Abstract
Pre-mRNA splicing is catalyzed through the activity of the spliceosome, a dynamic enzymatic complex. Forcing aberrant interactions within the spliceosome can reduce splicing efficiency and alter splice site choice; however, it is unknown whether such alterations are naturally exploited mechanisms of splicing regulation. Here we demonstrate that hnRNP L represses CD45 exon 4 by recruiting hnRNP A1 to a sequence upstream of the 5’ splice site. Together, hnRNP L and A1 induce extended contacts between the 5’ splice site-bound U1 snRNA and neighboring exonic sequences which, in turn, inhibit stable association of U6 snRNA and subsequent catalysis. Importantly, analysis of several exons regulated by hnRNP L shows a clear relationship between the potential for binding of hnRNP A1 and U1 snRNA, and the effect of hnRNP L on splicing. Together our results demonstrate conformational perturbations within the spliceosome are a naturally occurring and generalizable mechanism for controlling alternative splicing decisions.
Keywords: alternative splicing, U1 snRNA, tri-snRNP, spliceosome assembly, hnRNP L, hnRNP A1
Introduction
The removal of introns and appropriate joining of exons is an essential step in the biogenesis of eukaryotic mRNAs. In addition to being required for the expression of all intron-containing genes, splicing can be regulated to alter the open reading frame or the presence of cis-regulatory elements in a resultant mRNA. Such alternative splicing occurs in the vast majority of human genes and is a primary determinant of protein diversity and gene expression.
The machinery that accomplishes exon joining, the spliceosome, is one of the largest and most dynamic enzymatic complexes in the cell. The catalytically active form of the spliceosome contains 3 small nuclear RNAs (snRNAs) and at least 50 proteins; however an additional 2 snRNAs and tens to hundreds of additional proteins are required for assembly steps leading up to the final active conformation (Wahl et al., 2009). The general assembly of the spliceosome begins with binding of the U1 snRNP to the 5’ splice site (ss). Subsequent ATP-dependent binding of the U2 snRNP to the branchpoint sequence completes initial recognition of the splice sites and results in formation of “A complex”. The U4, U5 and U6 snRNPs are then recruited to the pre-mRNA as a preformed tri-snRNP complex. Stable association of the tri-snRNP and the multi-protein Nineteen complex (NTC) with the substrate defines “B complex”, which is then extensively remodeled resulting in the loss of the U1 and U4 snRNPs, rearrangement of RNA interactions, and the formation of catalytically active “C complex”.
The ordered assembly of the spliceosome is driven by interactions between the pre-mRNA substrate and the protein and RNA components of the snRNPs, as well as through protein and/or RNA interactions between the spliceosomal components themselves (Wahl et al., 2009). Importantly, the molecular interactions that pull together the spliceosome also function as decision points to determine which sequences of the pre-mRNA are to be retained in the final message, by first “defining” exons and then bringing specific exons together to be ligated in the final catalytic core. Therefore, understanding the details of the assembly of the spliceosome and how molecular decisions are made and regulated is a critical component of understanding the mechanisms of alternative splicing.
Recent studies have led to a growing appreciation for the role of kinetic effects in spliceosome assembly. Specifically, the current view of spliceosome assembly posits that the molecular interactions that drive transitions between each assembly step are in dynamic equilibrium, such that strengthening one interaction will effectively repress alternative assembly states (Smith et al., 2008); referred to here as the equilibrium model). For example, sequence mutations that hyperstabilize basepairing of U1 with the 5’ss repress the subsequent association of U6 snRNA with the 5’ss (Staley and Guthrie, 1999), while hyperstabilizing the U6-5’ss interaction blocks rearrangements required for the exon ligation reaction (Konarska et al., 2006). Because the spliceosome disassembles and releases the mRNA after catalysis, even relatively modest favoring of one assembly pathway over another can lead to irreversible decisions of what sequences are spliced together. Consistent with this prediction, a recent study demonstrated that regulatory sequences that perturb a non-rate limiting step in spliceosome assembly can shift the relative use of two competing alternative splice sites (Yu et al., 2008). However, despite general acceptance of the potential importance of kinetic traps, we currently lack any examples in which such traps have been shown to be leveraged as a naturally occurring mechanism of splicing regulation.
