Abstract
Long noncoding RNAs (lncRNAs) have emerged as an important class of molecules that regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels through a wide array of mechanisms. This regulation is of particular importance in the central nervous system (CNS), where precise modulation of gene expression is required for proper neuronal and glial production, connection and function. There are relatively few functional studies that characterize lncRNA mechanisms, but possible functions can often be inferred based on existing examples and the lncRNA’s relative genomic position. In this review, we will discuss mechanisms of lncRNAs as predicted by genomic contexts and the possible impact on CNS development, function, and disease pathogenesis. There is no doubt that investigation of the mechanistic role of lncRNAs will open a new and exciting direction in studying CNS development and function.
Recent advances in sequencing technology and genome-wide sequencing projects have overturned the protein-centric view of the cell by revealing that there is extensive non-protein coding transcription throughout eukaryotic genomes (Carninci et al., 2005; Katayama et al., 2005; Birney et al., 2007; Bernstein et al., 2012; Djebali et al., 2012). In fact, 80% of the human genome is likely linked to a biochemical function, with many functional regions assigned outside of traditional protein-coding loci (Bernstein et al., 2012). Only about 1.5% of DNA is protein-coding, indicating that the remaining transcriptional activity contributes to a large population of noncoding RNAs (ncRNAs) (Bertone et al., 2004; Kapranov et al., 2007). These transcripts can take several forms – housekeeping RNAs, such as ribosomal RNAs and transfer RNAs; small ncRNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs; and long ncRNAs (lncRNAs). While the housekeeping and small ncRNAs have been characterized, the mechanisms by which lncRNAs regulate gene expression and cellular functions remain poorly understood. In this review, we will highlight new discoveries of lncRNA functions in gene expression regulation with an emphasis on how these functions can influence the development of the central nervous system (CNS) and contribute to neurological disorders.
Features of lncRNAs
lncRNAs are classified as transcripts >200 nucleotides (nt) in length that show little or no evidence of an open reading frame (ORF) (Kapranov et al., 2007). They can be transcribed from either strand and can be classified as sense, antisense, bidirectional, intronic, or intergenic with respect to nearby protein-coding genes (Huang et al., 2012). Many, but not all, lncRNAs are processed as typical mRNAs including 5’ capping, polyadenylation, and histone modifications (Okazaki et al., 2002; Carninci et al., 2005; Cheng et al., 2005; Kapranov et al., 2007; Ponjavic et al., 2007; Derrien et al., 2012). Splicing of lncRNAs is less efficient than that of protein coding genes and may not occur at all in some cases (Tilgner et al., 2012). There is a heavy bias toward transcripts with two exons (42% of lncRNAs compared to 6% of protein-coding genes) (Derrien et al., 2012). Some lncRNAs are exported from the nucleus and perform important functions in the cytoplasm, but the majority are found in the nucleus, particularly associated with chromatin (Derrien et al., 2012). Though there is evidence for numerous lncRNA loci, only a fraction have been functionally characterized (Carninci et al., 2005; Katayama et al., 2005; Birney et al., 2007; Harrow et al., 2012). Those that have been characterized participate in a striking diversity of cellular processes (Ponting et al., 2009; Clark and Mattick, 2011; Wang and Chang, 2011; Qureshi and Mehler, 2012; Rinn and Chang, 2012).
There was initial debate that lncRNAs simply represented transcriptional noise, as evidenced by their lack of sequence conservation and the low fidelity of RNA polymerase II transcription initiation (Ponjavic et al., 2007; Struhl, 2007). In addition, even though there are many lncRNA transcripts identified through sequencing and array projects, they are transcribed at much lower levels than individual mRNAs (Mattick and Makunin, 2006). However, the low sequence conservation of lncRNAs has been shown to maintain lncRNA secondary structure rather than primary sequence (Ulitsky et al., 2011), which would also help illuminate how diverse lncRNAs can interact with the same protein complexes (Khalil et al., 2009; Guttman et al., 2011; Moran et al., 2012b). Furthermore, part of the ENCODE project probed the possibility that lncRNAs contain very short ORFs that encode small peptides, and showed that the vast majority of transcripts classified as lncRNAs are not translated, supporting a role for these transcripts as ncRNA rather than protein precursors (Banfai et al., 2012; Derrien et al., 2012). Finally, though direct functional studies are lacking, lncRNAs have been shown to be developmentally regulated (Blackshaw et al., 2004; Rinn et al., 2007), expressed in specific cell types (Mercer et al., 2008), localized to specific subcellular compartments (Sone et al., 2007; Bond and Fox, 2009; Ip and Nakagawa, 2011), associated with chromatin signatures indicating transcriptional regulation (Guttman et al., 2009), under evolutionary constraint (Pollard et al., 2006; Ponjavic et al., 2007; Guttman et al., 2009), and associated with diseases (Qureshi and Mehler, 2011; Wapinski and Chang, 2011), all of which support a meaningful role of lncRNAs.
lncRNAs in the central nervous system
The vertebrate CNS contains an enormous diversity of neuronal and glial cell types that differentiate and form networks through an intricate developmental program that must coordinate intrinsic and external stimuli to achieve proper form and function. Cells in the CNS are highly transcriptionally active and exhibit strong expression of ncRNAs, with 5,458 of a total 9,747 lncRNA transcripts detected in the human brain using a microarray, including specific expression of ~40% of the most highly and differentially expressed lncRNAs (Derrien et al., 2012). In mouse embryonic stem cells, shRNA-mediated knockdown of 93% of long intergenic ncRNAs (lincRNAs) resulted in significant gene expression changes, implying that the majority of lincRNAs are functional (Guttman et al., 2011). In addition, lncRNAs can work through various mechanisms to exert precise control over both individual genes and large gene networks, allowing the spatiotemporal precision necessary to execute complex neurobiological processes.
Though ncRNAs are present in species from bacteria to humans, expansion of ncRNAs has been linked to evolutionary complexity. The study by Taft et al. provides a beautiful illustration of the link between the proportion of non-protein coding DNA sequence in the genome and evolutionary complexity, with vertebrates containing the highest proportion of non-protein coding sequence (Taft et al., 2007). miRNAs have already been linked to brain complexity and have an important role in brain cellular diversity (Berezikov et al., 2006); as lncRNAs continue to be characterized they will doubtless follow suit.
Large-scale studies surveying lncRNAs in the nervous system have provided further evidence of their importance. In situ hybridization has shown that many lncRNAs are expressed in specific anatomical regions, cell types, or subcellular compartments in the mouse brain (Mercer et al., 2008). In a study of intergenic lncRNAs, those in the brain were found to be highly conserved between mouse and humans, enriched in predicted RNA secondary structure, and preferentially located near protein-coding genes that are also implicated in nervous system development or function (Ponjavic et al., 2009). lncRNAs have also been shown in both human and mouse cells to influence pluripotency or differentiation specifically in neural cells (Mercer et al., 2010; Lin et al., 2011; Ng et al., 2012). In addition, lncRNAs have been implicated in neurodevelopmental, neurodegenerative, neuroimmunological, and neuro-oncological disorders, underlining their importance in CNS development and function (Table 1) (Mehler and Mattick, 2007; Qureshi et al., 2010; Lin et al., 2011; Qureshi and Mehler, 2011; Ziats and Rennert, 2012). In this review, we will highlight the roles of lncRNAs in nervous system development by connecting their genomic context with their possible cellular functions.
Table 1.
