylation-mimetic mutant display enhanced cellular proliferation and tumorigenesis in vivo. Moreover, acetylation of Skp2 in the nuclear localization signal (NLS) promotes its cytoplasmic retention, and cytoplasmic Skp2 enhances cellular migration through ubiquitination and destruction of E-cadherin. Thus, our study identifies an acetylation-dependent regulatory mechanism governing Skp2 oncogenic function and provides insight into how cytoplasmic Skp2 controls cellular migration.
INTRODUCTION
The F-box protein S-phase kinase associated protein 2 (Skp2) forms an SCF-type E3 ubiquitin ligase complex by associating with Cullin-1, Skp1, and Rbx1 (Frescas and Pagano, 2008). Previous studies have shown that Skp2 plays an important role in governing cell cycle progression and cell survival by promoting the destruction of numerous tumor suppressor proteins, including p27, p21, p57, p130, and FOXO1 (Cardozo and Pagano, 2004), thereby functioning as a proto-oncogene. Notably, overexpression of Skp2 induces low-grade carcinomas in the mouse prostate (Shim et al., 2003) and facilitates the transformation of Rat1 cells (Gstaiger et al., 2001). Moreover, Skp2 overexpression has been detected in various types of cancers, including lymphomas (Lim et al., 2002) and prostate (Yang et al., 2002) and breast carcinomas (Traub et al., 2006), and it has been associated with poor prognosis as well as tumor metastasis (Li et al., 2004). In agreement with a critical role for Skp2 in tumor progression, Skp2−/− mice are resistant to tumor development induced by loss of p53 or PTEN (Lin et al., 2010). Therefore, a more complete understanding of how Skp2 activity is regulated would benefit not only basic cancer research but also clinical diagnosis and, ultimately, cancer therapy.
We and others have identified APC/Cdh1 as the upstream E3 ligase that promotes Skp2 destruction (Bashir et al., 2004; Wei et al., 2004). However, loss of Cdh1 is not a frequent event in human cancers, whereas elevated phosphoinositide 3-kinase (PI3-K)/Akt signaling is considered a hallmark of more aggressive cancers (Luo et al., 2003). Furthermore, a correlation between Skp2 overexpression and elevated Akt activity has been reported in many carcinomas (Mamillapalli et al., 2001). In agreement with this model, we and others have demonstrated that Akt1, but not Akt2, phosphorylates Skp2 at Ser72, protecting Skp2 from Cdh1-mediated degradation and localizing a pool of Skp2 to the cytoplasm by impairing its nuclear localization signal (NLS) function (Gao et al., 2009; Lin et al., 2009). However, Ser72 in human Skp2 is not conserved in mice and some other species, suggesting the existence of alternative pathways in the regulation of Skp2 function.
It has been reported that Skp2−/− mouse embryonic fibroblasts (MEFs) have significantly impaired migratory capacity compared to their wild-type counterparts (Lin et al., 2009). Moreover, ectopically expressed Skp2 harboring a nuclear export signal (NES) rescues the migration defect, whereas p27 degradation is unaffected (Lin et al., 2009). These results indicate that cytoplasmic Skp2 may control cell migration in a manner that is independent of its role in cell-cycle regulation. In support of this notion, Skp2 cytoplasmic localization has been observed in many clinical tumor samples and is correlated with aggressive malignancy and poor prognosis (Drobnjak et al., 2003; Radke et al., 2005; Signoretti et al., 2002). However, the mechanisms by which Skp2 controls cell migration and potentially tumor metastasis remain unclear.
RESULTS
Skp2 Is Acetylated by p300 at Both K68 and K71 within Its NLS Region
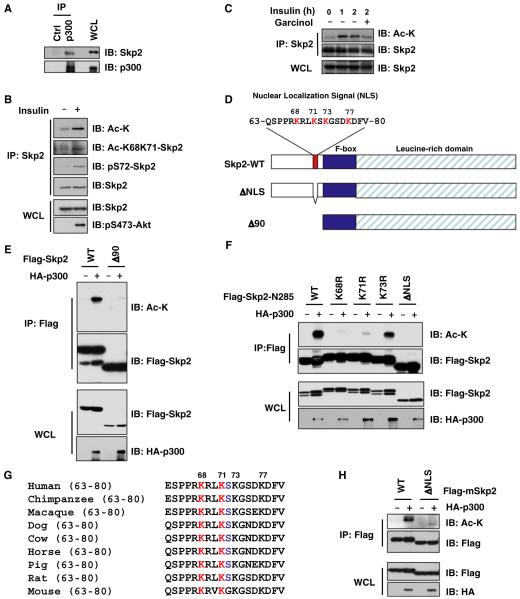
Skp2 was reported to interact with p300 to influence the tumor suppressor function of p53 (Kitagawa et al., 2008). This prompted us to determine whether Skp2 function is modulated by p300, whose acetyl transferase activity is activated by Akt-mediated phosphorylation (Huang and Chen, 2005). In agreement with a previous report (Kitagawa et al., 2008), an interaction between p300 and Skp2 (Figures 1A and S1A available online) is readily detected (Figure 1A). Furthermore, acetylation of Skp2 is detected after ectopic expression of p300, but not other acetyl transferases, which include CBP and GCN5 (Figure S1B). Moreover, acetylation of endogenous Skp2 is observed after induction of p300 acetyl transferase activity through activation of PI3K/Akt (Huang and Chen, 2005) by insulin (Figure 1B). On the other hand, inhibition of p300 enzymatic activity by Garcinol (Balasubramanyam et al., 2004) suppressed insulin-induced Skp2 acetylation (Figure 1C), indicating a critical role for p300 in Skp2 acetylation.
Figure 1. Skp2 Is Acetylated by p300 at K68 and K71.
(A) Immunoblot (IB) analysis of 293T whole-cell lysates (WCL) and anti-p300 immunoprecipitates (IP). Rabbit IgG was used as a negative control for the immunoprecipitation procedure.
(B and C) IB analysis of WCL derived from HeLa cells that were serum starved for 24 hr and then collected after 1 hr following addition of insulin to activate the PI3K/Akt signaling pathway. Cells were pretreated with TSA (2 μM) and NAM (10 mM) for 1 hr before the addition of insulin.
(D) Schematic representation of the Skp2 deletion mutants used in (E) and (F).
(E and F) IB analysis of WCL and anti-Flag IP derived from 293T cells transfected with HA-p300 and various Flag-human Skp2 constructs.
(G) Sequence alignment of the putative acetylation sites K68 and K71 in Skp2 from different species.
(H) IB analysis of WCL and anti-Flag IP derived from 293T cells transfected with HA-p300 and the indicated Flag-mouse Skp2 constructs.
See also Figure S1.