A well studied example of regulated alternative splicing is variable inclusion of exons 4–6 in the human CD45 gene. These CD45 variable exons (4, 5 and 6) are predominantly skipped in human T cells to regulate the activity of the encoded protein tyrosine phosphatase and maintain T cell homeostasis (Hermiston et al., 2002). In resting T cells, repression of each of the variable exons is independently regulated through the activity of hnRNP L, which binds to an common exonic silencer motif (ESS1) located in each exon (Rothrock et al., 2005; Tong et al., 2005). In our previous work we have demonstrated that hnRNP L-dependent repression of CD45 exon 4 occurs at a step following the ATP-dependent binding of U1 and U2 to the splice sites on either side of the exon (House and Lynch, 2006).
Strikingly, we demonstrate here that association of hnRNP L with CD45 exon 4 promotes altered interactions of the spliceosome with exonic sequences upstream of the 5’ splice site. Specifically, hnRNP L induces novel basepairing interactions between U1 snRNA and exon 4 sequences extending upstream of the 5’ splice site. This is achieved, at least in part, through the hnRNP L-dependent recruitment of hnRNP A1 also to the 3’ end of exon 4, and is necessary for exon repression both in vitro and in cells. We further show that the hnRNP L-induced aberrant U1/exon interactions repress stable tri-snRNP integration and NTC association by hindering the exchange of U1 for U6 snRNP. Together, these data provide a unique example of regulation of splicing through an hnRNP-induced conformation and/or kinetic trap in spliceosome assembly (Smith et al., 2008). Importantly, the sequence hallmarks involved in binding of hnRNP A1 and the U1 snRNA are present in at least several exons we have previously shown to be regulated by hnRNP L. Moreover, the predicted free-energy of potential basepairing