Summary of long noncoding RNAs with characterized functions in the central nervous system.
| Name | Type | Role | Species | References |
|---|---|---|---|---|
| Otx2OS | bidirectional | Expressed in the developing retina in a partially overlapping pattern with Otx2; expression pattern suggests repression of Otx2 | Mus musculus | Alfano et al., 2005 |
| Six3OS | bidirectional | Acts as a scaffold to direct histone modifiers to Six3 target genes; binds coregulators of Six3 to modulate target gene expression | Homo sapiens, Mus musculus, Gallus gallus | Alfano et al., 2005; Geng et al., 2007; Rapicavoli et al., 2011 |
| Vax2os1 | bidirectional | Retina-specific, overexpression delays cell cycle of photoreceptor progenitors during development | Mus musculus | Alfano et al., 2005; Meola et al., 2012 |
| FMR4 | bidirectional | Primate-specific, silenced in fragile-X syndrome patients; knockdown in HEK-293T and HeLa cells results in delayed cell cycle and increased apoptosis; overexpression promotes proliferation | Homo sapiens, Macaca mulatta | Khalil et al., 2008 |
| Sox8OT | bidirectional | Concordant expression with Sox8, may promote oligodendrocyte lineage commitment | Homo sapiens, Rattus norvegicus, Mus musculus | Mercer et al., 2010 |
| Lncat (Ube3a-ATS) | imprinted | Silences the paternal allele of Ube3a, involved in Prader-Willi and Angelman Syndromes | Homo sapiens, Mus musculus | Le Meur et al., 2005; Meng et al., 2012 |
| MEG3 | imprinted | Part of the imprinted DLK1-MEG3 locus; knockout leads to increased microvessel formation in the brain; loss may contribute to tumor formation | Homo sapiens, Mus musculus, Bos taurus | Hagan et al., 2009; Gordon et al., 2010; Wang et al., 2012 |
| Gomafu | lincRNA | Competes for splicing factors to inhibit splicing reactions; represses amacrine and Müller glial differentiation in the retina | Homo sapiens, mus musculus, Gallus gallus, Xenopus tropicalis | Sone et al., 2007; Rapicavoli et al., 2010; Ip and Nakagawa, 2011 |
| MALAT1 | lincRNA | Expressed late in neuron and oligodendrocyte differentiation; promotes synapse formation and dendrite growth | Homo sapiens, Mus musculus | Bernard et al., 2010; Mercer et al., 2010 |
| Neat1 | lincRNA | Architectural component of paraspeckles; promotes differentiation and maturation of neurons and oligodendrocytes | Homo sapiens, Mus musculus | Mercer et al., 2010; Ip and Nakagawa, 2011 |
| TUG1 | lincRNA | Required for photoreceptor formation in the developing retina, promotes rod differentiation and inhibits cone differentiation | Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Canis lupus familiaris | Young et al., 2005 |
| HAR1F | NAT | Primate-specific lncRNA expressed in Cajal-Retzius neurons in the developing neocortex; predicted to promote cortical neuron specification and migration | Homo sapiens | Pollard et al., 2006 |
| BC1/BC200 | NAT | Brain-specific transcript that represses translation in dendrites to allow spatial translation regulation in postsynaptic microdomains; dysregulated and implicated in Alzheimer’s disease | Homo sapiens, Mus musculus | Ohashi et al., 2000; Wang et al., 2002; Mus et al., 2007 |
| Sox4AS | NAT | Expressed with and independently of Sox4 throughout brain development; no function known | Mus musculus | Ling et al., 2009 |
| NrgnAS | NAT | Forms cytoplasmic dsRNA aggregates with Nrgn; may be involved in LTP | Mus musculus | Ling et al., 2011 |
| Camk2n1AS | NAT | Forms cytoplasmic dsRNA aggregates with Camk2n1; may be involved in LTP | Mus musculus | Ling et al., 2011 |
| BACE1-AS | NAT | Upregulated in Alzheimer’s brains; promotes BACE1 stability by blocking a miRNA binding site | Homo sapiens | Faghihi et al., 2008; Modarresi et al., 2011 |
| BDNF-AS | NAT | Coexpressed with BDNF in the brain; forms dsRNA duplexes to repress BDNF expression | Homo sapiens Homo | Pruunsild et al., 2007; Modarresi et al., 2012 |
| Emx2OS | NAT | In differentiating neural precursors, high levels destabilize Emx2, while low levels are required for proper Emx2 transcription | sapiens, Mus musculus | Spigoni et al., 2010 |
| Nkx2.2AS | NAT | Cytoplasmic localization, can increase Nkx2.2 mRNA levels; overexpression in NSCs promotes differentiation along the oligodendrocyte lineage | Homo sapiens, Mus musculus | Tochitani and Hayashizaki, 2008 |
| Disc2 | NAT | Translocation breakpoint segregates with susceptibility for schizophrenia; may regulate Disc1 sense gene expression | Homo sapiens | Millar et al., 2004; Brandon et al., 2009 |
| ATXN8OS | NAT | Repeat expansion associated with spinocerebellar ataxia type 8 causes accumulation of RNA foci and sequestration of Mbnl1, causing dysregulation of Mbnl1 target pathways | Homo sapiens, Mus musculus | Ikeda et al., 2008; Daughters et al., 2009 |
| anti-NOS2A | NAT | Undergoes reciprocal changes to NOS2A during hESC differentiation into neurons, indicating repression of NOS2A | Homo sapiens, Lymnaea stagnalis | Korneev et al., 2008 |
| SCAANT1 | NAT | Represses ataxin-7 expression in cis; implicated in spinocerebellar ataxia type 7 | Homo sapiens | Sopher et al., 2011 |
| Evf2 | overlapping | Influences expression of Dlx5, Dlx6, and Gad1 in the developing ventral forebrain; promotes generation of GABAergic interneurons in the hippocampus and dentate gyrus, loss results in reduced synaptic inhibition | Homo sapiens, Mus musculus | Feng et al., 2006; Bond et al., 2009 |
| Sox2OT | overlapping | May regulate Sox2 in neurogenesis to promote neural differentiation, also has independent roles | Homo sapiens, Mus musculus, Gallus gallus, Danio rerio | Amaral et al., 2009 |
| Svet1 | sense intronic | Expressed early in development in the proliferating cells of the SVZ and later in upper layer neurons, function unknown | Mus musculus | Tarabykin et al., 2001; Sasaki et al., 2008 |
lncRNAs associated with genomic contexts
It has become increasingly clear that eukaryotic genomic organization is highly complex, involving convergent, divergent, overlapping, and antisense transcription of protein-coding and ncRNAs (Carninci et al., 2005; Katayama et al., 2005; Kapranov et al., 2007; Derrien et al., 2012; Djebali et al., 2012). Many lncRNAs exhibit multiple splicing isoforms and transcription start sites, use of which may contribute to cell type- or developmental stage-specific events (Liu et al., 2006; Amaral et al., 2009; Spigoni et al., 2010). While several methods of classifying lncRNAs based on function have been proposed (Wilusz et al., 2009; Wang and Chang, 2011), this review will continue to classify lncRNAs based on genomic context as this information is easily accessible, while many cellular functions need to be further revealed.
lincRNAs
lncRNAs that are not found near any protein-coding locus (within <10kb) are considered long intergenic noncoding RNAs (lincRNAs) (Guttman et al., 2009; Ponting et al., 2009), the most numerous of the lncRNA types (Derrien et al., 2012) (Fig. 1). lincRNAs have been shown to have effects both in cis (Orom and Shiekhattar, 2011) and in trans gene expression regulation (Guttman et al., 2009; Guttman et al., 2011).
