Abstract
Non homologous end joining (NHEJ) is an important process that repairs double strand DNA breaks (DSBs) in eukaryotic cells. Cells defective in NHEJ are unable to join chromosomal breaks. Two different NHEJ assays are typically used to determine the efficiency of NHEJ. One requires NHEJ of linearized plasmid DNA transformed into the test organism; the other requires NHEJ of a single chromosomal break induced either by HO endonuclease or the I-SceI restriction enzyme. These two assays are generally considered equivalent and rely on the same set of NHEJ genes. PC4 is an abundant DNA binding protein that has been suggested to stimulate NHEJ. Here we tested the role of PC4's yeast homolog SUB1 in repair of DNA double strand breaks using different assays. We found SUB1 is required for NHEJ repair of DSBs in plasmid DNA, but not in chromosomal DNA. Our results suggest that these two assays, while similar are not equivalent and that repair of plasmid DNA requires additional factor(s) that are not required for NHEJ repair of chromosomal double-strand DNA breaks. Possible roles for Sub1 proteins in NHEJ of plasmid DNA are discussed.
Introduction
In cells, double strand DNA breaks (DSBs) can be induced by ionizing radiation, oxidation, DNA damaging chemicals, replication errors and others [1], [2]. Because a single DSB can cause chromosomal loss or translocation which may lead to cell death or cancerous transformation, repair pathways for DSBs are conserved in eukaryotes ranging from yeast to human and have been the focus of numerous studies [2], [3]. DSBs can be repaired by two distinct repair pathways: homology-directed repair (HR) and non homologous end joining (NHEJ) [4], [5]. HR is a repair process that finds homologous sequences in the genome and uses it as a template to repair the chromosomal break [6]–[8]. In contrast, NHEJ is a repair process that brings the ends of the broken DNA together and repairs the break by direct ligation [9]–[11]. NHEJ requires the KU complex, DNA ligase and other factors [11], [12]. Since the DSB ends joined in NHEJ may result in alterations of DNA bases at the junction, NHEJ is considered to be more error-prone [13].
Two experimental approaches have been widely used to introduce DSBs into cells to study the NHEJ repair pathway [11], [14]–[16]. The first approach is to induce DSBs in chromosomes in vivo. This can be achieved by treating cells with ionizing radiation or chemicals that generate DSBs randomly throughout the genome. Alternatively, meganucleases such as HO and I-SceI endonucleases have been used to induce site specific DSBs in the chromosome [14], [17]. Because induction of the meganucleases can be controlled and the genomic location of the DSB can be precisely engineered, many details of the molecular events in DSB repair have been obtained using this approach [18]–[20]. The other approach used to study NHEJ is to introduce DNA containing a specific break into the cells. This is usually achieved by linearizing a plasmid using a restriction enzyme and transforming the cells with the linearized DNA [21]. Because the sequence flanking the DSB has no homology in the host genome, cells can only maintain the plasmid by first repairing it. Therefore the efficiency of NHEJ repair can be determined by the transformation efficiency of the linear plasmid relative to that of the control circular plasmid. The plasmid repair assay has proven useful by studies that characterized several key NHEJ factors [21]–[23]. It is noteworthy that both approaches have been used to study NHEJ in human and yeast cells [24]–[31].
PC4 is a small nuclear protein produced by the human SUB1 gene. It can bind to double strand DNA and single strand DNA without sequence specificity [32], [33]. It was first isolated as a transcription coactivator in 1994, able to promote basal transcription at low concentrations [34], [35]. The yeast SUB1 gene is homologous to PC4 in sequence and activity. Both proteins bind DNA and function as transcription coactivators [36]–[39]. Additionally, Wang et al found that PC4 and its yeast homolog SUB1 suppress oxidative mutagenesis and confer oxidative resistance in yeast, suggesting a role for PC4 in DNA repair or DNA damage prevention [40]. Interestingly, Batta et al showed that PC4 promotes DNA ligation in vitro and suggested that it may stimulate repair of DNA breaks by NHEJ [41]. However, it was less certain if PC4 is required for NHEJ in vivo. In the present study we tested the role of PC4's yeast homolog SUB1 in NHEJ in vivo by using several different assays; HO-induced chromosomal breaks, I-SceI induced chromosomal breaks and restriction enzyme induced breaks in plasmid DNA prior to transformation. NHEJ was then assayed comparing yeast sub1 mutants with wild type and yku70 mutant cells. Unexpectedly, the yeast sub1Δ mutant displays differential repair capacities for DNA breaks in chromosomes and in linearized plasmids, showing a severe defect in the repair of transformed, linearized plasmid DNA breaks, while being fully capable of repairing chromosomal DNA breaks.
Materials and Methods
Yeast strains
Yeast strains used in this study are listed in Table 1. The PCR based gene replacement method was used to create the yeast knock out strains [42], [43].