of these exons with U1 snRNA is sufficient to explain the functional effect of hnRNP L on splicing. This observation, together with several recent reports demonstrating widespread splicing repression by hnRNP A1 when bound immediately upstream of a 5’ splice site (Huelga et al., 2012; Yu et al., 2008), suggests that extended interactions of U1 snRNA with exon substrates may be a common mechanism for regulating splicing.
Results
ESS1 induces novel interactions of both U1 snRNA and hnRNP A1 upstream of the 5’ splice site of exon 4
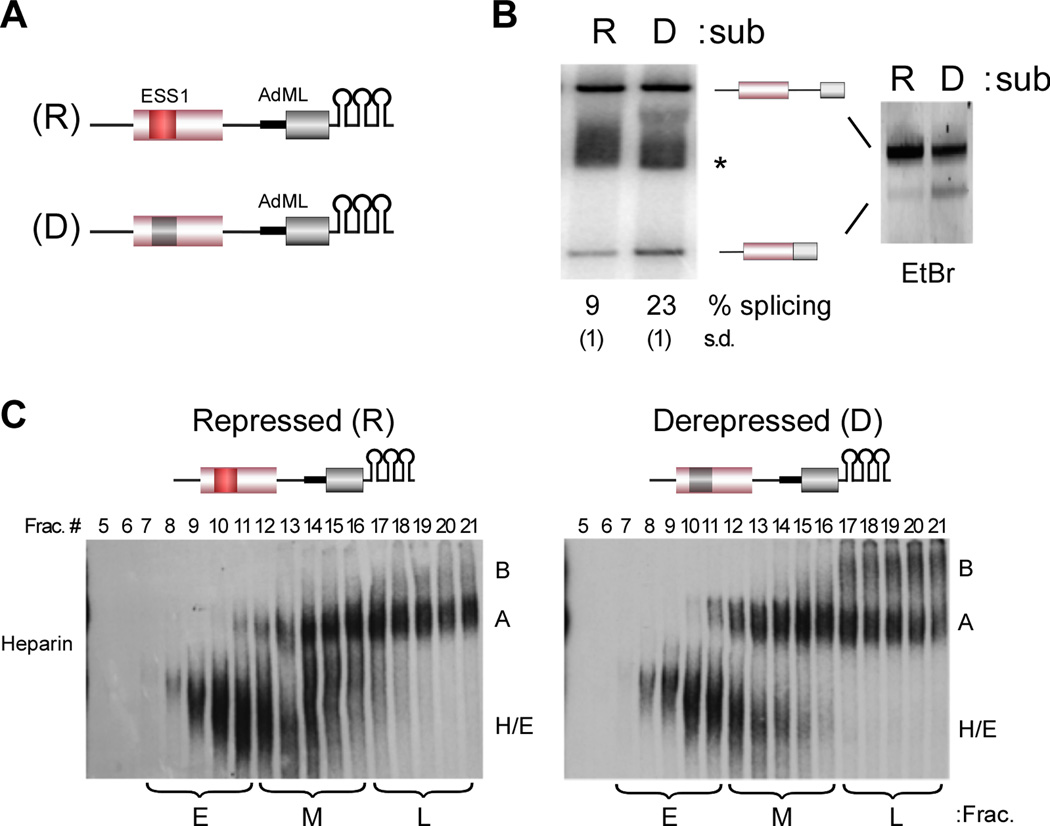
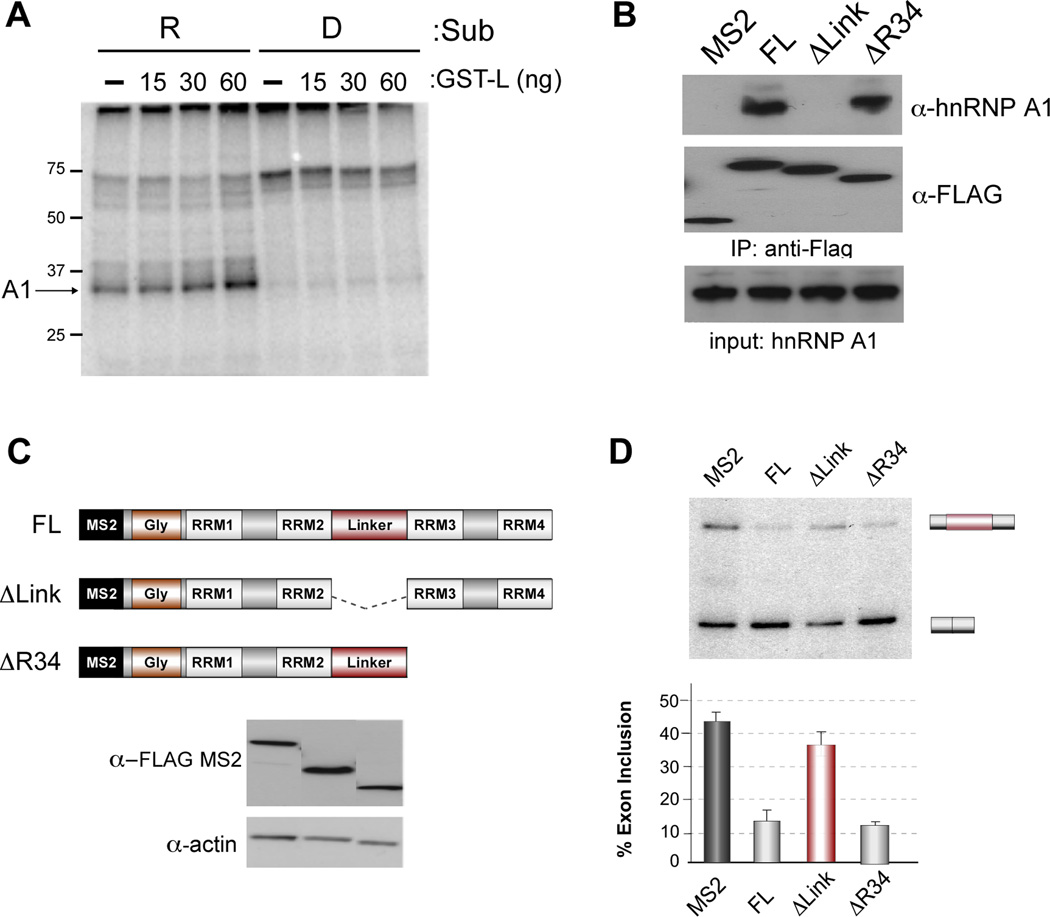
In previous studies we have demonstrated that binding of hnRNP L to the ESS1 exonic silencer sequence within CD45 exon 4 represses exon inclusion both in vivo and in vitro (House and Lynch, 2006; Rothrock et al., 2005). In order to facilitate a more complete characterization of the mechanism of hnRNP L-mediated repression of exon 4 we fused either the wildtype repressed (R) exon, or a derepressed (D) version that contains mutations in ESS1 which abrogate hnRNP L binding, to a well characterized ADML exon (Figure 1A). This splicing module is followed by three copies of the high-affinity hairpin sequence binding site for the MS2 viral coat protein to permit purification of the pre-mRNA substrate using an MS2-MBP fusion protein (Jurica et al., 2002; Zhou et al., 2002). Importantly, repression of exon 4 in the resulting constructs exhibits all of the hallmarks we have previously observed, including dependence on the ability of the ESS1 repressor element to bind hnRNP L, a requirement for flanking introns, the formation of a stalled A complex and a lack of B complex formation on the repressed substrate in the presence of heparin (Figure 1B–C, S1 and data not shown).
Figure 1. The ESS1 represses formation of a stable B complex and exon inclusion.