Using deletion constructs, we determined that Skp2 acetylation sites are primarily located within the NLS sequence that contains four lysine residues (Figures 1D, 1E, S1C, and S1D). We next used mutagenesis to demonstrate that Skp2 acetylation occurs at K68 and K71, which are evolutionarily conserved (Figures 1F, 1G, and S1E). K68 acetylation was also confirmed by mass spectrometry (Figure S1F). Moreover, mouse Skp2 is also acetylated by p300 within the conserved NLS (Figure 1H).
SIRT3 Specifically Interacts with and Deacetylates Skp2
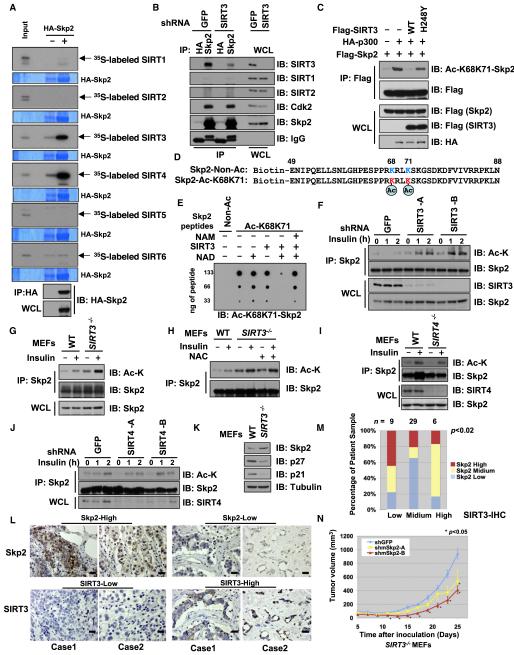
Recent studies have shown that acetylation of numerous proteins, such as FOXO3a (Brunet et al., 2004; Motta et al., 2004), PPARγ (Picard et al., 2004), and p53 (Vaziri et al., 2001), is a dynamic process that can be catalytically reversed by specific deacetylase(s). Because the Sirtuin family of deacetylases has been demonstrated to play critical roles in tumorigenesis and cellular metabolism (Haigis and Sinclair, 2010), we tested whether members of the Sirtuin family could specifically deacetylate Skp2. Among the various Sirtuin family members examined, only SIRT3 and SIRT4 specifically interacted with Skp2 (Figure 2A). As SIRT3 has been recently characterized as a tumor suppressor protein (Finley et al., 2011a; Kim et al., 2010) that specifically interacts with Skp2 (Yang et al., 2011), we further examined a potential role for SIRT3 in regulation of Skp2 acetylation. We detected a specific interaction between Skp2 and SIRT3 (Figures 2B and S2A). Furthermore, ectopic expression of WT-SIRT3, but not a catalytic-inactive mutant (H248Y) of SIRT3 (Schwer et al., 2002), led to a significant decrease of in vivo Skp2 acetylation (Figure 2C) detected by an antibody that can recognize acetylation of Skp2 at K68 and K71 (Figure S1G). Using the anti-Ac-K68K71-Skp2 antibody, we further showed that recombinant SIRT3 efficiently deacetylates Skp2 in vitro, a process that is blocked by nicotinamide (NAM), a pan-Sirtuin family inhibitor (Figures 2D and 2E). In support of a physiological role for SIRT3 in Skp2 deacetylation, depletion of SIRT3 (Figures 2F and 2G), but not SIRT4 (Figures 2I and 2J), led to an increase in acetylation of endogenous Skp2. Furthermore, inhibition of reactive oxygen species (ROS) production with N-acetyl-L-cysteine (NAC) (Finley et al., 2011a) did not cause any noticeable changes in the elevated acetylation of Skp2 in SIRT3−/− cells, arguing that SIRT3 may directly regulate Skp2 acetylation status instead of through modulating ROS production (Figures 2H and S2B) (Bell et al., 2011; Jing et al., 2011). Finally, depletion of SIRT3 (Figures 2K, S2C, and S2D), but not SIRT4 (Figures S2E–S2G), led to a moderate increase in Skp2 abundance. These results indicate that enhanced acetylation stabilizes Skp2. In support of this notion, we observed an inverse correlation between Skp2 and SIRT3 immunohistochemical staining in an array of breast cancer clinical samples (Figures 2L and 2M). This result suggests that loss of SIRT3 may lead to elevated Skp2 expression in breast cancers. In support of a tumor suppressor function for SIRT3 (Finley et al., 2011a; Kim et al., 2010), depletion of SIRT3 facilitates in vivo tumorigenesis in a xenograft model (Figures 2N and S2M). Furthermore, depletion of endogenous Skp2 severely retarded the in vivo tumorigenesis of SIRT3−/− cells (Figures 2N and S2J–S2L), but not the transformed WT-MEFs (Figures S2M and S2N). Together, these results indicate that the Skp2 signaling pathway may be a major route through which SIRT3 executes its tumor suppressor function.
Figure 2. SIRT3 Interacts with and Deacetylates Skp2.
(A) Autoradiography of 35S-labeled Sirtuins bound to HA-Skp2 immunoprecipitated from the transfected 293T cells. Empty vector (EV) transfection was used as a negative control.
(B) IB analysis of WCL and anti-Skp2 IP derived from HeLa cells that were infected with the indicated lentiviral vector. Anti-HA IgG was used as a negative control for the immunoprecipitation procedure.
(C) IB analysis of WCL and anti-Flag IP derived from 293T cells transfected with HA-p300, Flag-Skp2, and the various Flag-SIRT3 constructs.
(D) Schematic representation of the various biotinylated peptides used in (E). Where indicated, the K68 and/or K71 residue is acetylated.
(E) 2 μg of indicated peptides were incubated in the presence or absence of recombinant SIRT3 at 37°C for 2 hr. Where indicated, 1.5 mM NAD or 20 mM NAM were added into the reaction. The indicated amount of peptides after reaction was spotted on nitrocellulose membrane and immunoblotted with the Ac-K68K71-Skp2 antibody.
(F) IB analysis of WCL and anti-Skp2 IP derived from HeLa cells that were infected with the indicated lentiviral vector. Before Skp2 immunoprecipitation, the indicated cells were serum starved for 24 hr and then collected at the indicated time periods following the addition of insulin. Cells were pretreated with TSA (2 μM) for 1 hr before the addition of insulin.
(G–I) IB analysis of WCL and anti-Skp2 IP derived from WT, SIRT3−/− (G–H), or SIRT4−/− (I) MEFs. Before Skp2 immunoprecipitation, the indicated cells were serum starved for 24 hr and then collected 2 hr following addition of insulin. Cells were pretreated with TSA (2 μM) for 1 hr before the addition of insulin. Where indicated, 10 mM ROS inhibitor NAC were added for 16 hr before harvesting for IP (H).