Table 1. Yeast strains used in this study.
| Strain | Original name, genotype (annotation) | Reference |
| MVY101 | FY833, Mata ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 | [57] |
| MVY105 | MVY101 with sub1Δ::hisG | [57] |
| MVY601 | MVY101 with yku70Δ::HIS3 | this study |
| MVY603 | MVY101 with sub1Δ::hisG yku70Δ::HIS3 | this study |
| MVY610 | JKM179, MATα hoΔ hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2,3-112 lys5 trp1Δ::hisG ura3-52 ade3::GAL-HO | [52] |
| MVY614 | MVY610 with yku70Δ::URA3 | [52] |
| MVY617 | MVY610 with sub1Δ::TRP1 | this study |
| MVY625 | MVY610 with yku70Δ::URA3 sub1Δ::TRP1 | this study |
| MVY665 | YW714, MATα, ade2::SD2-::URA3 his3D1 Leu2D0 LYS2 MET15 ura3D0 | [48] |
| MVY666 | YW713, MATα, ade2::SD2-::URA3 his3D1 Leu2D0 LYS2 MET15 ura3D0 yku70::kanMX4 | [48] |
| MVY667 | MVY665 with sub1::KanMX4 | this study |
| MVY696 | MVY665 with sub1::KanMX4 yku70::HIS3 | this study |
Transformation
The yeast transformation procedure is as described by Knop et al. [44]. Briefly, cells are grown to early log phase, sonicated briefly, made transformation-competent and used immediately or stored at −80°C. 100–200 ng DNA is used in each transformation. After transformation, cells are plated directly onto minimal media lacking the selected nutrient or incubated in YPD medium (1% yeast extract, 2% peptone, 2% glucose) for 2 hours before plating onto YPD containing 200 µg/ml G418 for KanMX selection.
Plasmid NHEJ repair assay
The plasmid repair assay is similar to the procedures described elsewhere [21], [45]. Linearized plasmid DNA is produced by digesting the plasmid with the specified restriction enzyme, followed by gel-purification of linear DNA. Both linearized and circular plasmids are used to transform the yeast cells. Colonies are counted 3–4 days after transformation. The plasmid repair efficiency is calculated as the transformation efficiency of the linearized plasmid divided by the transformation efficiency of the circular plasmid.
To quantify the ratio of mutagenic ligation events, yeast cells are transformed with NcoI-linearized pMV1328, which cuts within the KanMX6 coding sequence, then selected on plates lacking leucine. After incubation, Leu+ transformants are streaked on YPD agar medium containing 200 µg/ml G418 to test KanMX function. Colonies that are Leu+but G418-sensitive are counted as mutagenic ligation events.
HO induction and cell survival
Wild type and the mutant cells are incubated in YEP-raffinose (1% yeast extract, 2% peptone, 2% raffinose) to log phase (OD600<0.5) and sonicated briefly. Half of each culture is supplemented with 2% galactose to induce the HO endonuclease for the indicated times. Both the induced and uninduced cells are diluted in water and plated onto YPD agar medium to count the number of viable cells. The presence of glucose in the YPD medium suppresses HO expression.
The induction of DSBs is measured by the PCR based method as described [46]. Briefly, at the indicated times after adding 2% galactose to the cell culture, an aliquot of cells is removed and genomic DNA is extracted using the phenol and glass beads method [47]. Real time PCR is performed on a ViiA7 QPCR machine using the primers SG2285 (AATATGGGACTACTTCGCGCAACA) and SG2286 (CGTCACCACGTACTTCAGCATAA) to amplify MATα, which contains the HO site, and the primers SG525 (TTGGATTTGGCTAAGCGTAATC) and SG526 (CTCCAATGTCCCTCAAAATTTCT) to amplify the SMC2 control PCR products as described [46]. The decline in the ratio between the PCR products of MATα and SMC2 represents the increase in the number of the cells with a DSB in the MATα locus. DSB induction is normalized to the respective 0 time point.
Suicide deletion assay
The suicide deletion assay is performed as described in Karathanasis and Wilson [48]. Briefly, wild type and mutant cells are incubated in medium lacking uracil for 3 days to reach stationary phase and keep the I-SceI-Ura3 insert under selection. Cells are then sonicated briefly and diluted in water. Dilutions are plated on synthetic, complete agar medium with uracil to allow loss of the insert and galactose as the carbon source (SC-galactose) to induce I-SceI. After I-SceI induction, survival requires loss of the insert and repair of the DSB. Surviving colonies growing on SC-galactose exhibit a white color if the ADE2 gene is restored by NHEJ repair, or a red color if the repair is inaccurate and the ADE2 gene is disrupted. The frequency of mutagenic ligation is the number of red colonies divided by the total number of colonies on the SC-galactose plates.