(A) Schematic of splicing substrates used in the study; CD45 exon 4 (pink box) and downstream intron (thin line) fused to the AdML exon (light grey box) and upstream intron (thick line), followed by three copies of the MS2 binding site hairpin. The repressed (R) construct contains the wildtype exon 4 including the ESS1 element (red box). The derepressed substrate (D) contains three point mutations within ESS1 that abolish hnRNP L binding and silencer activity (dark grey box). (B) In vitro splicing of R and D substrates in JSL1 nuclear extract. (left) RT-PCR of splicing reactions using radiolabeled primer, resolved on a denaturing gel and quantified by phosphorimager as done previously (House and Lynch, 2006; Melton et al., 2007). % splicing and standard deviation (in parentheses) here and in following figures is calculated as the average from at least three independent experiments. * denotes a non-specific build up of radiolabel on a salt-front not observed on an ethidium-bromide stained gel (right). (C) In vitro splicing reactions as in (B) were separated by glycerol gradient and then individual fractions run on a native gel in the presence of heparin. Early (E), middle (M) and late (L) fractions pooled for subsequent experiments are indicated. All experiments throughout were repeated at least three times with equivalent results. See also Figure S1.
Previous analyses of the stalled A complex simply confirmed the presence of the U1 and U2 snRNP components (House and Lynch, 2006). To better investigate whether the nature of the snRNP interactions within the repressed versus derepressed complexes differs, we interrogated RNA interactions by psoralen crosslinking. Following glycerol gradient separation, complexes in the middle fractions were pooled and incubated with AMT psoralen and UV light to induce covalent crosslinks between base-paired regions of RNA. Sites of crosslinks within the substrate were then visualized as psoralen/UV-dependent stops in a primer extension reaction using three primers to interrogate distinct regions of the substrate (Figure 2A). We focused on the complexes in the middle fractions of the glycerol gradient as these fractions exhibit similar mobility between the repressed and derepressed substrates (Figure 1C), yet are committed to different fates.
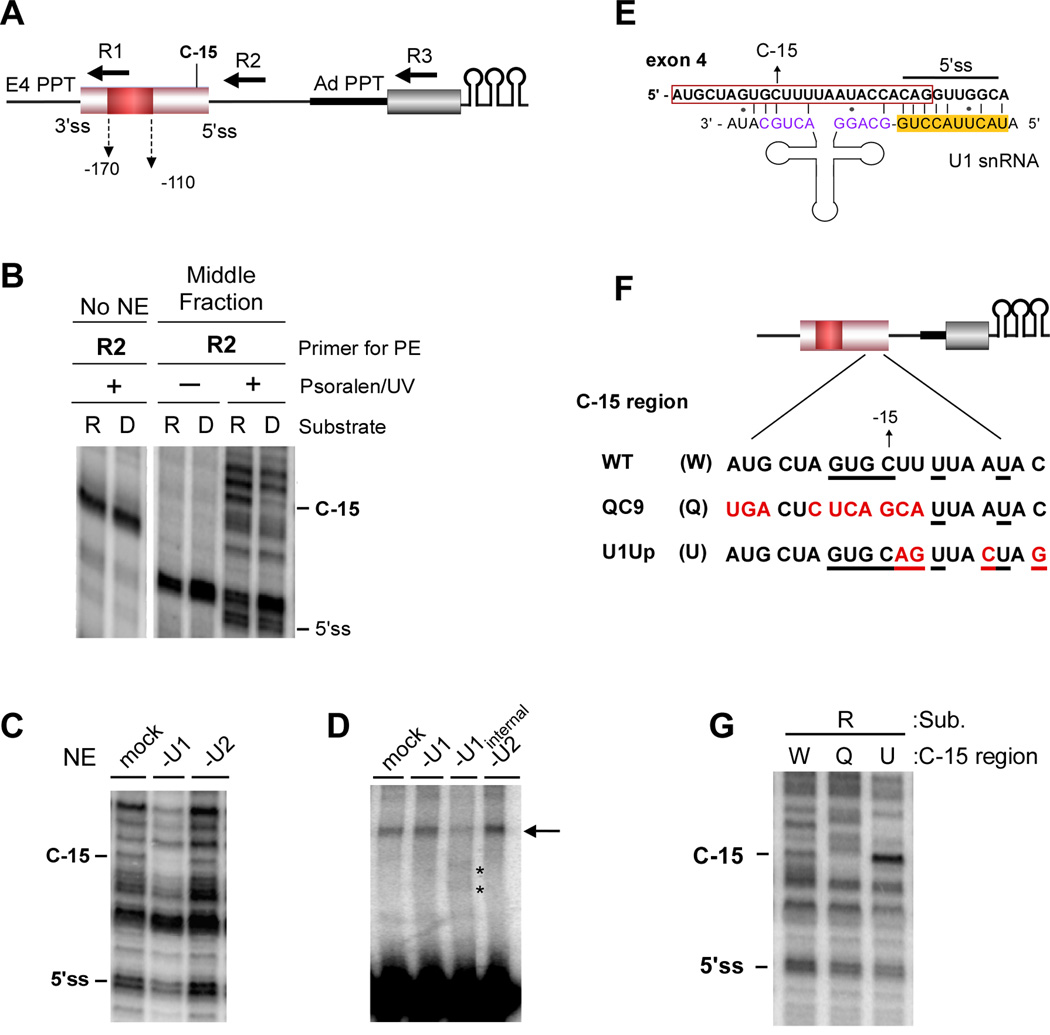
Figure 2. U1 makes extended basepair interactions with the 3’ end of exon 4 in an ESS1-dependent manner.