(J) IB analysis of WCL and anti-Skp2 IP derived from HeLa cells that were infected with the indicated lentiviral vector. Before Skp2 immunoprecipitation, the indicated cells were serum starved for 24 hr and then collected at the indicated time periods following the addition of insulin. Cells were pretreated with TSA (2 mM) for 1 hr before addition of insulin.
(K) IB analysis of WCL derived from WT or SIRT3−/− MEFs.
(L and M) Representative images of SIRT3 and Skp2 expression in breast tumor cells as assessed by immunohistochemistry (L). Both Skp2 and SIRT3 levels were classified as low, medium, or high based on the intensities of the IHC staining, and the percentages of patients classified in each category are depicted in the histogram in (M).
(N) Growth curves for the xenograft experiments with the indicated tumor cells that were inoculated subcutaneously. In each flank of nine nude mice, 8 × 106 cells were injected. The visible tumors were measured at the indicated days. Error bars represent ±SEM, and *p < 0.05 (Student’s t test).
See also Figure S2.
Skp2 Protein Is Regulated by p300-Mediated Acetylation
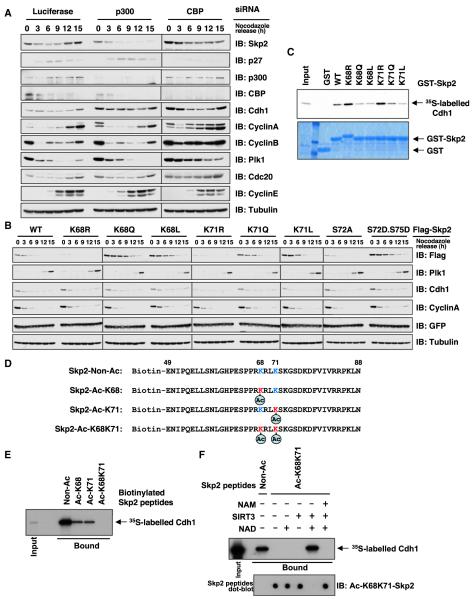
Our previous studies indicated that a region within Skp2 between residues 46 and 90, which contains the Ser72 Akt phosphorylation site as well as the two acetylated lysines K68 and K71, is both necessary and sufficient for its interaction with Cdh1 (Bashir et al., 2004; Wei et al., 2004). Moreover, phosphorylation of Skp2 at Ser72 by Akt impairs Cdh1 interaction with Skp2, leading to its stabilization (Gao et al., 2009; Lin et al., 2009). Thus, we next examined whether Skp2 acetylation at K68 and K71 also interferes with Cdh1-mediated Skp2 degradation. Consistent with a previous report (Turnell et al., 2005), depletion of CBP leads to stabilization of most of the examined Cdh1 downstream substrates, including Cyclin A, Cyclin B, Plk1, and Cdc20 (Figure 3A). Notably, depletion of p300 leads to a significant decrease in Skp2 abundance (Figure S3A), especially in mid to late G1 (Figure 3A). Interestingly, this phenotype seems to be unique to Skp2 because p300 depletion did not cause significant changes in other Cdh1 targets (Turnell et al., 2005). This result is further supported by a reciprocal set of experiments in which Skp2 was stabilized by coexpression of p300 (Figure S3B).
Figure 3. p300-Dependent Acetylation of Skp2 Impairs Cdh1-Mediated Skp2 Proteolysis Pathway.
(A) IB analysis of HeLa cells transfected with the indicated siRNA oligos after synchronization with nocodazole and release at the indicated time periods.
(B) IB analysis of HeLa cells transfected with limited amount of the indicated Flag-Skp2 constructs, along with a green fluorescent protein (GFP) as a transfection control. HeLa cells were synchronized in the M phase with nocodazole and then released into G1 for the indicated time periods.
(C) Autoradiography of 35S-labeled Cdh1 bound to the indicated GST fusion proteins.
(D) Schematic representation of the various biotinylated peptides used in Figures 3E, 3F, 5D, and 5E, which are derived from the Cdh1-interaction motif of Skp2. Where indicated, the K68 and/or K71 residue is acetylated.
(E) Autoradiography of 35S-labeled Cdh1 bound to the indicated biotinylated peptides.
(F) Autoradiography of 35S-labeled Cdh1 bound to the indicated biotinylated peptides that have been subject to SIRT3 in vitro deacetylation assays as described in Figure 2E.
See also Figure S3.
To further understand the role of p300-mediated acetylation in Skp2 stability, various Skp2 constructs were transfected into HeLa cells, and their expression levels were monitored during cell-cycle progression. A K→R replacement was used to generate acetylation-deficient mutants while both K→L (Rea et al., 2000; Schwer et al., 2002) and K→Q replacements (Karanam et al., 2007; Kemper et al., 2009) were used to mimic lysine-acetylation. WT-Skp2 levels are decreased in early- to mid-G1 phase where APC/Cdh1 is highly active. Similar to the phosphomimetic mutant (S72D/S75D), acetylation-mimetic mutants (K68Q, K68L, K71Q, and K71L) are degraded with significantly slower kinetics than WT-Skp2. Similar to S72A, K68R and K71R mutants are degraded more rapidly than WT-Skp2 (Figure 3B). However, it should be noted that the transcription of transfected Skp2 constructs is driven by the cytomegalovirus (CMV) promoter, which is different from the transcription of endogenous Skp2 that can be activated by E2F1 after cells enter the S phase (Figure 3A) (Zhang and Wang, 2006). Thus, they cannot faithfully phenocopy endogenous Skp2 expression in the S phase (Figures 3A and 3B). Using glutathione S-transferase (GST) pull-down analysis, we showed that replacement of K68 or K71 by acetylation-mimetic amino acids disrupts the interaction between Skp2 and Cdh1 (Figure 3C). To eliminate the possible nonspecific effect derived from amino acid substitution, we performed in vitro pull-down experiments with biotinylated peptides derived from the Cdh1-interaction motif of Skp2 (aa 49–88; Figure 3D) to illustrate that acetylation of K68 or K71 significantly reduced the interaction between Cdh1 and Skp2 in a synergistic manner (Figure 3E). Importantly, we further showed that SIRT3 could catalytically remove the acetylation of K68 and K71 from the acetylated Skp2 peptides, restoring the interaction between Skp2 and Cdh1 in vitro (Figure 3F). These results support the notion that p300-mediated acetylation of Skp2 impairs interaction between Skp2 and Cdh1, allowing evasion of Cdh1-mediated proteolysis pathways and subsequent Skp2 stabilization.