Statistics
All experiments were performed at least three times (n>3). Error bars represent standard errors. Two tailed student t-test is used to calculate the P values.
Results
Reduced plasmid repair in the sub1Δ mutant
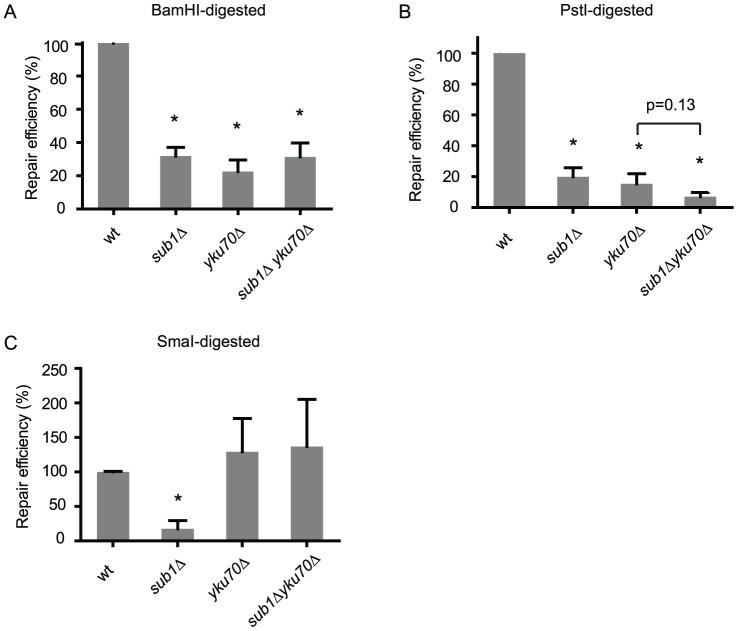
PC4 has been shown to stimulate the ligation of DNA in vitro, suggesting that PC4 plays a role in NHEJ [41]. Here we tested if the yeast homolog SUB1 is required for NHEJ in yeast. We first used the plasmid repair assay to test if the sub1 mutant is impaired to rejoin a double strand DNA break that is introduced into the plasmid by restriction digestion and transformed into the cell [21]. BamHI linearizes plasmid pRS315 with 5' sticky ends in a region that has no homology in the yeast genome. We found that the sub1Δ mutant exhibits significantly reduced repair capacity, comparable to the level seen in the yku70Δ mutant, which is known to be deficient in NHEJ (Figure 1A). The sub1Δ yku70Δ double mutant does not exhibit a further reduction in NHEJ efficiency, indicating that Sub1 functions in the KU dependent repair pathway.
Figure 1. SUB1 is required for repair of dsDNA breaks in plasmid DNA.
Repair efficiency is the ratio of the number of stable transformants obtained when transformed with linearized versus circular pRS315 plasmid DNA. Data are normalized to the ligation efficiency of wild type. Asterisks * indicate a significant difference from wild type (P<0.05). All strains are derivatives of the wild type (MVY101), sub1Δ (MVY105), yku70 (MVY601), sub1Δ yku70 (MVY603). A. Repair efficiencies of the BamHI-linearized plasmid. BamHI produces a unique DSB in pRS315 with 5' end overhangs. B. Repair efficiencies of the PstI-linearized plasmid. PstI produces a unique DSB in pRS315 with 3' end overhangs. C. Repair efficiencies of the SmaI-linearized plasmid. SmaI produces a unique DSB in pRS315 with blunt DNA ends.
Next we tested if SUB1 is required for repair of DNA breaks with different DNA end structures. We used the restriction enzyme PstI to generate DSB in pRS315 with 3' sticky ends. Similar to the BamHI-generated DSB, the sub1Δ mutant has a reduced repair efficiency (Figure 1B). The sub1Δ yku70Δ double mutant does not have a significant further reduction in repair efficiency (P = 0.13), supporting previous results that Sub1 and Yku70 function in the same NHEJ pathway.
DSBs with blunt ends may be repaired by NHEJ in a KU independent fashion with low efficiencies [21], [49]–[51]. We used SmaI to generate a blunt-end DSB in pRS315. Surprisingly, the sub1Δ mutant exhibits a reduced repair efficiency for blunt-end DSBs, while the yku70Δ mutant is highly proficient (Figure 1C). This suggests that Sub1 is required for repair of blunt-end DSBs but not Yku70. The sub1Δ yku70Δ double mutant has a repair capacity similar to the yku70Δ mutant, raising the possibility that repair of blunt-end DSBs is channeled into the Yku70 independent pathway. In this pathway Sub1 is dispensable, or alternatively Yku70 may inhibit blunt end ligations in the absence of Sub1.