Figure 1. Genomic contexts of lncRNAs.
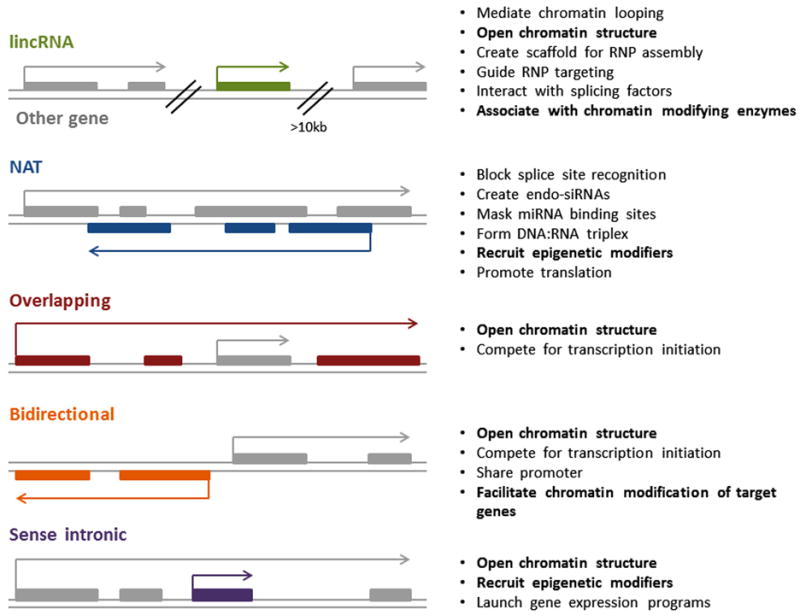
Schematic diagram demonstrating the various genomic positions of lncRNA types (in color) with respect to other genes (in grey). Possible mechanisms of each type are shown on the right, with thematic functions in bold.
lincRNAs in cis regulation
In cis, lincRNAs can have enhancer-like functions (Orom et al., 2010). Knockdown of these enhancer lincRNAs (also termed activating ncRNAs or ncRNA-a) with siRNAs resulted in decreased expression of neighboring genes, indicating that the enhancer-like effect was ncRNA-a-dependent (Orom and Shiekhattar, 2011). While the exact mechanism by which enhancer lincRNAs perform their function is unknown, the working model is that the ncRNA transcribed from the enhancer locus acts as a scaffold to mediate chromatin looping and assembly of transcriptional machinery (Orom and Shiekhattar, 2011). A second class of lincRNAs functioning in cis, called eRNA, is transcribed from the enhancer region of a gene and promotes transcription of nearby genes (Kim et al., 2010). Although the exact mechanism of eRNA function is unknown, the expression of eRNAs may promote transcription of nearby genes by opening chromatin structure, recruiting RNA polymerase II to the promoter, or by facilitating associations between enhancer and promoter regions (Kim et al., 2010).
MALAT1 (also known as NEAT2) has been shown to interact with splicing factors and is implicated in nervous system development as it is expressed during later stages of neuronal and oligodendrocyte development (Mercer et al., 2010). Knockdown of MALAT1 in cultured hippocampal neurons results in decreased synaptic density, while overexpression has opposite effects (Bernard et al., 2010). Contrary to in vitro studies, a Malat1 knockout mouse was generated and showed no general effects on transcription or splicing (Zhang et al., 2012). However, some genes located near the Malat1 locus were upregulated in mutant cortices, supporting a role for Malat1 in cis regulation (Zhang et al., 2012). The authors attribute the cis effect of Malat1 to transcriptional interference or histone modification, but do not eliminate the possibility that the Malat1 locus contains DNA elements that contribute to cis regulation (Zhang et al., 2012). Due to the identity of Malat1 as a lincRNA (>10kb from other loci), it is unlikely that Malat1 works through transcriptional interference, as other examples of transcriptional interference occur over much shorter distances (Bumgarner et al., 2012; Hongay et al., 2006; Petruk et al., 2006).
In the CNS, a study of transcription in the mouse neocortical layers revealed a set of 66 lincRNA loci that were patterned across the layers (Belgard et al., 2011). These transcripts were more likely to be correlated with the expression of their protein coding neighbors than unpatterned lincRNAs, supporting a role for some of these lincRNAs in cis regulation (Belgard et al., 2011). Further, these lincRNAs were enriched for transcription across enhancers, providing support for eRNA functions in cis. Specific lincRNAs functioning in cis in the CNS remain to be characterized.
lincRNAs in trans regulation
lincRNAs have also been shown through short hairpin RNA (shRNA) knockdown methods to influence global gene expression in trans (Guttman et al., 2011). Several studies have shown that many lincRNAs physically associate with chromatin modifiers, including readers, writers, and erasers, to influence gene expression at various loci (Khalil et al., 2009; Guttman et al., 2011). Similar to lincRNA action in cis, the proposed mechanism for lincRNAs acting in trans describes the ncRNA as a scaffold that aids in the assembly of large ribonucleoprotein complexes and their targeting to specific loci to execute cell-type-specific gene expression programs (Guttman et al., 2011).
Several lincRNAs that work in trans have been identified with important roles in the nervous system. Gomafu (also known as RNCR2 or MIAT) is expressed in differentiating neurons and oligodendrocytes (Mercer et al., 2010) as well as mitotic and postmitotic retinal precursors (Rapicavoli et al., 2010) and forms a punctate pattern of subnuclear domains that are not associated with any known nuclear bodies (Sone et al., 2007). The mechanism of Gomafu action is unknown, but it has been shown to interact with splicing factors and may influence alternative splicing patterns to drive differentiation (Tsuiji et al., 2011).
The lincRNA Neat1 is an architectural component of paraspeckles, a nuclear body involved in edited RNA retention (Fox and Lamond, 2010). Neat1 has been shown to be downregulated in neural and oligodendrocyte progenitors and upregulated in their progeny in mice (Mercer et al., 2010). Knockdown of Neat1 disrupts paraspeckles, causing export of the retained RNA to the cytoplasm (Chen and Carmichael, 2009). Because its nuclear localization and function in RNA retention are independent of genomic location, Neat1 works in trans.
In the developing mouse retina, lincRNA TUG1 is important for photoreceptor development (Young et al., 2005). Overexpression of TUG1 in the rat retina showed no obvious phenotype, but siRNA-mediated knockdown resulted in mislocalization and improper formation of rod photoreceptor cells (Young et al., 2005). Cells electroporated with the TUG1 siRNA constructs exhibited a decrease in rod-specific rhodopsin and an increase in cone-specific peanut agglutinin, as well as complementary changes in gene expression (Young et al., 2005). Microarray analysis of wild type and TUG1 knock down retinas revealed global down regulation of the rod-specific gene expression program in trans, indicating that the normal role of TUG1 is to promote photoreceptor differentiation into rod cells (Young et al., 2005).
Natural antisense transcripts
Natural antisense transcripts (NATs) are lncRNAs that are transcribed overlapping with and in the antisense orientation to protein coding genes or ncRNAs (Fig. 1). While the importance of NATs has been known for many years (Knee and Murphy, 1997), it was not until recently that studies by the FANTOM consortium discovered that antisense transcription is widespread in the mammalian genome and presents a powerful means of regulation (Carninci et al., 2005; Katayama et al., 2005). Further studies in human cells have also found that antisense transcription occurs in genomic contexts distinct from sense transcription, nonrandomly across the genome, and differs among cell types, indicating its importance in gene regulation (He et al., 2008).