Acetylation-Dependent Regulation of Skp2 Oncogenic Functions
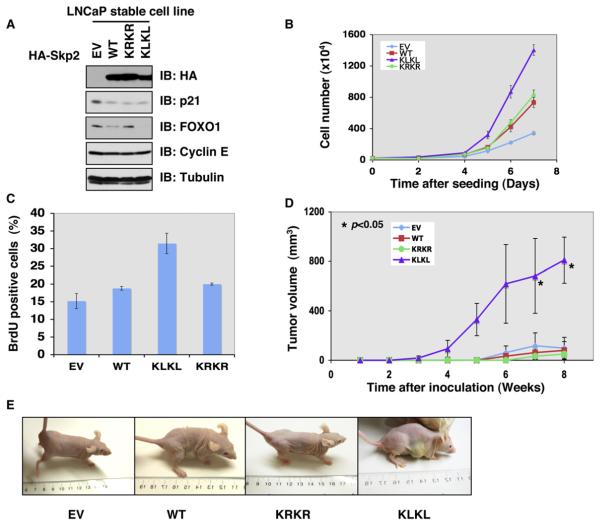
Because overexpression of Skp2 has been associated with tumorigenesis, we next evaluated whether acetylation of Skp2 by p300 also governs Skp2 oncogenic activity. We generated LNCaP cell lines stably expressing various Skp2 constructs (Figure 4A). Because both KQ and KL acetylation-mimetic mutants behave similarly in biochemical and cellular assays (Figures 3B–C and 5B-C), we utilized KL as an acetylation-mimetic and KR as an acetylation-deficient mutant. As illustrated in Figures 4A and S4A, ectopic expression of both the acetylation-deficient (K68R/K71R, denoted as KRKR) and acetylation-mimetic (K68L/K71L, denoted as KLKL) mutants promoted destruction of the Skp2 substrate p21 as efficiently as wild-type Skp2. Interestingly, the acetylation-mimetic mutant has an enhanced capacity in promoting the destruction of FOXO1 (Figure 4A). Furthermore, KLKL-expressing LNCaP cell lines grow more rapidly than other LNCaP stable cell lines (Figure 4B). However, further studies are required to determine whether other substrates, in addition to FOXO1, contribute to this phenotype. Regardless, this phenotype could be due to increased cell cycle progression (Figure S4B), as KLKL-expressing LNCaP cells display elevated bromodeoxyuridine (BrdU) staining (Figure 4C). In support of this, depletion of SIRT3, which results in increased acetylation of Skp2 (Figures 2F and 2G), also leads to increased incorporation of BrdU (Figures S4C and S4D). Ectopic expression of Skp2-KLKL mutant also significantly promotes in vivo tumorigenesis in a xenograft mouse model (Figures 4D and 4E). These results indicate that acetylation of Skp2 positively regulates its oncogenic function, partly through modulating its stability.
Figure 4. Acetylation of Skp2 Positively Regulates Skp2 Oncogenic Functions.
(A) IB analysis of LNCaP cell lines stably transfected with the indicated HA-Skp2 constructs.
(B) Cell growth curves of the various LNCaP cell lines stably expressing the indicated HA-Skp2 constructs. Results were presented as mean ±SD from three independent experiments.
(C) Various LNCaP cell lines stably expressing the indicated HA-Skp2 constructs were pulsed with BrdU for 30 min, and the BrdU incorporation rate was measured. Results were presented as mean ±SD from three independent experiments.
(D and E) LNCaP cells stably transfected with the indicated HA-Skp2 constructs (with empty vector as a negative control) were injected subcutaneously into nude mice (n = 5 for each group) and examined over time for in vivo tumorigenesis. Pictures in (E) were taken 6 weeks after injection. Results were presented as mean ±SD and *p < 0.05 (Student’s t test).
See also Figure S4.
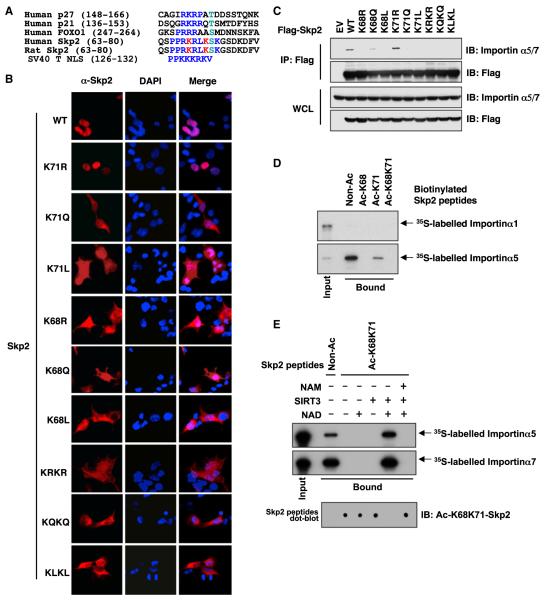
Figure 5. Acetylation of Skp2 by p300 Promotes Skp2 Cytoplasmic Localization.
(A) Sequence alignment of the Skp2 NLS with p21, p27, and FOXO1 NLS.
(B) Immunofluorescence and DAPI staining of 293T cells transfected with the indicated Flag-Skp2 constructs.
(C) IB analysis of WCL and anti-Flag IP derived from 293T cells transfected with the indicated Flag-Skp2 constructs.
(D) Autoradiography of 35S-labeled importin α1 or importin α5 bound to the indicated biotinylated peptides.
(E) Autoradiography of 35S-labeled importin α5 or importin α7 bound to the indicated biotinylated peptides, which have been subject to SIRT3 in vitro deacetylation assays as described in Figure 2E.
See also Figure S5.
p300 Promotes Skp2 Translocation into the Cytoplasm
Because acetylation of lysines within an NLS often influences cellular localization (di Bari et al., 2006; Dietschy et al., 2009), we hypothesized that acetylation of the Skp2 NLS (Figure 5A) may influence its localization. Indeed, cytoplasmic relocalization was observed when wild-type Skp2 was cotransfected with p300 (Figure S5A). Importantly, the same phenotype was observed with S72A-Skp2, implying that p300-mediated acetylation governs Skp2 cellular localization independent of Akt-mediated Skp2 phosphorylation (Figure S5A).