(A) Schematic of the R substrate showing relative location of primers used for mapping of psoralen crosslinks (R1–3), exon 4 splice sites (ss), PPT from exon 4 and AdML, primary differential crosslink (C-15), and the ESS1 (red box). Substrate D is identical except for mutation of the ESS1 described in Figure 1. (B) Primer extension with the R2 primer following gradient separation (Figure 1C) and treatment with (+) or without (−) psoralen and 365 nm UV light. No NE is an RNA-only control. (C) R2 primer extension of psoralen-crosslinked 5’ss region of exon 4 (see Figure S2D) incubated in nuclear extract (NE) depleted of U1 or U2 activity by oligo-directed RNAse H cleavage (see Figure S1B). (D) Super-shift analysis of psoralen-crosslinked 5’ss region of exon 4 in which U1 or U2 snRNAs are cleaved following crosslinking. “U1 internal” cleaves within the snRNA instead of the 5’ end (see Experimental Procedures). Asterisks mark new species following cleavage. (E) Putative extended basepairing interaction between U1 snRNA and exon 4. Location of 5’ss and C-15 are indicated on exon 4. Red box indicates exon. 5’ss cognate sequences in U1 snRNA are highlighted in yellow. Purple sequences form helix H in U1. (F) Mutant sequences used in subsequent studies. Nucleotides that differ from wildtype exon 4 (W) are indicated in red. Underlined nucleotides can pair with U1 in the scenario shown in panel E and S2E. (G) R2 Primer extension as in panel (B) with the indicated substrates. See also Figure S2.
Primer extension of psoralen-crosslinked repressed and derepressed RNAs did not detect any differences in RNA base-pairing across most regions of the substrates, including the 3’ splice site region upstream of either exon 4 or the AdML exon, and the intron downstream of exon 4 (Figure S2A; R1, R3). Moreover, the crosslinking pattern at the 5’ss itself is unchanged between the repressed and derepressed substrates (Figure 2B; R2, 5’ss). Therefore, the splice sites of exon 4 are themselves appropriately engaged by their cognate snRNAs in the presence of the ESS1, consistent with our earlier studies (House and Lynch, 2006).
Surprisingly, however, we detect novel ESS1-dependent RNA interactions upstream of the 5’ splice site in the repressed but not derepressed substrates (Figure 2B; R2, C-15). By running sequencing reactions with primer R2 in parallel with the crosslink analysis we were able to precisely map the prominent repression-specific crosslink to a cytosine residue 15 nucleotides upstream of the exon 4 5’ splice site (Figure S2B; Figure 2A,B, C-15). Importantly, the C-15 crosslink falls within a region of exon 4 that we have previously shown to influence the efficiency of repression. Specifically, mutation of residues −24 to −13 reduces exon skipping in vivo, although not quite to the extent of mutations within the ESS1 ((Lynch and Weiss, 2001) and see below).
The repression specific C-15 crosslink is dependent on the presence of nuclear extract (Figure 2B), suggesting an intermolecular interaction with a distinct RNA species. Given the proximity of the C-15 residue to the 5’ splice site, which binds U1 snRNA, we considered this snRNA to be a likely cause of the crosslink at C-15. Consistent with this prediction, using a truncated version of the exon 4 5’ss substrate we find that depletion of the U1 snRNA by RNase H cleavage (Figure 2C) or blocking with an antisense oligo (Figure S2C) abolishes the C-15 crosslink, while depletion of U2 snRNAs has no effect (Figure 2C). We also observe a psoralen-induced super-shift of radiolabeled substrate that is dependent on the C-15 region of the substrate and U1 snRNA (Figure S2D). Importantly, migration of this super-shift is altered by RNAse H-mediated cleavage of an internal region of U1, but not U2 snRNA, confirming that this species is indeed comprised of the U1 snRNA crosslinked to the substrate (Figure 2D). The fact that we observe little change in the super-shifted species when the first 11 nucleotides of U1 are cleaved is consistent with the notion that this particular super-shift is not reporting on the standard U1:5’ss interaction (Figure 2D and S2D).