NATs in cis regulation
Most NATs that have been described work in cis through various mechanisms. Splicing of the sense mRNA can be regulated by binding of the complementary NAT to block recognition of a splice site (Enerly et al., 2005; Werner and Sayer, 2009). NATs can create endo-siRNAs by binding the sense mRNA to generate regions of double stranded RNA (Tam et al., 2008; Watanabe et al., 2008). These endo-siRNAs can take either the sense or the antisense orientation to silence the coding mRNA or the NAT (Werner et al., 2009). More recent work has suggested a further mechanism for NATs in which they bind to the sense mRNA and mask miRNA binding sites, thus blocking miRNA-induced gene repression (Faghihi et al., 2010). NATs can also use their sequence complementarity to form a DNA:RNA triplex, creating a roadblock for the transcriptional machinery, which results in downregulation of the sense gene expression (Shearwin et al., 2005). Furthermore, NATs can recruit epigenetic modifiers to the locus from which they are transcribed to cause silencing or activation of neighboring genes (Morris, 2011; Magistri et al., 2012). Finally, NATs can help their complementary protein-coding mRNA associate with active polysomes to promote translation (Carrieri et al., 2012).
In the nervous system, many important genes have antisense transcription (Mercer et al., 2008). Among these genes is BDNF, which is involved in the developing and adult nervous system in processes such as cell survival and differentiation, axon growth and pathfinding, and synaptic transmission as well as several neuropsychiatric and neurodegenerative diseases (Bibel and Barde, 2000; Binder and Scharfman, 2004). Upon examination of the BDNF locus in humans, it was found that there are 12 isoforms of a lncRNA transcribed antisense to the BDNF mRNA (Pruunsild et al., 2007). At least 4 of these BDNF-AS transcripts are coexpressed with BDNF in the brain, where they form RNA:RNA duplexes (Pruunsild et al., 2007). A recent study found that inhibition of BDNF-AS results in upregulated BDNF mRNA, decreased chromatin silencing marks at the BDNF locus, increased BDNF protein, and increased neurite outgrowth and neuronal differentiation (Modarresi et al., 2012). The formation of RNA:RNA duplexes suggests that BDNF-AS could act through generation of endo-siRNAs or by blocking splicing. Moreover, the observation of changed chromatin marks suggests that BDNF-AS could also regulate its sense partner through recruitment of chromatin modifiers.
The transcription factor Emx2 has been implicated in arealization, precursor proliferation, and lamination in the developing cerebral cortex (Mallamaci et al., 1998; Bishop et al., 2000; Heins et al., 2001). Antisense transcription from the Emx2 locus produces several antisense isoforms, one of which (Emx2OS) is expressed in the developing CNS (Spigoni et al., 2010). In differentiating neural precursors, high levels of Emx2OS cause destabilization of Emx2 mRNA by Dicer1, while low levels are necessary for proper Emx2 transcription (Spigoni et al., 2010). It was also found that Emx2 sense transcription is required for proper Emx2OS expression (Spigoni et al., 2010).
Analysis of the locus containing Nrgn and Camk2n1 revealed multiple overlapping and antisense transcripts that were found to be developmentally regulated during corticogenesis (Ling et al., 2011). These NATs, Nrgn-AS and Camk2n1-AS, were localized in the cytoplasm and formed double stranded RNA complexes with their sense transcripts. The detailed mechanism is unknown, but the formation of dsRNA may prevent translation of their sense counterparts or serve as sources of endo-siRNAs and regulate the spatiotemporal expression of Nrgn and Camk2n1 during corticogenesis (Ling et al., 2011).
The BACE1 protein is critical in Alzheimer’s disease and the BACE1 mRNA has a highly conserved antisense transcript (BACE1-AS) that is upregulated in the disease state (Faghihi et al., 2008a). BACE1-AS stabilizes its sense partner by blocking the binding site for a miRNA, resulting in upregulated BACE1 expression (Faghihi et al., 2010). Knockdown of BACE1-AS causes reduced levels of beta-amyloid, suggesting that the antisense transcript could be used as both an early biomarker and a therapeutic target (Modarresi et al., 2011).
The gene Uchl1 encodes a deubiquitinating enzyme that plays roles in brain function as well as Parkinson’s disease and Alzheimer’s disease pathogenesis (Setsuie and Wada, 2007). In a search for sense-antisense gene pairs in the brain, antisense Uchl1 was identified, which overlaps with the first two exons of Uchl1 (Carrieri et al., 2012). Through knockdown and overexpression experiments, this antisense lncRNA was found together with Uchl1 in the cytoplasm and promote its association with active polysomes to increase translation (Carrieri et al., 2012).
Other genes with NATs include Nkx2.2, a transcription factor known to direct differentiation of neural stem cells into the oligodendrocyte lineage, and Sox4, a transcription factor important for specification and differentiation of neurons and glia in the developing CNS (Cheung et al., 2000; Tochitani and Hayashizaki, 2008; Ling et al., 2009). Another interesting study found that some sense-antisense transcript pairs are coexpressed in synaptoneurosomal preparations, indicating that they are coexpressed in the dendrites of pyramidal neurons and may be involved in regulation of synaptic plasticity (Smalheiser et al., 2008).
NATs in trans regulation
NATs can also work in trans, meaning they are transcribed from a genomic locus different from the one they target, but maintain sequence complementarity (Wang et al., 2006). A study of ESTs across many species estimates that there are 2,896 transcriptional units in humans that are involved in trans sense/antisense pairs, one fourth of which (>700) involve noncoding RNAs (Li et al., 2008). Of the trans sense/antisense pairs involving ncRNAs, a majority were correlated to inhibition or stabilization of complementary sense transcripts, while the remainder could be involved in regulation of translation, splicing, RNA editing, trafficking, or subcellular localization, similar to their cis-acting counterparts (Li et al., 2008). For example, the mouse Msh4 meiotic recombination gene forms a dsRNA complex with Hspa5 mRNA and ultimately causes cell death (Hirano and Noda, 2004).
In the human nervous system, a duplicated and inverted locus produces an RNA transcript with antisense homology to the NOS2A mRNA (Korneev et al., 2008). This trans-NAT undergoes expression changes reciprocal to the NOS2A mRNA during differentiation of human embryonic stem cells into neurospheres, suggesting it may negatively regulate its sense partner and play an important role in neuronal differentiation (Korneev et al., 2008). This antisense transcript was also shown to be expressed in human brain tumors, including glioblastomas and meningiomas, indicating a possible role in tumor formation (Korneev et al., 2008).
In the human genome, several regions have been identified to show significant evolutionary acceleration compared to the chimpanzee genome, many of which are associated with genes involved in neurodevelopment (Chimpanzee Sequencing and Analysis Consortium, 2005; Nielsen et al., 2005). One of these regions, HAR1, encodes a lncRNA, HAR1F, which is antisense to another lncRNA, HAR1R, and is expressed in the developing human cortex (Pollard et al., 2006). HAR1F is coexpressed with reelin in Cajal-Retzius neurons and forms a stable secondary structure, suggesting a role in establishing the lamination of the cortex (Pollard et al., 2006). However, whether HAR1F influences reelin or reelin receptor expression is unknown. HAR1R is expressed at lower levels and later stages in brain development and may have a role in HAR1F downregulation by antisense inhibition (Pollard et al., 2006).