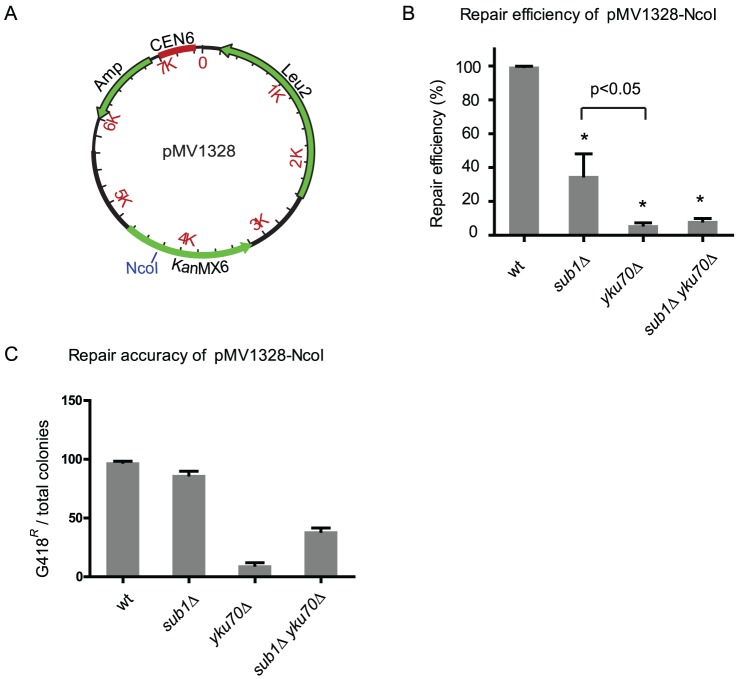
When NHEJ efficiency is reduced, the remaining ligation events can become mutagenic, as is the case in the KU mutant yku70 [21]. To test if the residual NHEJ seen in the sub? mutant is mutagenic, we inserted the KanMX6 gene into pRS315 to make the plasmid pMV1328 (Figure 2A). When the NcoI restriction enzyme is used to cut the single NcoI site that lies within the KanMX6 gene, a functional KanMX6 gene will be inherited only if NHEJ is accurate. This can be detected by testing Leu+transformants for resistance to G418. We first examined the mutants for their abilities to repair the NcoI-linearized pMV1328. As shown in Figure 2B, the sub? mutant has a reduced repair efficiency compared to wild type, confirming our previous results obtained with plasmid pRS315. However, the yku70Δ mutant exhibits a lower repair efficiency than the sub1Δ mutant (P<0.05), indicating that the DNA sequence flanking the DNA break may affect the efficiency of NHEJ [50]. Nonetheless, the sub1Δ yku70Δ double mutant does not exhibit an additive deficiency, suggesting Sub1's role in NHEJ depends on Yku70. Interestingly, most cells that repair the NcoI-cut in pMV1328 are resistant to G418 in both wild type and the sub1Δ mutant (Figure 2C), suggesting that NHEJ in the sub1Δ mutant is highly accurate as is the case in wild type cells. In contrast, repair in the yku70Δ mutant is highly mutagenic (Figure 2C) as previously reported [21]. This suggests Sub1 and Yku70 may have different roles in the same NHEJ pathway. More puzzling, the double mutant exhibits a higher repair accuracy than the yku70Δ mutant, but lower than the sub1Δ mutant. This raises the possibility that the fidelity of NHEJ results from the interplay of multiple factors and the mutagenicity of the yku70 mutant can be partially masked by depleting Sub1.
Figure 2. Ligation accuracy of the plasmid repair.
. Strains, same as in Figure 1. A. Plasmid map of pMV1328. The unique NcoI restriction site within the KanMX6 gene is indicated. B. Repair efficiencies of the mutants when the NcoI linearized pMV1328 is used in the repair assay. Asterisks * indicate a significant difference from wild type (P<0.05). C. Accuracy of plasmid ligation is measured by testing Leu+transformants for G418 resistance (G418R), which requires accurate religation of the NcoI site within the KanMX6 gene.