We next evaluated the contribution of acetylation of individual lysine residues for Skp2 cytoplasmic localization. Interestingly, the K71R mutant behaves similar to WT-Skp2, primarily localizing to the nucleus, whereas the acetylation-mimetic K71Q, K71L K68L, K68Q, K68L/K71L, and K68Q/K71Q mutants are largely retained in the cytoplasm (Figure 5B). Curiously, the acetylation-deficient K68R or K68R/K71R mutants also localize mainly in the cytoplasm (Figure 5B). This might be due to the fact that the first lysine residue of a given NLS is critical for its recognition by the importin complex, and even replacement of this first lysine abolishes NLS function (Fabbro and Henderson, 2003; Poon and Jans, 2005). Consistent with this notion, both K68R and K68R/K71R mutants display decreased interaction with the importin α5/7 subunit (Figures 5C and S5B). On the other hand, substitution of the second lysine (K71R) does not affect its recognition by the importin α5/7 subunit (Figures 5C and S5B). Most importantly, all acetylation-mimetic mutants do not interact with the importin complex (Figures 5C and S5B). To provide a better understanding of how acetylation of Skp2 affects its recognition by the importin complex, we performed pull-down assays with biotinylated peptides that contain the canonical NLS (Figure 3D). Importin α5, but not α1 (Figure 5D), interacts with Skp2 in vitro. Moreover, acetylation of K71 leads to a significant decrease, whereas acetylation of K68 results in an almost complete loss of recognition by importin α5 in vitro (Figure 5D). Furthermore, SIRT3 efficiently attenuates acetylation of Skp2 at K68 and K71, which leads to a complete restoration of importin α5 and α7 interactions with Skp2 in vitro (Figure 5E). These results indicate that acetylation of the first lysine (K68) and, to a lesser extent, the second lysine (K71), within the NLS disrupts its recognition by the importin complex, offering a molecular mechanism for acetylation-dependent regulation of Skp2 cytoplasmic localization.
Importantly, consistent with previous reports (Bao et al., 2010; Cooper and Spelbrink, 2008; Hirschey et al., 2010; Jing et al., 2011; Schwer et al., 2002), we found that SIRT3 exclusively localized in the mitochondria (Figures S5D and S5E). Moreover, WT-Skp2 mainly localized in the nucleus (Figures S5C and S5F), whereas the acetylation-mimetic KLKL-Skp2 is primarily localized in the cytoplasm (Figures S5C and S5G). Interestingly, inactivating SIRT3 with the pan-Sirtuin inhibitor, NAM, led to increased mitochondrial localization of Skp2 (Figure S5D). Moreover, there is a significant enrichment of mitochondrial localization for the acetylation-mimetic Skp2 mutant, KLKL-Skp2 (Figures S5C and S5G). These results indicate that the acetylated form of Skp2 might specifically interact with SIRT3 in the mitochondria where the deacetylation of Skp2 may occur. In keeping with this notion, purified mitochondria from WT, but not SIRT3−/−, MEFs could deacetylate Skp2 in vitro (Figures S5H and S5I).
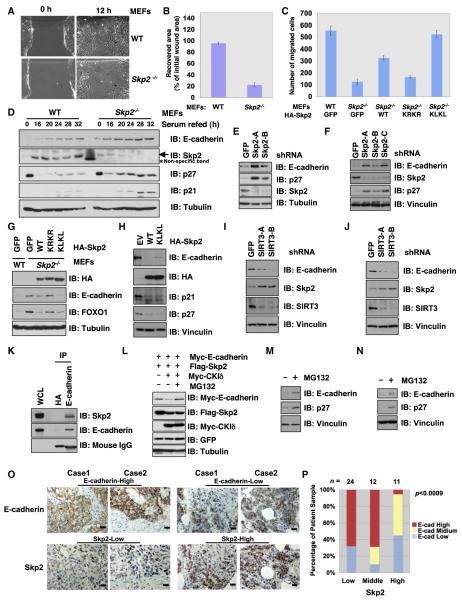
Skp2 Plays an Important Role in Regulating Cell Migration
In agreement with a previous report (Lin et al., 2009), Skp2−/− MEFs exhibit decreased ability to close a wounded area in monolayer cultures (Figures 6A and 6B) as well as to migrate toward chemoattractants in Transwell assays (Figure 6C). Reexpression of Skp2 in Skp2−/− MEFs rescued these deficiencies, demonstrating that Skp2 can directly modulate cell migration (Figure 6C). Moreover, expression of the KLKL acetylationmimetic Skp2 mutant in Skp2−/− MEFs further enhances migration when compared to cells expressing wild-type-Skp2 or the acetylation-deficient KRKR-Skp2 (Figure 6C). Similarly, LNCaP cells expressing KLKL-Skp2 also display increased cell migration compared to wild-type Skp2 (Figures S6A–S6D). Interestingly, we detected an increased expression of E-cadherin in Skp2−/− MEFs (Figure 6D), as well as in Skp2-depleted normal human fibroblasts (Figure 6E). More importantly, a marked increase of E-cadherin expression is observed after depletion of endogenous Skp2 in various epithelial carcinoma cell lines, including SKOV3 (Figure 6F), DU145 (Figure S6H), and HeLa cells (Figure S6I). In support of a role of E-cadherin in Skp2-mediated migration, an inverse correlation was observed between cell migration ability (Figure 6C) and E-cadherin expression levels in Skp2−/− MEFs reconstituted with either wild-type, KLKL-Skp2, or KRKR-Skp2 (Figure 6G). In keeping with these results, ectopic expression of WT- or KLKL-Skp2 in various other epithelial carcinoma cell lines, including DU145 (Figure 6H), MCF7 (Figures S6E and S6L), T47D (Figure S6F), and ZR75 (Figure S6G), led to a severe reduction of E-cadherin abundance. Conversely, depletion of endogenous SIRT3 in MCF7 (Figure 6I), ZR75 (Figure 6J), and DU145 (Figure S6J) resulted in a dramatic elevation of Skp2 expression and a subsequent reduction of E-cadherin abundance. Altogether, these results support the notion that the SIRT3/Skp2 signaling pathway is a major physiological regulator that controls the stability of the E-cadherin tumor suppressor. Importantly, E-cadherin mRNA levels are not increased in Skp2−/− MEFs (Figure S6K), indicating that Skp2 regulates E-cadherin abundance primarily through posttranslational modifications. Consistent with this, a physiological interaction between endogenous Skp2 and E-cadherin (Figure 6K) is detected. Furthermore, ectopically expressed Skp2 promotes E-cadherin degradation, which can be rescued with the proteasome inhibitor MG132 (Figure 6L). Additionally, MG132 treatment in multiple cell lines led to a marked increase in endogenous E-cadherin (Figures 6M, 6N, and S6M). These results suggest the involvement of the 26S proteasome pathway in regulating E-cadherin abundance. We also observed an inverse correlation between the Skp2 and E-cadherin expression in breast cancer clinical samples (Figures 6O and 6P). Thus, these data altogether suggest that the SIRT3/Skp2 signaling axis may promote cellular migration in part by promoting E-cadherin destruction.
Figure 6. Cytosolic Skp2 Plays a Critical Role in Cellular Migration.
(A and B) Skp2 is required for cell migration. WT and Skp2−/− MEFs were plated for in vitro wound healing assays (A). Results in (A) were quantified in (B) and presented as mean ±SD from three independent experiments.
(C) WT and Skp2−/− MEFs were infected with the indicated viral constructs before plating for transwell assay. The results were quantified and presented as mean ±SD from three independent experiments.