The rodent-specific BC1 and the primate-specific BC200 are highly expressed NATs in the brain, where they repress translation initiation in dendrites in an activity-dependent manner (Muslimov et al., 1998; Wang et al., 2002). Because the 200 nt cut off of lncRNAs is arbitrary and meant to distinguish lncRNAs from small ncRNAs, BC1 is treated here as a lncRNA, even though it produces a transcript of 152 nt, due to its function in trans regulation of translation initiation. In addition, BC200 was found to be upregulated in Alzheimer’s Disease brains, and may contribute to the complex etiology of the disease (Mus et al., 2007).
In summary, it appears that the NAT lncRNAs work through multiple mechanisms, both in cis and in trans, to ensure proper expression levels of its sense partner in both CNS development and function.
Overlapping transcripts
One additional type of lncRNA is an overlapping transcript, which is transcribed in the same direction as and contains a protein-coding gene (Fig. 1). Possible mechanisms of overlapping transcript regulation include cis-acting promoter competition (Conte et al., 2002) or synergistic expression through opening of chromatin structure and deposition of histone marks that activate downstream transcription (Wagner and Carpenter, 2012).
lncRNA Sox2ot, which overlaps with Sox2 within an intron, has several isoforms that are dynamically regulated and expressed both in conjunction with Sox2 and independently, indicating a possible role in regulation of Sox2 (Amaral et al., 2009). In situ hybridization for Sox2ot in the adult mouse brain revealed a striking overlap of expression with the Sox2 protein-coding gene in areas of adult neurogenesis (Mercer et al., 2008).
The overlapping lncRNA Evf2 is transcribed in the opposite direction of Dlx6 and contains Dlx6 within an intron (Feng et al., 2006). From this genomic location, Evf2 can recruit positive and negative transcription factors to the nearby enhancer to regulate the expression of Dlx5 and Dlx6 (Bond et al., 2009). Evf2 has been shown to form a stable complex with the transcription factor Dlx2 in vivo, indicating that it has direct influence on transcriptional activity (Feng et al., 2006). Mice mutant in Evf2 had decreased numbers of postnatal GABAergic interneurons and reduced synaptic inhibition in the adult hippocampus, which may be implicated in disorders such as schizophrenia, autism, Tourette’s syndrome, and epilepsy (Bond et al., 2009). Knockdown of Evf2 by insertion of an early polyadenylation signal resulted in increased transcription of both Dlx5 and Dlx6, but ectopic Evf2 could only rescue the Dlx5 level, indicating that the lincRNA works through different mechanisms to regulate transcription of each of the genes (Bond et al., 2009).
As 52 overlapping sense lncRNAs have been identified in the human genome (Derrien et al., 2012), the importance of this class of lncRNAs is emerging.
Bidirectional lncRNAs
Bidirectional or divergent lncRNAs have transcription start sites in close proximity to a protein-coding gene but are transcribed in the opposite direction (Fig. 1). Transcription of bidirectional lncRNAs is often concordant with expression of the nearby protein-coding gene, suggesting a cis-regulatory role of the lncRNAs, possibly in maintaining an open chromatin structure through pervasive expression (Wei et al., 2011) or through shared promoters (Ho et al., 2012). However, some lncRNAs have a discordant expression relationship with the nearby protein-coding gene (Mercer et al., 2008), implying a repressive role of the lncRNA in cis, possibly through promoter competition (Trinklein et al., 2004), or an independent role in trans (Engstrom et al., 2006). For the majority of bidirectional pairs, the orientation is conserved between human and mouse, implying functional importance in evolution (Trinklein et al., 2004).
In humans, a primate-specific bidirectional lncRNA, FMR4, is silenced in fragile-X syndrome patients and appears to have an anti-apoptotic function (Khalil et al., 2008). The Sox8 locus is important in oligodendrocyte lineage specification and also hosts a bidirectional lncRNA, Sox8OT (Mercer et al., 2010). A biochemical function for Sox8OT has not been identified, but it shows concordant expression with its sense partner and may share a promoter (Mercer et al., 2010). The bidirectional lncRNA Six3OS has also been found to work in trans to regulate retinal differentiation (Alfano et al., 2005; Geng et al., 2007; Rapicavoli et al., 2011). Knockdown or overexpression of Six3OS does not affect Six3 expression in the mouse retina, indicating there is not an interaction in cis on the host gene (Rapicavoli et al., 2011). However, as discovered through protein microarray and confirmed by RNA immunoprecipitation, Six3OS does interact with histone modification enzymes, suggesting that Six3OS can regulate Six3 function by facilitating chromatin modifications of Six3 target genes (Rapicavoli et al., 2011).
Other genes important in retinal development that have bidirectionally transcribed lncRNAs include Otx2 and Vax2 (Alfano et al., 2005; Meola et al., 2012). Functionality of Otx2OS has not been confirmed, but its expression pattern suggests possible repression of Otx2 (Alfano et al., 2005). Vax2OS1 represses cell cycle progression and differentiation in photoreceptor progenitors (Meola et al., 2012). The most likely mechanism for Vax2OS1 is in trans since it is expressed in different cell types than Vax2 and does not affect Vax2 transcription (Meola et al., 2012).
Until now, there are 3,417 bidirectional lncRNAs annotated in the human genome (Derrien et al., 2012) and 51 identified in the mouse brain (Mercer et al., 2008). Fully functionally characterization of bidirectional lncRNAs remains an active research direction.
Sense intronic lncRNAs
A final class of lncRNAs is long intronic RNAs that are transcribed in the same direction as their protein-coding host gene (Fig. 1). Unlike their NAT counterparts, these sense intronic lncRNAs do not have antisense complementarity to their host gene and therefore must work through different mechanisms. The most well-known example of a sense intronic lncRNA is COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), which is transcribed from an intron of FLOWERING LOCUS C (FLC) (Heo and Sung, 2011). COLDAIR epigenetically regulates the FLC locus by recruiting the polycomb repressive complex 2 (PRC2) to silence FLC expression (Heo and Sung, 2011). Though this example comes from Arabidopsis, the mechanism may be conserved as lncRNAs in the nervous system are known to interact with PRC2 and other epigenetic regulators (Johnson et al., 2009; Ng et al., 2012). Another mechanism of sense intronic lncRNAs is coordination of gene expression programs through trans activation of diverse genes (Hill et al., 2006). In the mouse brain, sense intronic lncRNAs were found both co-expressed with their host genes and expressed independently, indicating that they could have an activating or repressive effect on the host locus or could work in trans (Mercer et al., 2008). One possible sense intronic lncRNA, Svet1, has been identified, but its function is unknown. Svet1, which maps to an intron of the Unc5d putative Netrin receptor gene, is expressed early in development in the subventricular zone and later in the upper layer neurons of the mouse cortex (Tarabykin et al., 2001). Sasaki et al. attributed Svet1 to a splicing product of the Unc5d mRNA as it is only found in the nucleus, but did not eliminate the possibility that Svet1 is an independent transcript (Sasaki et al., 2008).