Efficient joining of chromosomal breaks in the sub1Δ mutant
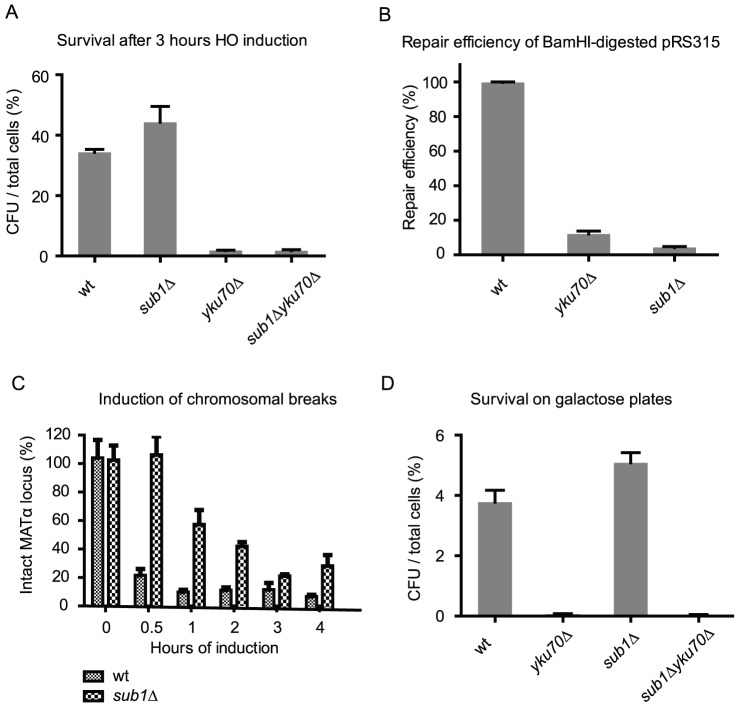
Our results suggest that Sub1 is important for repair of DSBs in transformed plasmid DNA. We next asked if Sub1 is required for repair of chromosomal breaks. We first used the inducible HO endonuclease system to test if the sub1Δ mutant can repair the induced chromosomal breaks efficiently. After transcriptional induction of the HO endonuclease a single chromosomal break is produced in the cell. After HO expression is again repressed, cells that successfully repair the DNA break will survive. We used the strain JKM179, which lacks the homologous sequences needed to repair HO-induced breaks by HR, and can only use NHEJ to carry out the repair needed for survival [52]. Unexpectedly, as shown in Figure 3A, the sub1Δ mutant survives the HO induced chromosomal breaks even better than wild type while the yku70Δ mutant and the sub1Δyku70Δ double mutant succumb to the induced chromosomal breaks.
Figure 3. SUB1 is not required for repair of chromosomal breaks.
All strains used are derived from the ‘donorless’ strain JKM179 (MVY610): sub1Δ (MVY617), yku70 (MVY614), sub1Δ yku70 (MVY625). CFU: Colony Forming Units. A. Cell survival after HO induction for 3 hours followed by repression by glucose. B. Plasmid repair assay is performed in the JKM179 background. Repair efficiencies are normalized to wild type. C. Quantification of DSBs after HO induction. The yeast MATα locus is cleaved by HO endonuclease and the fraction of remaining, intact MATα DNA is determined by real time PCR. D. Cell survival when HO is constantly induced by galactose. Cells are plated on galactose-containing plates to count surviving cells and glucose-containing plates to count the total number of cells.
We tested if the sub1Δ mutant in the JKM179 background is also deficient in repair the plasmid-borne DSB. Results in figure 3B show that NHEJ repair in the sub1Δ mutant and the yku70Δ mutant is greatly reduced, suggesting that the NHEJ deficiency in the sub1Δ mutant in repairing plasmid DSB is not restricted to one yeast strain background.
Sub1 has been found as a transcription cofactor that can activate basal transcription [34], [35]. Therefore we tested if HO induction is compromised in the sub1Δ mutant. Quantitative PCR analysis of DSB induction [46] showed that the sub1Δ mutant does have fewer DSBs after HO induction (Figure 3C). However, 3 hours after HO induction, the difference between wild type and the sub1Δ mutant is only 10%. After subtracting this difference from the survival ratio in Figure 3A, the sub1 mutant appears not to have reduced survival after chromosomal breaks are induced.
Furthermore, we plated the mutants directly on galactose plates to continuously express the HO nuclease. In the constant presence of HO nuclease, the cycles of break production and repair will continue until, either HO induction is lost, or the HO recognition site is mutated during repair. As shown in Figure 3D, the continuously induced chromosomal break is highly lethal to the yku70Δ mutant. However, the sub1Δ mutant does not exhibit reduced survival, suggesting chromosomal breaks are repaired efficiently by NHEJ in the sub1Δ mutant. The double mutant has a survival rate similar to the yku70Δ mutant, further indicating a dispensable role for Sub1 in repairing chromosomal breaks.