Sequence analysis reveals a potential base-pairing interaction between the U1 snRNA and the 3’ end of exon 4 that could co-exist with the standard U1 snRNA-5’ss interaction and extend into the exon, providing sufficient base-pairing at C-15 to confer psoralen reactivity (Figure 2E). Importantly, the previously-identified functionally-defective mutation in the −24 to −13 region reduces this potential basepairing interaction (Figure 2F, S2E; QC9) and disrupts the psoralen-induced U1-dependent supershift of the substrate (Figure S2D). We also mutated several residues downstream of C-15 to increase the base-pairing in the potential interface between the U1 snRNA and exon 4 (Figure 2F, S2E; U1Up). Consistent with the predicted interaction model, the U1Up mutation greatly increases the C-15 crosslink, whereas the QC9 mutation markedly reduces it (Figure 2G). Notably, neither of these mutations significantly impacts crosslinking of U1 to the 5’ splice site itself, demonstrating that interaction of U1 with the exon does not impact the standard basepairing between U1 and the intron sequences at the 5’ss.
As a complementary approach, we attempted to map the psoralen crosslinks within the U1 snRNA itself. Unfortunately, the nucleotides which we predict to pair with exon 4 in the extended conformation are normally predicted to be paired in an intramolecular stem in the free U1 snRNP (helix H; Pomeranz Krummel et al., 2009). Therefore, while we do observe psoralen crosslinks at the appropriate nucleotides in the U1 snRNA, we cannot distinguish intra vs intermolecular binding. However, the 5’ portion of helix H is predicted to be less basepaired in the extended conformation than in the canonical helical form (Figure 2E). Consistent with our model, we observe that the psoralen crosslinking of the 5’ portion of helix H is reduced in U1 snRNA bound to the wildtype C-15 substrate relative to the U1 snRNA bound to the QC9 control, in which helix H should be in the canonical form (Figure S2F). Therefore, while we cannot fully rule out that other regions of U1 may interact with the C-15 region, the model in Figure 2E of extended pairing between U1 and the 3’ end of exon 4 is consistent with all of our data.
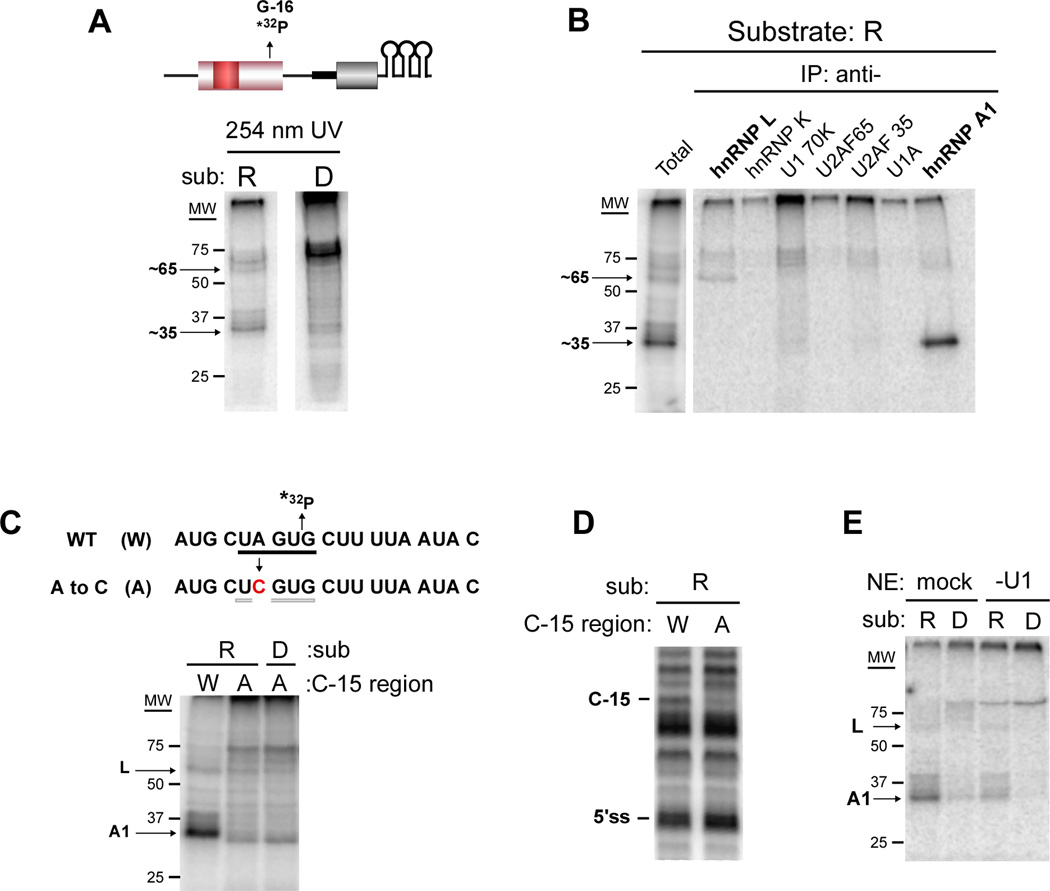
We next engineered a single radiolabeled phosphate at position G-16 within the repressed and derepressed substrates to investigate the presence of proteins in the vicinity of the U1-exon interaction. Spliceosomal complexes assembled on these substrates were subject to UV crosslinking, in which proteins are crosslinked to RNA by shortwave UV light followed by RNase digestion. Interestingly, we observe a ~35 kD protein that crosslinks to the radiolabeled G-16 in the repressed substrate but not the derepressed version (Figure 3A). We also observe weak crosslinking to a species of ~ 65 kD that is likewise dependent on the ESS1 element (Figure 3A). Immunoprecipitation with antibodies to candidate proteins of 35 and 65 kD demonstrates the 35 kD protein to be hnRNP