There are 593 sense intronic lncRNAs annotated in the human genome (Derrien et al., 2012) and 182 shown to be expressed in the adult mouse brain (Mercer et al., 2008); while less prevalent than other types of lncRNAs, sense intronic lncRNAs may still be shown to have interesting and important functions in the nervous system.
lncRNAs independent of genomic context
Pseudogenes
Previously thought of as junk DNA or evolutionary relics, pseudogenes have been shown to have functionality in recent studies (Tam et al., 2008; Watanabe et al., 2008; Hawkins and Morris, 2010; Muro and Andrade-Navarro, 2010; Poliseno et al., 2010; Han et al., 2011). Pseudogenes can be formed through spontaneous mutation that prevents transcription or translation, gene duplication due to uneven crossing over, or reintegration of an mRNA transcript (reviewed in Pink et al., 2011). The recent GENCODE project has annotated 11,224 pseudogenes in the human genome, of which 863 were transcribed and associated with active chromatin marks (Pei et al., 2012). There are several proposed mechanisms of pseudogene action. First, they can regulate the expression of their parent gene by competing for mRNA stability factors (Chiefari et al., 2010). Pseudogenes can also be a source of transNATs (Hawkins and Morris, 2010; Muro and Andrade-Navarro, 2010; Muro et al., 2011). Several studies have shown that endo-siRNAs originating from pseudogenes binding their parent mRNA can regulate mRNA expression through RNA interference (Tam et al., 2008; Watanabe et al., 2008; Poliseno et al., 2010). Finally, an additional function of pseudogenes is providing decoy miRNA binding sites, resulting in increased expression of the parent mRNA (Poliseno et al., 2010).
While a large proportion of transcribed pseudogenes were found to be expressed in the brain (Pei et al., 2012), few functional studies have been performed. An RNA-seq study conducted during the differentiation of induced pluripotent stem cells into neurons found that expression of 1,371 pseudogenes changed significantly between the two cell types (Lin et al., 2011). Mentioned above, the NAT to NOS2A is derived from a pseudogene of NOS2A and may suppress the parent gene expression (Korneev et al., 2008). Aside from this example, the functions and mechanisms of pseudogenes in the nervous system are yet to be characterized.
Imprinted lncRNAs
The best known lncRNAs involved in imprinting are inactive X-specific transcript (Xist) and X (inactive)-specific transcript, antisense (TsiX). These lncRNAs are required for X chromosome inactivation in females, an epigenetic event that maintains equal X chromosome expression between the mammalian sexes (Leeb et al., 2009). More generally, lncRNAs are frequently found within imprinted clusters and are often implicated in the cis regulation of imprinted genes (Santoro and Barlow, 2011). The most widely studied mechanism of lncRNA-driven silencing is through recruitment of chromatin-modifying enzymes to the locus from which the lncRNA is transcribed (Mohammad et al., 2009). Another possible mechanism is transcriptional interference, in which constant transcription from a lncRNA promoter can block transcription from a coding gene promoter on the opposite strand (Callen et al., 2004; Pauler et al., 2007).
The Prader-Willi syndrome/Angelman syndrome locus in humans is differentially imprinted on the maternal and paternal chromosome in neurons, perturbation of which results in neurodevelopmental disorders (Buiting, 2010). The UBE3A gene encodes a ubiquitin E3 ligase that is expressed maternally, while the paternal allele expresses a lncRNA antisense transcript, UBE3A-ATS (LNCAT in mouse) (Le Meur et al., 2005). The causal link between UBE3A-ATS expression and UBE3A paternal silencing in neurons has recently been demonstrated, though it is still unknown whether the silencing is mediated through the RNA itself or through transcriptional interference (Meng et al., 2012).
The lncRNA MEG3 is part of the imprinted DLK1-MEG3 locus that is expressed from the maternal allele but suppressed at the paternal allele in humans and mice (Hagan et al., 2009). Knockout of Meg3 in mice followed by microarray analysis revealed that Meg3 is involved in many processes in brain development, including calcium, Notch, and Wnt signaling, long-term potentiation, and angiogenesis (Gordon et al., 2010). In the mouse brain, knockout of Meg3 resulted in increased microvessel formation (Gordon et al., 2010). Meg3 activates the tumor suppressor p53 in vitro and is lost in several tumor types, indicating that it works as a tumor suppressor as well as playing roles in brain development, though the biochemical mechanism is unknown (Gordon et al., 2010; Wang et al., 2012).
lncRNAs in neurological diseases
Many lncRNAs are implicated in a wide variety of neurological diseases. In neurodevelopmental disorders, Ube3a-ATS is transcribed from the Angelman Syndrome and Prader-Willi Syndrome locus and FMR4 is silenced in Fragile-X Syndrome (Khalil et al., 2008; Meng et al., 2012). In addition, the DISC locus, including DISC1 and its NAT DISC2, is disrupted in schizophrenia and affective disorders (Millar et al., 2004). DISC1 regulates neuron proliferation, migration, and neurite outgrowth during cortical development (Kamiya et al., 2012). Like many NATs, DISC2 is thought to regulate the expression of its sense partner, thereby contributing to disease etiology (Brandon et al., 2009).
BACE1-AS and BC1/BC200 have been implicated in Alzheimer’s disease, while ATXN8OS is associated with neurodegenerative spinocerebellar ataxia type 8 (Mus et al., 2007; Faghihi et al., 2008b; Daughters et al., 2009). In addition, spinocerebellar ataxia type 7 is a neurological disorder characterized by cerebellar and retinal degeneration caused by a repeat expansion in the ataxin-7 gene (Martin et al., 1994). A convergently transcribed and slightly overlapping NAT, SCAANT1 has been found to repress ataxin-7 transcription in cis (Sopher et al., 2011). Repeat expansion of ataxin-7 reduces SCAANT1 promoter activity, thereby derepressing ataxin-7 and contributing to the disease state (Sopher et al., 2011). Moreover, anti-NOS2A is upregulated in human brain tumors and can influence the efficacy of chemotherapeutic drugs, while MEG3 is lost in several tumor types (Broholm et al., 2003; Gordon et al., 2010; Zhou et al., 2012).
As more lncRNAs associated with neurological disorders are identified and functions of more lncRNAs are characterized, lncRNAs may become a new therapeutic target for the treatment of neurological diseases.
Perspectives
With the release of the latest lncRNA information from GENCODE (Derrien et al., 2012; Djebali et al., 2012; Pei et al., 2012), one of the greatest obstacles in lncRNA biology – annotation – has been largely overcome. The field must now move toward detailed functional and mechanistic studies. As evidenced by the current dearth of functionally defined lncRNAs, this is not an easy task.
Generation of knockout models of lncRNAs may be instructive to learn about lncRNA function, but the results must be analyzed with care as knockout of lncRNAs that work in cis could produce effects beyond the action of the lncRNA itself. Some lncRNAs may also function as precursors for small RNAs, such as miRNAs or small nucleolar RNAs (snoRNAs), which could complicate functional studies (Derrien et al., 2012). It will be important to demonstrate recovery of phenotype with the lncRNA in trans in these cases to determine if the transcript functions as a lncRNA or if the lncRNA is merely a precursor. A risk of generating lncRNA knockout animal models is the lack of identifiable phenotypes due to its fine-tuned regulation of gene expression and unknown functions. In addition, fusion of lncRNAs to an IRES-GFP sequence is a complementary approach to knockdown lncRNAs, as these constructs mislocalize to the cytoplasm and produce a dominant negative effect (Rapicavoli et al., 2010). Again, these results must be interpreted with care as cytoplasmic localization of some lncRNAs may have off-target effects.