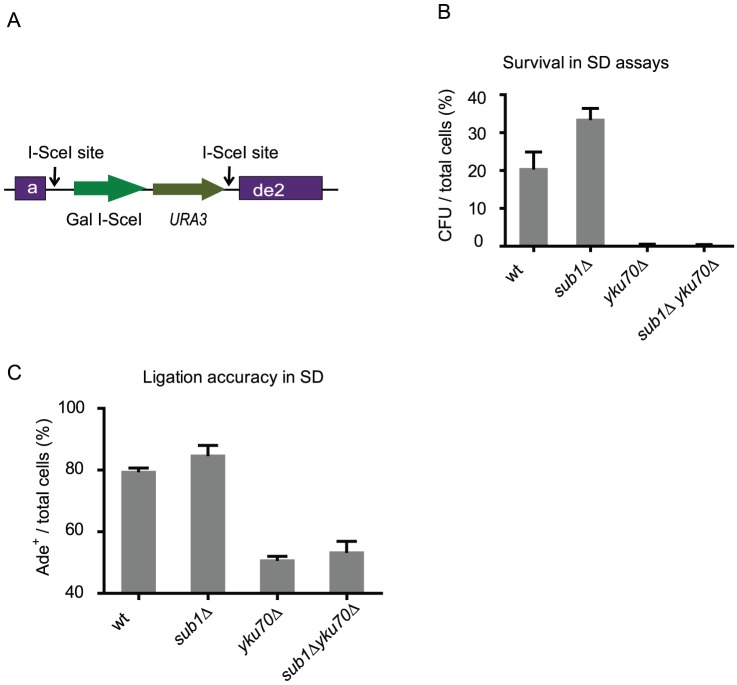
The HO survival assays do not measure the mutagenic ligation events occurring at the chromosomal breaks. Therefore we employed the suicide deletion assay to measure the accuracy of the chromosomal repair in the mutants [48]. In this assay the ADE2 gene is disrupted by the insertion of a cassette containing the galactose-inducible I-SceI gene and the selectable URA3 gene between two flanking I-SceI sites (Figure 4A) [48]. Induction of I-SceI causes removal of the I-SceI-URA3 cassette and, if ligation is accurate, it restores a functional ADE2 gene, which renders the resultant colonies white. If NHEJ is not accurate, mutagenesis of ADE2 results in red colonies. We first tested sensitivity of the mutants to the I-SceI induced chromosomal breaks. As shown in Figure 4B, the sub1Δ mutant has a slightly increased survival compared to wild type, while the yku70Δ mutant and the double mutant have similar reduced survival rates. Further analysis of the surviving cells reveals comparable frequencies of accurate repair when wild type and the sub1 mutant are compared (Figure 4C). In contrast, repair accuracy is much lower in the yku70Δ mutant (Figure 4C), as reported previously [21]. The double mutant exhibits a high rate of mutagenic ligation similar to the yku70Δ mutant, suggesting deletion of YKU70 results a dominant mutagenic phenotype in chromosomal NHEJ repair.
Figure 4. Sub1 is not required for maintaining repair accuracy of chromosomal breaks.
Strain used: wild type (MVY665), yku70Δ (MVY666), sub1Δ (MVY667), sub1Δ yku70Δ (MVY696). A. Schematic drawing of the suicide deletion system. The I-SceI gene, which is expressed from a Gal-inducible promoter, and the URA3 gene with its own promoter are inserted between two I-SceI sites in the ADE2 gene [48]. After Gal induction, the DNA fragment containing I-SceI and URA3 is lost and the functional ADE2 gene is restored only if NHEJ is accurate. B. Percent of surviving cells when cells are plated on galactose-containing plates. Total number of cells is determined by plating cells on YPD plates. CFU: Colony Forming Units. C. Ligation accuracies of chromosomal breaks induced in the suicide deletion assay. Ligation accuracy is determined by dividing the number of red colonies to the total number of surviving cells times 100.
Discussion
We have shown that SUB1 is required for NHEJ repair of DNA breaks in plasmids, but not in chromosomes. The fact that deletion of SUB1 does not reduce accuracy of NHEJ indicates that Sub1 functions differently from the KU complex, which is required for accurate ligations. In fact, the dispensable role of Sub1 in chromosomal repair suggests that Sub1 is not a core component of NHEJ, since KU and other NHEJ factors, but not Sub1, are absolutely required for NHEJ repair of chromosomal breaks.
Previously a microarray-based screen has been performed by Ooi et al to identify new genes that are required for repairing plasmid DSBs [22]. While the screen has been proven useful, successfully identifying NEJ1 as a novel component in NHEJ, SUB1 was not reported. However, several mutants including rad9Δ, rad17 Δ, rad24 Δ, and srs2Δ that are known to have reduced NHEJ efficiencies were also not identified in the microarray-based screen [22]. Furthermore, the authors reported that 13% of haploid mutants were not analyzed due to high signal noise. Thus, it is possible that SUB1 was missed in this screen.
It is interesting to note that the sub1Δ mutant, unlike the yku70Δ mutant, is deficient in repairing the blunt-end DSB in plasmid DNA. It has long been found that blunt-end DSB is repaired at a low efficiency and that knockout mutations of Yku70 or Yku80 can increase the efficiency of repair [21], [49]. Furthermore, Westmoreland et al found that yeast cells inefficiently survive PvuII-induced chromosomal breaks and this survival is not affected by deletion of RAD52 or DNL4 [51], suggesting repair of blunt-end DSBs is independent of HR or canonical NHEJ. It remains elusive how blunt-end DSBs are repaired independently of KU. Our results show that deletion of SUB1 reduces the efficiency of the already-inefficient repair of blunt-end DSBs. However, in the absence of YKU70, repair of blunt-end DSBs is higher than wild type, regardless of whether Sub1 is functional or not. We speculate that when repair is channeled into the KU-independent pathway, Sub1 no longer plays a role.