(D) IB analysis of WT and Skp2−/− MEFs, synchronized by serum starvation for 72 hr and then released by readdition of serum for the indicated periods.
(E and F) IB analysis of normal human fibroblasts (E) or SKOV3 epithelial cancer cell line (F) infected with the indicated lentiviral shRNA vectors.
(G) IB analysis of WT and Skp2−/− MEFs infected with the indicated viral constructs.
(H) IB analysis of DU145 cells infected with the indicated viral constructs.
(I and J) IB analysis of MCF7 (I) or ZR75 epithelial cancer cell line (J) infected with the indicated lentiviral shRNA vectors.
(K) IB analysis of MCF7 WCL and anti-E-cadherin IP. Anti-HA IgG was used as a negative control for the immunoprecipitation procedure.
(L) IB analysis of WCL of HeLa cells transfected with the indicated plasmids. Cells were treated 20 hr posttransfection with the proteasome inhibitor MG132 overnight before harvesting.
(M and N) IB analysis of WCL derived from DU145 (M) or SKOV3 (N) cells treated with 10 μM MG132 for 12 hr before harvesting.
(O and P) Representative images of E-cadherin and Skp2 expression in breast tumor cells as assessed by immunohistochemistry (O). Both Skp2 and E-cadherin levels were classified as low, medium, or high based on the intensities of the IHC staining, and the percentages of patients classified in each category are depicted in the histogram in (P).
See also Figure S6.
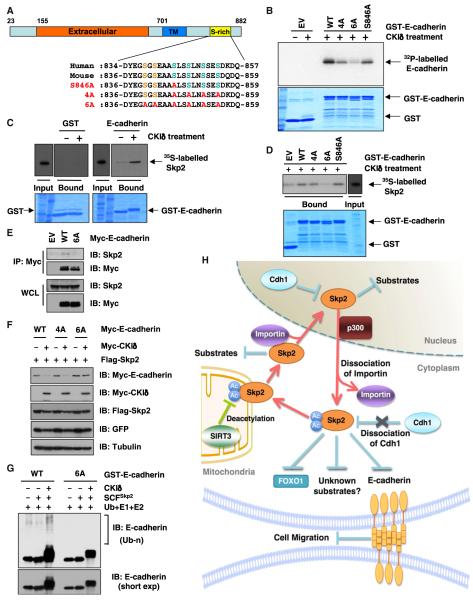
Skp2 Promotes the Ubiquitination and Destruction of E-Cadherin in a CKI-Dependent Manner
We next explored the mechanism by which Skp2 promotes E-cadherin destruction. E-cadherin is phosphorylated by Casein Kinase I (CKI) (Dupre-Crochet et al., 2007), and this negatively regulates E-cadherin function. Consistent with this, Skp2 expression promotes E-cadherin degradation in a CKI-dependent manner (Figure 6L), which is in agreement with the well-established mechanism whereby Skp2 requires prior phosphorylation of its targets for destruction (Frescas and Pagano, 2008). Moreover, mutation of Ser846, Ser849, Ser852, and Ser855 (4A) in the serine-rich region of E-cadherin significantly attenuates phosphorylation by CKI in vitro (Figures 7A and 7B). However, residual phosphorylation of E-cadherin remains even in the 4A mutant, suggesting the existence of additional potential CKI sites (Figure 7B). A closer examination of the mouse E-cadherin sequence reveals that Ser840 and Ser842 are possible CKI consensus motifs (Figure 7A). In support of this, additional mutation of Ser840 and Ser842 (6XA mutant) leads to a further decrease in CKI-mediated E-cadherin phosphorylation in vitro (Figure 7B). Also, E-cadherin phosphorylation by CKI promotes its interaction with Skp2 (Figure 7C), but this is significantly impaired in the 6A mutant, both in vitro (Figure 7D) and in cells (Figure 7E). Mutation of the CKI sites also abolishes Skp2-mediated E-cadherin degradation (Figures 7F and S7A) and impairs the ability of Skp2 to promote E-cadherin ubiquitination in a CKI-dependent manner (Figure 7G). Consistent with a tumor suppressor role for E-cadherin, ectopic expression of the nondegradable version (6XA) of E-cadherin in LNCaP cells (Figure S7B) resulted in a significant reduction in cellular migration ability (Figure S7C), which correlates with a significant decrease in their ability to form tumors in vivo (Figures S7D–S7G). These results indicate that misregulation of the SIRT3/Skp2 signaling pathway might result in aberrant expression of the E-cadherin tumor suppressor, thus facilitating tumorigenesis.
Figure 7. Skp2 Promotes the Ubiquitination and Destruction of E-Cadherin in a CKI-Dependent Manner.
(A) Sequence alignment of the putative CKI phosphorylation sites in Skp2 from various species.
(B) Indicated GST-fusion proteins were incubated with CKIδ and [γ-32P]ATP. The kinase reaction products were resolved by SDS-PAGE, and phosphorylation was detected by autoradiography.
(C and D) Autoradiography of 35S-labeled Skp2 bound to the indicated GST-fusion proteins. Where indicated, GST-fusion proteins were pretreated with CKId before pull-down assays were performed.
(E) IB analysis of WCL and anti-Myc IP derived from HeLa cells transfected with the indicated Myc-E-cadherin constructs.
(F) IB analysis of WCL from HeLa cells transfected with the indicated plasmids.
(G) SCFSkp2 E3 ligase complex promotes E-cadherin ubiquitination in vitro. Where indicated, GST-E-cadherin proteins were pretreated with CKI before the in vitro ubiquitination assays.
(H) Proposed model for how acetylation of Skp2, which subsequently regulates Skp2 stability and cellular localization to influence its oncogenic functions, is governed by both p300 and SIRT3.
See also Figure S7.
DISCUSSION
Deregulation of the Skp2/p27 pathway, including overexpression of Skp2 or loss of p27, is frequently observed in numerous human tumors, but the underlying molecular mechanisms remain largely undescribed. Although phosphorylation-mediated regulatory mechanisms control the activity of multiple F-box proteins, including Skp2 (Gao et al., 2009; Lin et al., 2009) and Fbx4 (Barbash et al., 2008), regulation by acetylation has not been previously linked to F-box control. Our results demonstrate that p300-mediated acetylation of Skp2 affects its stability and cytoplasmic localization, which, in turn, can influence its oncogenic activity (Figure 7H). These results suggest that acetylation-mediated posttranslational modifications might also control the function of other F-box proteins.