As many lncRNAs have been implicated in epigenetic regulation (Khalil et al., 2009; Tsai et al., 2010; Magistri et al., 2012; Ng et al., 2012), it will be helpful to know where in the genome the lncRNA is binding. Chromatin isolation by RNA purification (ChIRP) allows genome-wide mapping of lncRNA occupancy based on the lncRNA sequence (Chu et al., 2011; Chu et al., 2012). This technique is especially powerful because it depends only on the lncRNA sequence, not the structural or functional domains.
In addition to learning where in the genome lncRNAs are binding, it will be important to know what proteins they are interacting with. RNA coimmunoprecipitation followed by deep sequencing (RIP-seq) is a method used to determine all of the lncRNAs that are interacting with a single protein, which can be used to infer lncRNA function (Moran et al., 2012a). A complementary method for pulldown of a specific lncRNA involves insertion of a sequence that creates a stem-loop structure at the 3’ end of the lncRNA and is recognized by the bacteriophage MS2 coat-binding protein (Gong et al., 2012). Finally, lncRNAs can be hybridized to protein microarrays to identify lncRNA:protein interactions (Hu et al., 2009; Rapicavoli et al., 2011). Subsequent analysis of the proteins and RNAs that co-purify with the lncRNA will give clues to the lncRNA function.
Another obstacle to determining lncRNA function is the lack of methods to determine secondary structure. Parallel analysis of RNA structure (PARS) is a technique that has been used in yeast to profile the genome-wide secondary structure of RNAs (Kertesz et al., 2010). The technique involves treatment of RNA with structure-specific enzymes followed by deep sequencing to identify the resulting fragments. PARS has the potential to be modified for use with lncRNAs and could give insights to conservation of lncRNA secondary structure across different species or allow categorization of secondary structures capable of interacting with different proteins.
Finally, many strategies currently used to target mRNAs or small ncRNAs could be applied to lncRNAs, such as antisense and catalytic oligonucleotides. For example, shRNA and siRNA knockdown of lncRNAs have become successful research tools and could present a potential lncRNA-targeting therapeutic strategy (Bernard et al., 2010; Orom et al., 2010; Poliseno et al., 2010; Rapicavoli et al., 2010; Kotake et al., 2011; Flockhart et al., 2012; Modarresi et al., 2012). Catalytic oligonucleotides, such as antagomirzymes (also called LNAzymes) have been used in vitro and in vivo to inhibit miRNA function through sequence complementarity and a catalytic core that cleaves the target RNA (Jadhav et al., 2009; Suryawanshi et al., 2012). For both of these tools, a targeted delivery method would be required to observe specific effects of lncRNA knockdown in the CNS.
Conclusions
It has become abundantly clear that noncoding transcription plays an important role in the functional genome. lncRNAs are a pervasive class of ncRNAs transcribed from various genomic orientations, providing a wide array of regulatory mechanisms that can work through fine-scale modulation of gene expression or large-scale regulation of developmental programs. The precise temporal and spatial control of lncRNAs is particularly important in the CNS, where lncRNAs have been implicated in every stage of neurodevelopment as well as a growing number of CNS pathologies (Table 1). However, despite their apparent importance, a strikingly small number of lncRNAs have been mechanistically characterized due to their diverse roles and often multiple target loci. With the latest ENCODE annotation of lncRNAs and new tools for precipitation, purification, and knockdown of lncRNAs, it is time to move toward functional characterization of individual lncRNAs. As we learn more about how lncRNAs are working, we can further our understanding of the complex molecular interactions of the extensive non-protein-coding components of our genome in regulating the development and function of the CNS.
Highlights.
About 98% of the human genome contains transcripts of noncoding RNAs.
Many long noncoding RNAs (lncRNAs) are expressed in the central nervous system.
lncRNAs can be classified as sense, antisense, bidirectional, intronic, intergenic
lncRNAs regulate coding gene expression using many mechanisms in cis and trans.
Dysregulation of lncRNAs is associated with human neurological disorders.
Acknowledgments
This work was supported by the Hirschl/Weill-Caulier Trust (T. S.), an NPRP grant (09-1011-3-260) from the Qatar National Research Fund and an R01-MH083680 grant from the NIH/NIMH (T. S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, McInnes RR, Auricchio A, Banfi S. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14:913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. Rna. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Research. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques Ana C, Oliver Peter L, Abaan HO, Sirey Tamara M, Hoerder-Suabedissen A, García-Moreno F, Molnár Z, Margulies Elliott H, Ponting Chris P. A Transcriptomic Atlas of Mouse Neocortical Layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nature Genetics. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The EMBO Journal. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde Y-A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes & Development. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, O’Leary DDM. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biology. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm H, Rubin I, Kruse A, Braendstrup O, Schmidt K, Skriver EB, Lauritzen M. Nitric oxide synthase expression and enzymatic activity in human brain tumors. Clinical Neuropathology. 2003 Nov-Dec;:273–281. [PubMed] [Google Scholar]
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Molecular Brain Research. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Chiefari E, Iiritano S, Paonessa F, Le Pera I, Arcidiacono B, Filocamo M, Foti D, Liebhaber SA, Brunetti A. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat Commun. 2010;1:40. doi: 10.1038/ncomms1040. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial squence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012 doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Seminars in Cell & Developmental Biology. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Conte C, Dastugue B, Vaury C. Promoter competition as a mechanism of transcriptional interference mediated by retrotransposons. The EMBO Journal. 2002;21:3908–3916. doi: 10.1093/emboj/cdf367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genetics. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerly E, Sheng Z, Li K-B. Natural Antisense as Potential Regulator of Alternative Initiation, Splicing and Termination. In Silico Biology. 2005;5:367–377. [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, Kunarso G, Ng EL, Batalov S, Wahlestedt C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Wells C, Bajic VB, Orlando V, Reid JF, Lenhard B, Lipovich L. Complex Loci in human and mouse genomes. PLoS Genetics. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biology. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Development. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Research. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Lavado A, Lagutin OV, Liu W, Oliver G. Expression of Six3 Opposite Strand (Six3OS) during mouse embryonic development. Gene Expr Patterns. 2007;7:252–257. doi: 10.1016/j.modgep.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Popp MW, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012 doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, O’Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YJ, Ma SF, Yourek G, Park YD, Garcia JG. A transcribed pseudogene of MYLK promotes cell proliferation. Faseb J. 2011;25:2305–2312. doi: 10.1096/fj.10-177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The Antisense Transcriptomes of Human Cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Cremisi F, Malatesta P, Gangemi RM, Corte G, Price J, Goudreau G, Gruss P, Gotz M. Emx2 promotes symmetric cell divisions and a multipotential fate in precursors from the cerebral cortex. Mol Cell Neurosci. 2001;18:485–502. doi: 10.1006/mcne.2001.1046. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hill AE, Hong JS, Wen H, Teng L, McPherson DT, McPherson SA, Levasseur DN, Sorscher EJ. Micro-RNA-like effects of complete intronic sequences. Frontiers in Bioscience. 2006;11:1998–2006. doi: 10.2741/1941. [DOI] [PubMed] [Google Scholar]