Given the pronounced effect of SUB1 on repair all types of DSBs in plasmids, it is surprising to find that Sub1 is dispensable for repair of chromosomal breaks, whether they are induced by HO or I-SceI. While the reasons remain to be determined, this is reminiscent of the work by Batta et al. They found that PC4 enhances DNA ligation in vitro but human cells with PC4 knocked down are not sensitive to DSB inducing reagents [41]. In this study it was shown by atomic force microscopy that PC4 has activity that bridges DNA ends [41]. Thus the proposed model has been that PC4 facilitates ligation of plasmid DSBs by bridging the free DNA ends. Indeed, in yeast the ends of chromosomal breaks have been shown to be held in place by chromatin structures after chromosomal breaks are induced [53]–[55], whereas the ends of plasmid DNA are produced prior to transformation and therefore not held in close proximity. This distinct difference between ends of chromosomal breaks and plasmid breaks may underlie the differential requirement of SUB1 in repair of plasmid DSBs versus chromosomal DSBs.
We do not rule out other possible reasons that may explain the differential requirement of SUB1 in repairing plasmid DNA and chromosomal DNA. For example, as a DNA binding protein, Sub1 could potentially affect DNA resection. DNA end resection can have different outcomes in long chromosomes versus short DNA of plasmids: resection at both ends of the plasmid will destroy it, resulting in an apparent low repair efficiency. Alternatively, the effects of Sub1 can be indirect as Sub1 is a transcription factor. However, the plausible targets of transcriptional regulation are not expected to be any known NHEJ factors that are required for repair of chromosomal breaks, since the consequences of SUB1 deficiency is different and unique compared to the consequences due to loss of known NHEJ factors.
The plasmid repair assay has been widely used to test the ability of the cells to repair DNA breaks [15], [56]. Often the conclusions are extended to implicate the ability of the cells to repair chromosomal breaks. Here we provide an example that such implications may not be warranted, since the strong genetic requirement for Sub1 in NHEJ of transformed plasmid DNA does not extend to repair of endonuclease induced chromosomal double-strand DNA breaks. In summary, our results clearly demonstrate that repair of transformed DNA and chromosomal or chromatin associated DNA is quite different in their requirement for Sub1. While many of the genetic requirements are identical, the ability to genetically separate the two methods of assessing NHEJ clearly demonstrates that the two substrates are not repaired in an identical manner.
Acknowledgments
We thank Michael Hampsey, James Haber, Craig Peterson, Thomas Wilson, and Jen-Yeu Wang for yeast strains, Xiufeng Wu for technical help, and James Haber and Michael Resnick for discussions.
Funding Statement
The authors are partially funded by National Institutes of Health (NIH) CA100122 and the University of Massachusetts Medical School. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Xu Y, Price BD (2011) Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10: 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hiom K (2010) Coping with DNA double strand breaks. DNA Repair (Amst) 9: 1256–1263. [DOI] [PubMed] [Google Scholar]
- 3. Stracker TH, Petrini JHJ (2011) The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohnishi T, Mori E, Takahashi A (2009) DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat Res 669: 8–12. [DOI] [PubMed] [Google Scholar]
- 5. Helleday T, Lo J, Van Gent DC, Engelward BP (2007) DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 6: 923–935. [DOI] [PubMed] [Google Scholar]
- 6. Hartlerode AJ, Scully R (2009) Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van den Bosch M, Lohman PHM, Pastink A (2002) DNA double-strand break repair by homologous recombination. Biol Chem 383: 873–892. [DOI] [PubMed] [Google Scholar]
- 8. Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271. [DOI] [PubMed] [Google Scholar]
- 9. Weterings E, Chen DJ (2008) The endless tale of non-homologous end-joining. Cell Res 18: 114–124. [DOI] [PubMed] [Google Scholar]
- 10. Lewis LK, Resnick MA (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. . Mutat Res 451: 71–89. [DOI] [PubMed] [Google Scholar]
- 11. Daley JM, Wilson TE (2005) Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol 25: 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lieber MR, Ma Y, Pannicke U, Schwarz K (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol 4: 712–720. [DOI] [PubMed] [Google Scholar]
- 13. Jackson SP (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23: 687–696. [DOI] [PubMed] [Google Scholar]
- 14. Haber JE (2002) Uses and abuses of HO endonuclease. Meth Enzymol 350: 141–164. [DOI] [PubMed] [Google Scholar]
- 15. Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39: 431–451. [DOI] [PubMed] [Google Scholar]
- 16. Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, et al. (2001) NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae . Nature 414: 666–669. [DOI] [PubMed] [Google Scholar]
- 17. Sugawara N, Haber JE (2006) Repair of DNA double strand breaks: in vivo biochemistry. Meth Enzymol 408: 416–429. [DOI] [PubMed] [Google Scholar]
- 18. Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, et al. (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boulton SJ, Jackson SP (1996) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J 15: 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 22. Ooi SL, Shoemaker DD, Boeke JD (2001) A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae . Science 294: 2552–2556. [DOI] [PubMed] [Google Scholar]