Skp2 has been reported to interact with p300 and to negatively affect the ability of p300 to modulate apoptosis induced by p53 (Kitagawa et al., 2008). Our data provide an additional mechanism for the Skp2/p300 interaction, whereby p300 can reciprocally govern Skp2 function in an acetylation-dependent manner. Our studies also point to a potential crosstalk between the Akt and p300 pathways in the regulation of Skp2. In this regard, Akt activates p300 acetyl-transferase activity (Huang and Chen, 2005) to influence Skp2 acetylation (Figures 1B and 1C). Therefore, in humans and other mammals in which Ser72 is conserved, Akt could potentially regulate Skp2 function by two related mechanisms, one through direct Ser72 phosphorylation and the other through activation of p300 to promote Skp2 acetylation (Figure S7H). However, in mice, Skp2 Ser72 is replaced with a Gly; therefore, additional studies are required to examine whether Akt indirectly affects Skp2 function through a p300-dependent manner (Figure S7I). As the crosstalk between histone phosphorylation and acetylation is well established (Cheung et al., 2000; Lo et al., 2000), we evaluated whether Akt phosphorylation of Skp2 affects acetylation at K68 and K71. Acetylation-mimetic mutants displayed a moderate decrease in Ser72 phosphorylation, indicating that Skp2 acetylation at K68 or K71 might negatively affect Ser72 phosphorylation (Figure S1I). However, phosphorylation of either Ser72 or Ser75 does not affect the ability of p300 to acetylate Skp2 (Figure S1J). Because both Akt-mediated phosphorylation of Ser72 and p300-mediated acetylation at K68 and K71 serve to promote Skp2 cytoplasmic localization as well as to stabilize Skp2, activation of either pathway promotes Skp2 oncogenic signaling. It is possible that p300 and Akt are activated in response to distinct upstream signals to modulate Skp2 in specific settings.
In addition to Skp2, many of Skp2 downstream substrates, including p21 (Zhou et al., 2001), p27 (Liang et al., 2002; Shin et al., 2002; Viglietto et al., 2002), and FOXO1 (Brunet et al., 1999), have been characterized as Akt phosphorylation targets. Interestingly, Akt-mediated phosphorylation all resulted in a relocation of these proteins to the cytosol (Figure S5S). Especially for FOXO1, Akt-mediated phosphorylation of FOXO1 at Ser256 is required for subsequent ubiquitination by Skp2 (Huang et al., 2005), suggesting that Akt drives both FOXO1 and Skp2 to the cytosol where phosphorylated FOXO1 is actively degraded by SCFSkp2 (Zhang, 2010). Similar to Skp2, FOXO1 is actively acetylated by p300 (Figure S5Q) at multiple lysine residues within its NLS (Matsuzaki et al., 2005; Perrot and Rechler, 2005). Furthermore, p27, but not p21, could be acetylated by p300 (Figures S5J and S5K). Interestingly, to mimic acetylation, replacement of K153 to Q within the NLS of p27 led to a significant increase in the cytoplasmic localization of p27 (Figure S5L) that correlates with reduced interaction with the importin complex subunits (Figures S5M–S5O). These results suggest that both Akt-mediated phosphorylation and p300-mediated acetylation could cooperatively regulate the cellular localization of Skp2 as well as its various downstream ubiquitin substrates. However, further studies are warranted to better understand the biological significance of this complicated regulation of the Skp2 signaling pathway.
More importantly, we also identified SIRT3 as an Skp2 deacetylase. SIRT3 has been recently shown to function as a tumor suppressor by regulating both genomic stability and tumor cell metabolism (Bell et al., 2011; Finley et al., 2011a; Kim et al., 2010). Moreover, SIRT3 deletion increases cell proliferation, although the underlying molecular mechanisms remain unknown (Bell et al., 2011; Finley et al., 2011a; Kim et al., 2010). Interestingly, we detected an inverse correlation between Skp2 and SIRT3 levels in breast cancer clinical samples (Figures 2L and 2M), which suggests that reduced SIRT3 expression may have a causal effect on Skp2 overexpression in breast and other cancers.
Interestingly, our results suggest that acetylation of Skp2 might differentially regulate the destruction of its downstream targets, such as p21 and FOXO1 (Figure 4A). As dimerization has been recently characterized as a key regulatory mechanism for the function of several F-box proteins (Barbash et al., 2008; Tang et al., 2007b; Welcker and Clurman, 2007), and as p300-mediated acetylation has been reported to influence protein dimerization (Tang et al., 2007a; Yuan et al., 2005), we further explored whether acetylation of Skp2 affects dimerization. We found that p300-mediated Skp2 acetylation promotes Skp2 dimerization, and this is impaired upon deletion of the NLS (Figure S5U). Mutation of either K68 or K71 disrupts the ability of Skp2 to dimerize, whereas an acetylation-mimetic mutant (KLKL) forms dimers even in the absence of p300 (Figures S5V–S5Y). Our results therefore suggest that dimerization might affect the Skp2 substrate spectrum.
Skp2 cytoplasmic staining correlated with metastasis and high-grade breast and prostate cancers (Dowen et al., 2003; Drobnjak et al., 2003; Radke et al., 2005; Signoretti et al., 2002), though the underlying molecular mechanisms are not known. Here, we found that Skp2 promotes E-cadherin degradation in a CKI-dependent manner (Figure 7), which is consistent with the established role of E-cadherin as a critical cell adhesion protein whereby E-cadherin loss promotes cell migration (Giehl and Menke, 2008; Schmalhofer et al., 2009). Although promoter methylation or loss of the E-cadherin genetic locus can account for reduced E-cadherin expression, it has also been reported that the E-cadherin is an unstable protein such that enhanced proteolysis contributes to decreased E-cadherin levels (Yang et al., 2006). Furthermore, we detected an inverse correlation between Skp2 and E-cadherin expression in an array of breast cancer clinical samples (Figures 6O and 6P). These results suggest that the effects of Skp2 on cellular migration could be partly mediated by modulation of E-cadherin stability.
Skp2−/− mice are resistant to tumorigenesis that is induced by overexpression of the BCR-Abl oncoprotein (Agarwal et al., 2008) or by loss of the tumor suppressors p53 or PTEN (Lin et al., 2010). Consistent with this, inhibition of Skp2 in MCF7 breast cancer cells suppresses cell-cycle progression (Sun et al., 2007). These results support the idea of Skp2-targeted therapy, given the central role of the Skp2/p27 pathway in breast cancer initiation and metastasis. However, the critical role of Skp2 in malignancy has not yet been fully elucidated. Our studies provide a mechanism whereby acetylation of Skp2 by p300 regulates protein stability and cellular localization, thus modulating oncogenic signaling, and our studies also provide further rationale for the development of Skp2 antagonists for cancer therapy.
EXPERIMENTAL PROCEDURES
Transwell Cell Migration Assay and In Vitro Wound Healing Assay
Cell migration and wound healing assays were performed as described previously (Lin et al., 2009).
Skp2 Binding Assays
Binding to immobilized GST proteins and biotinylated peptides was performed as described previously (Wei et al., 2004; Wei et al., 2005).