- Hirano M, Noda T. Genomic organization of the mouse Msh4 gene producing bicistronic, chimeric and antisense mRNA. Gene. 2004;342:165–177. doi: 10.1016/j.gene.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Ho MR, Tsai KW, Lin WC. A unified framework of overlapping genes: Towards the origination and endogenic regulation. Genomics. 2012;100:231–239. doi: 10.1016/j.ygeno.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu N, Wang JP, Wang YQ, Yu XL, Wang ZB, Cheng XC, Zou Q. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012 doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Development, Growth & Differentiation. 2011 doi: 10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- Jadhav VM, Scaria V, Maiti S. Antagomirzymes: Oligonucleotide Enzymes That Specifically Silence MicroRNA Function. Angew Chem Int Ed. 2009;48:2557–2560. doi: 10.1002/anie.200805521. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, Buckley NJ, Stanton LW, Lipovich L. Regulation of neural macroRNAs by the transcriptional repressor REST. Rna. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Sedlak TW, Pletnikov MV. DISC1 Pathway in Brain Development: Exploring Therapeutic Targets for Major Psychiatric Disorders. Front Psychiatry. 2012;3:25. doi: 10.3389/fpsyt.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A Novel RNA Transcript with Antiapoptotic Function Is Silenced in Fragile X Syndrome. PLoS ONE. 2008:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee R, Murphy PR. Regualtion of Gene Expression by Natural Antisense RNA Transcripts. Neurochem Int. 1997;31:379–392. doi: 10.1016/s0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O’Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. Rna. 2008;14:2030–2037. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Developmental Biology. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Leeb M, Steffen PA, Wutz A. X chromosome inactivation sparked by non-coding RNAs. RNA Biology. 2009;6:94–99. doi: 10.4161/rna.6.2.7716. [DOI] [PubMed] [Google Scholar]
- Li JT, Zhang Y, Kong L, Liu QR, Wei L. Trans-natural antisense transcripts including noncoding RNAs in 10 species: implications for expression regulation. Nucleic Acids Research. 2008;36:4833–4844. doi: 10.1093/nar/gkn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. RNA-Seq of Human Neurons Derived from iPS Cells Reveals Candidate Long Non-Coding RNAs Involved in Neurogenesis and Neuropsychiatric Disorders. PLoS ONE. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KH, Hewitt CA, Beissbarth T, Hyde L, Banerjee K, Cheah PS, Cannon PZ, Hahn CN, Thomas PQ, Smyth GK, Tan SS, Thomas T, Scott HS. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biology. 2009;10:R104. doi: 10.1186/gb-2009-10-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KH, Hewitt CA, Beissbarth T, Hyde L, Cheah PS, Smyth GK, Tan SS, Hahn CN, Thomas T, Thomas PQ, Scott HS. Spatiotemporal regulation of multiple overlapping sense and novel natural antisense transcripts at the Nrgn and Camk2n1 gene loci during mouse cerebral corticogenesis. Cereb Cortex. 2011;21:683–697. doi: 10.1093/cercor/bhq141. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallamaci A, Iannone R, Briata P, Pintonello L, Mercurio S, Boncinelli E, Corte G. EMX2 protein in the developing mouse brain and olfactory area. Mechanisms of Development. 1998;77:165–172. doi: 10.1016/s0925-4773(98)00141-5. [DOI] [PubMed] [Google Scholar]
- Martin JJ, Van Regemorter N, Krols L, Brucher JM, de Barsy T, Szliwowski H, Evrard P, Ceuterick C, Tassignon MJ, Smet-Dieleman H. On an autosomal dominant form of retinal-cerebellar degeneration: an autopsy study of five patients in one family. Acta Neuropathol. 1994;88:277–286. doi: 10.1007/BF00310370. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA Editing in Brain Development, Functional Diversification, and Neurological Disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. Rna. 2012;18:111–123. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, James R, Brandon NJ, Thomson PA. DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric illness. Annals of Medicine. 2004;36:367–378. doi: 10.1080/07853890410033603. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. Knockdown of BACE1-AS Nonprotein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int J Alzheimers Dis. 2011;2011 doi: 10.4061/2011/929042. 929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- Moran VA, Niland CN, Khalil AM. Co-Immunoprecipitation of Long Noncoding RNAs. Methods Mol Biol. 2012a;925:219–228. doi: 10.1007/978-1-62703-011-3_15. [DOI] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Research. 2012b;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Modulation of gene-specific epigenetic states and transcription by non-coding RNAs. Clinical Epigenetics. 2011;2:433–437. doi: 10.1007/s13148-011-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Andrade-Navarro MA. Pseudogenes as an alternative source of natural antisense transcripts. BMC Evolutionary Biology. 2010;10:338. doi: 10.1186/1471-2148-10-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916–1921. doi: 10.1016/j.biochi.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Banker G, Brosius J, Tiedge H. Activity-dependent Regulation of Dendritic BC1 RNA in Hippocampal Neurons in Culture. Journal of Cell Biology. 1998;141:1601–1611. doi: 10.1083/jcb.141.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. The EMBO Journal. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, Sninsky JJ, Adams MD, Cargill M. A Scan for Positively Selected Genes in the Genomes of Humans and Chimpanzees. PLoS Biology. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Kobayashi S, Omori A, Ohara S, Omae A, Muramatsu T, Li Y, Anzai K. The Single-Stranded DNA- and RNA-Binding Proteins Pur α and Pur β Link BC1 RNA to Microtubules Through Binding to the Dendrite-Targeting RNA Motifs. J Neurochem. 2000;75:1781–1790. doi: 10.1046/j.1471-4159.2000.0751781.x. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Current Opinion in Genetics & Development. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends in Genetics. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu X, Harte R, Balasubramanian S, Tanzer A, Diekhans M, Reymond A, Hubbard TJ, Harrow J, Gerstein MB. The GENCODE pseudogene resource. Genome Biology. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genetics. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Research. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. 2011;17:337–346. doi: 10.1016/j.molmed.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Developmental Biology. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F, Barlow DP. Developmental control of imprinted expression by macro non-coding RNAs. Seminars in Cell & Developmental Biology. 2011;22:328–335. doi: 10.1016/j.semcdb.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Tabata H, Tachikawa K, Nakajima K. The cortical subventricular zone-specific molecule Svet1 is part of the nuclear RNA coded by the putative netrin receptor gene Unc5d and is expressed in multipolar migrating cells. Mol Cell Neurosci. 2008;38:474–483. doi: 10.1016/j.mcn.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Torvik VI, Mise N, Ikeda R, Abe K. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci Res. 2008;62:236–239. doi: 10.1016/j.neures.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, Thienes CP, Gaasterland T, Filippova GN, La Spada AR. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigoni G, Gedressi C, Mallamaci A. Regulation of Emx2 expression by antisense transcripts in murine cortico-cerebral precursors. PLoS ONE. 2010;5:e8658. doi: 10.1371/journal.pone.0008658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature Structural & Molecular Biology. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Suryawanshi H, Lalwani MK, Ramasamy S, Rana R, Scaria V, Sivasubbu S, Maiti S. Antagonism of microRNA function in zebrafish embryos by using locked nucleic acid enzymes (LNAzymes) Chembiochem. 2012;13:584–589. doi: 10.1002/cbic.201100789. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Research. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochemical and Biophysical Research Communications. 2008;372:691–696. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An Abundance of Bidirectional Promoters in the Human Genome. Genome Research. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chua NH, Wang XJ. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biology. 2006;7:R92. doi: 10.1186/gb-2006-7-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sononberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: Functional Role in Regulation of Translation Initiation. The Journal of Neuroscience. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biology. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- Werner A, Sayer JA. Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens. 2009;18:343–349. doi: 10.1097/MNH.0b013e32832cb982. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & Development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Current biology : CB. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. Journal of Molecular Endocrinology. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant Expression of Long Noncoding RNAs in Autistic Brain. Journal of Molecular Neuroscience. 2012 doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