- 23. Wilson TE, Grawunder U, Lieber MR (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498. [DOI] [PubMed] [Google Scholar]
- 24. Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, et al. (2010) Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 6: e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yun MH, Hiom K (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459: 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H, Rosidi B, Perrault R, Wang M, Zhang L, et al. (2005) DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 65: 4020–4030. [DOI] [PubMed] [Google Scholar]
- 27. Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, et al. (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14: 611–623. [DOI] [PubMed] [Google Scholar]
- 28. Van Heemst D, Brugmans L, Verkaik NS, Van Gent DC (2004) End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair (Amst) 3: 43–50. [DOI] [PubMed] [Google Scholar]
- 29. Windhofer F, Krause S, Hader C, Schulz WA, Florl AR (2008) Distinctive differences in DNA double-strand break repair between normal urothelial and urothelial carcinoma cells. Mutat Res 638: 56–65. [DOI] [PubMed] [Google Scholar]
- 30. Ahmad A, Robinson AR, Duensing A, Van Drunen E, Beverloo HB, et al. (2008) ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol 28: 5082–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monnat RJ Jr, Hackmann AF, Cantrell MA (1999) Generation of highly site-specific DNA double-strand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem Biophys Res Commun 255: 88–93. [DOI] [PubMed] [Google Scholar]
- 32. Kaiser K, Stelzer G, Meisterernst M (1995) The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J 14: 3520–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Werten S, Stelzer G, Goppelt A, Langen FM, Gros P, et al. (1998) Interaction of PC4 with melted DNA inhibits transcription. EMBO J 17: 5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M (1994) A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78: 525–534. [DOI] [PubMed] [Google Scholar]
- 35. Ge H, Roeder RG (1994) Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78: 513–523. [DOI] [PubMed] [Google Scholar]
- 36. Knaus R, Pollock R, Guarente L (1996) Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J 15: 1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 37. Henry NL, Bushnell DA, Kornberg RD (1996) A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem 271: 21842–21847. [DOI] [PubMed] [Google Scholar]
- 38. Koyama H, Sumiya E, Nagata M, Ito T, Sekimizu K (2008) Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells 13: 1113–1126. [DOI] [PubMed] [Google Scholar]
- 39. Calvo O, Manley JL (2005) The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J 24: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J-Y, Sarker AH, Cooper PK, Volkert MR (2004) The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol Cell Biol 24: 6084–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Batta K, Yokokawa M, Takeyasu K, Kundu TK (2009) Human transcriptional coactivator PC4 stimulates DNA end joining and activates DSB repair activity. J Mol Biol 385: 788–799. [DOI] [PubMed] [Google Scholar]
- 42. Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae . Nucleic Acids Res 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams A, Gottschling D, Kaiser C, Stearns T (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory.p
- 44. Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- 45. Schär P, Herrmann G, Daly G, Lindahl T (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev 11: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- 47. Papamichos-Chronakis M, Krebs JE, Peterson CL (2006) Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev 20: 2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karathanasis E, Wilson TE (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hegde V, Klein H (2000) Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res 28: 2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moscariello M, Florio C, Pulitzer JF (2010) Accurate repair of non-cohesive, double strand breaks in Saccharomyces cerevisiae: enhancement by homology-assisted end-joining. Yeast 27: 837–848. [DOI] [PubMed] [Google Scholar]
- 51. Westmoreland JW, Summers JA, Holland CL, Resnick MA, Lewis LK (2010) Blunt-ended DNA double-strand breaks induced by endonucleases PvuII and EcoRV are poor substrates for repair in Saccharomyces cerevisiae . DNA Repair (Amst) 9: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, et al. (1998) Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. [DOI] [PubMed] [Google Scholar]
- 53. Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, et al. (2004) DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14: 2096–2106. [DOI] [PubMed] [Google Scholar]
- 54. Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K (2004) Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112. [DOI] [PubMed] [Google Scholar]
- 55. Lisby M, Antúnez de Mayolo A, Mortensen UH, Rothstein R (2003) Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2: 479–483. [PubMed] [Google Scholar]
- 56. Tachibana A (2004) Genetic and physiological regulation of non-homologous end-joining in mammalian cells. Adv Biophys 38: 21–44. [PubMed] [Google Scholar]
- 57. Wu WH, Pinto I, Chen BS, Hampsey M (1999) Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]