In Vitro Deacetylation Assay
Recombinant human SIRT3 protein was purchased from BIOMOL. To perform the in vitro deacetylation assay, 2 μg of acetylated Skp2 peptides were incubated with recombinant SIRT3 in 30 μl of SDAC buffer (50 mM Tris-HCl [pH 9.0], 1 mM MgCl2, 50 mM NaCl, and 0.5 mM dithiothreitol [DTT]), with or without 1.5 mM nicotinamide adenine dinucleotide (NAD) at 37°C for 2 hr. Where indicated, 20 mM nicotinamide (NAM) were added. The reactions were spotted on nitrocellulose membrane to detect deacetylation of Skp2 peptides with anti-Ac-K68K71-Skp2 antibody. The rest of the reactions were subjected to subsequent in vitro peptide-protein binding assays.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lewis Cantley, Pengda Liu, and Qing Zhang for critical reading of the manuscript; Christoph Geisen, Yasuyujki Fujita, and Hui-Kuan Lin for providing reagents; and members of the Wei, Haigis, Pandolfi, and Toker labs for useful discussions. W.W. is an MLSC New Investigator and an American Cancer Society Research Scholar (W.W., RSG-12-096). This work was supported in part by the Susan G. Komen Breast Cancer Foundation (Y.R.C., 0706963), the Department of Defense Prostate Cancer Research Program (W.W., PC080377), and by grants from the National Institutes of Health (W.W., GM089763, GM094777; A.T., CA122099; H.I., AG041218).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.05.038.
REFERENCES
- Agarwal A, Bumm TG, Corbin AS, O’Hare T, Loriaux M, VanDyke J, Willis SG, Deininger J, Nakayama KI, Druker BJ, Deininger MW. Absence of SKP2 expression attenuates BCR-ABL-induced myeloproliferative disease. Blood. 2008;112:1960–1970. doi: 10.1182/blood-2007-09-113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr., Leclerc J, Nguyen P, et al. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J. Cell. Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem. J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- di Bari MG, Ciuffini L, Mingardi M, Testi R, Soddu S, Barilà D. c-Abl acetylation by histone acetyltransferases regulates its nuclear-cytoplasmic localization. EMBO Rep. 2006;7:727–733. doi: 10.1038/sj.embor.7400700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy T, Shevelev I, Pena-Diaz J, Hühn D, Kuenzle S, Mak R, Miah MF, Hess D, Fey M, Hottiger MO, et al. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009;122:1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- Dowen SE, Scott A, Mukherjee G, Stanley MA. Overexpression of Skp2 in carcinoma of the cervix does not correlate inversely with p27 expression. Int. J. Cancer. 2003;105:326–330. doi: 10.1002/ijc.11066. [DOI] [PubMed] [Google Scholar]
- Drobnjak M, Melamed J, Taneja S, Melzer K, Wieczorek R, Levinson B, Zeleniuch-Jacquotte A, Polsky D, Ferrara J, Perez-Soler R, et al. Altered expression of p27 and Skp2 proteins in prostate cancer of African-American patients. Clin. Cancer Res. 2003;9:2613–2619. [PubMed] [Google Scholar]
- Dupre-Crochet S, Figueroa A, Hogan C, Ferber EC, Bialucha CU, Adams J, Richardson EC, Fujita Y. Casein kinase 1 is a novel negative regulator of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 2007;27:3804–3816. doi: 10.1128/MCB.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1a destabilization. Cancer Cell. 2011a;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl K, Menke A. Microenvironmental regulation of E-cadherin-mediated adherens junctions. Front. Biosci. 2008;13:3975–3985. doi: 10.2741/2985. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. USA. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell. Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc. Natl. Acad. Sci. USA. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Lee SH, McCormick F. Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Mol. Cell. 2008;29:217–231. doi: 10.1016/j.molcel.2007.11.036. [DOI] [PubMed] [Google Scholar]
- Li JQ, Wu F, Masaki T, Kubo A, Fujita J, Dixon DA, Beauchamp RD, Ishida T, Kuriyama S, Imaida K. Correlation of Skp2 with carcinogenesis, invasion, metastasis, and prognosis in colorectal tumors. Int. J. Oncol. 2004;25:87–95. [PubMed] [Google Scholar]
- Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 2002;8:1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- Lim MS, Adamson A, Lin Z, Perez-Ordonez B, Jordan RC, Tripp S, Perkins SL, Elenitoba-Johnson KS. Expression of Skp2, a p27(Kip1) ubiquitin ligase, in malignant lymphoma: correlation with p27(Kip1) and proliferation index. Blood. 2002;100:2950–2956. doi: 10.1182/blood.V100.8.2950. [DOI] [PubMed] [Google Scholar]
- Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat. Cell Biol. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2) Curr. Biol. 2001;11:263–267. doi: 10.1016/s0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol. Endocrinol. 2005;19:2283–2298. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Radke S, Pirkmaier A, Germain D. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene. 2005;24:3448–3458. doi: 10.1038/sj.onc.1208328. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, Zhang H. Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate. Cancer Res. 2003;63:1583–1588. [PubMed] [Google Scholar]
- Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, Arteaga CL. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat. Med. 2002;8:1145–1152. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J. Clin. Invest. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun L, Cai L, Yu Y, Meng Q, Cheng X, Zhao Y, Sui G, Zhang F. Knockdown of S-phase kinase-associated protein-2 expression in MCF-7 inhibits cell growth and enhances the cytotoxic effects of epirubicin. Acta Biochim. Biophys. Sin. (Shanghai) 2007;39:999–1007. doi: 10.1111/j.1745-7270.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007a;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007b;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Traub F, Mengel M, Lück HJ, Kreipe HH, von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res. Treat. 2006;99:185–191. doi: 10.1007/s10549-006-9202-3. [DOI] [PubMed] [Google Scholar]
- Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Viglietto G, Motti ML, Bruni P, Melillo RM, D’Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 2002;8:1136–1144. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Ayala G, De Marzo A, Tian W, Frolov A, Wheeler TM, Thompson TC, Harper JW. Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin. Cancer Res. 2002;8:3419–3426. [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, Broglio K, Berry DA, Hung MC. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell. Biol. 2006;26:7269–7282. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chen KY, Tong Q. Murine Sirt3 protein isoforms have variable half-lives. Gene. 2011;488:46–51. doi: 10.1016/j.gene.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zhang H. Skip the nucleus, AKT drives Skp2 and FOXO1 to the same place? Cell Cycle. 2010;9:868–869. [PubMed] [Google Scholar]
- Zhang L, Wang C. F-box protein Skp2: a novel transcriptional target of E2F. Oncogene. 2006;25:2615–2627. doi: 10.1038/sj.onc.1209286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.