A1, while the 65 kD protein is hnRNP L (Figure 3B, S3A). Immunoprecipitation also revealed that the ~ 75 kD species which crosslinks with variable intensity to the derepressed construct is hnRNP M (Figure 3A, C, E and data not shown), a protein known to bind GU-rich sequences (Huelga et al., 2012). However, the presence of this species does not correlate with increased splicing efficiency (see Figure S4). Thus, we conclude that hnRNP M binds the 5’ss region when vacated by hnRNP A1 and the U1 snRNP, but does not impact splicing, and have not pursued this observation further.
Figure 3. HnRNP A1 interacts cooperatively with U1 to the 3’ end of exon 4 in an ESS1-dependent manner.
(A) Repressed (R) and derepressed (D) substrates containing a single radiolabeled phosphatase at G-16 (shown for R, top) were incubated with nuclear extract under splicing conditions and crosslinked with 254 nm light, followed by RNase digestion and resolution by SDS-PAGE. Migration of molecular weight markers are shown. (B) Crosslinking reaction with R substrate as in panel A (total) was then incubated in separate reactions with the antibodies indicated (IP: anti-). Immunoprecipitated proteins were resolved by SDS-PAGE. (C) (top) Sequence of C-15 region showing site of radiolabeled phosphate (*32P), weak consensus A1 binding site (underline) and mutation (red) that disrupts core A1 element (light underline). (bottom) UV crosslinking as in panel (A) with indicated substrates. (D) R2 primer extension of psoralen-crosslinked reactions as in Figure 2B with substrates from panel C. (E) UV crosslinking as in panel (A) using nuclear extract depleted of U1 by oligo-directed RNase H cleavage as in Figure 2C. See also Figure S3.
Analysis of the sequence at the 3’ end of exon 4 reveals a weak match to the hnRNP A1 consensus binding site spanning nucleotides −20 to −16 (UAGUG, consensus site is UAGRG (Martinez-Contreras et al., 2007); Figure 3C). We note that this sequence was disrupted in the QC9, but not the U1Up, mutant. Consistently, we find a decrease in hnRNP A1 crosslinking in the QC9 substrate relative to the wildtype or U1Up constructs (Figure S3B). To investigate the interplay between recruitment of hnRNP A1 and the psoralen-detected RNA interactions, we additionally made a substrate containing a single A to C change that is predicted to abolish the weak hnRNP A1 binding site, but not alter the putative basepairing with U1 snRNA (Figure 3C; S3C). Strikingly, this mutation not only disrupts crosslinking of hnRNP A1 to G-16 (Figure 3C), but also eliminates the U1 interaction with C-15 as monitored by both primer extension and supershift after psoralen crosslinking (Figure 3D, S3D). Conversely, depletion of U1 snRNA from the reaction results in decreased UV crosslinking of hnRNP A1 at G-16, while depletion of U2 snRNA has no effect (Figure 3E, S3E). Taken together, these results demonstrate that hnRNP A1 and the U1 snRNA associate cooperatively with the 3’ end of exon 4 in a manner that is dependent on the ESS1.
Recruitment of hnRNP A1 and the U1 snRNA to the 3’ end of exon 4 is required for hnRNP L dependent repression of exon 4
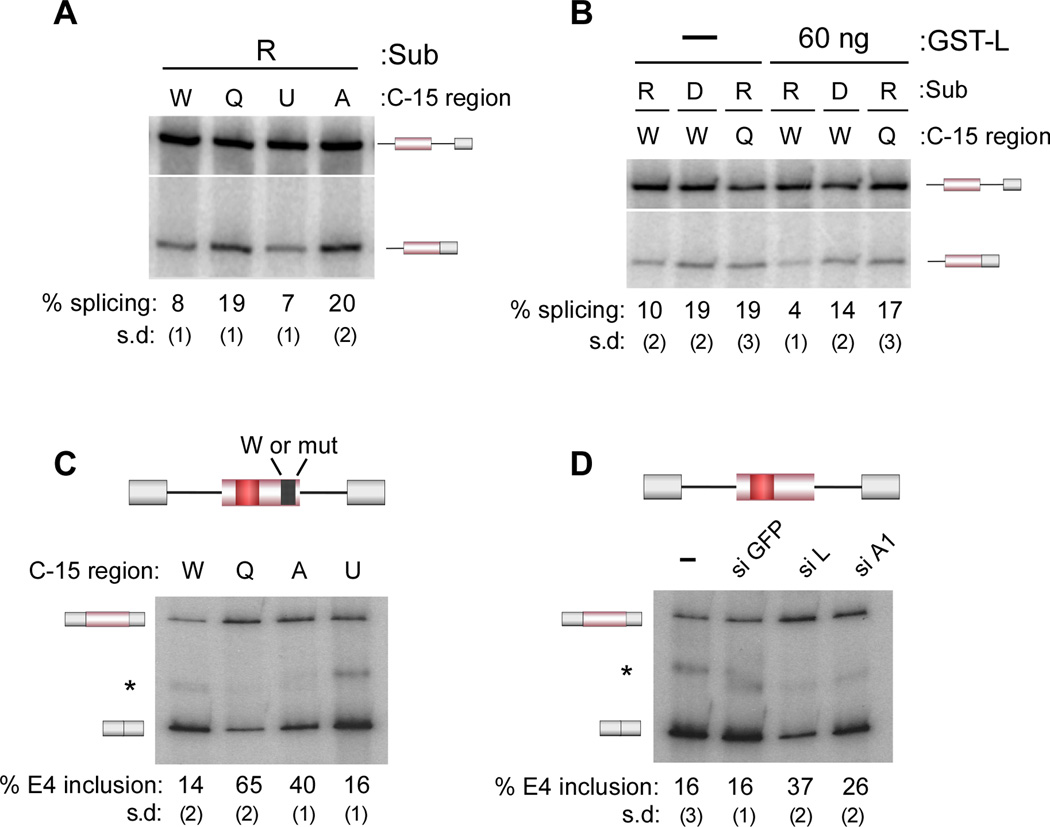
The data in Figures 2 and 3 establish a unique activity of an exonic silencer in promoting extraneous interactions between the spliceosome and the pre-mRNA substrate. Remarkably, the extended interactions between the 3’ end of exon 4 and the U1 snRNA and hnRNP A1 are required for the repressive activity of ESS1 as both the QC9 and A-to-C mutations disrupt exon 4 repression by 2–3 fold (Figure 4A). This loss of repression is directly linked to hnRNP L activity as the splicing of the QC9 mutant is essentially unaffected by excess hnRNP L even at a concentration sufficient to hyper-repress the wildtype substrate and marginally represses the ESS1-mutant D substrate, which has significantly decreased affinity for hnRNP L (Figure 4B). Finally, blocking association of both the U1 snRNA and hnRNP A1 with the 3’ end of exon 4 using an antisense 2’OMe oligonucleotides complementary to nucleotides −24 to −13 also results in a loss of ESS1-dependent exon silencing (anti-3’E4; Figure S4A–C). Therefore, interactions of hnRNP A1 and U1 with the 3’ end of exon 4 are required for both hnRNP L and ESS1-dependent exon repression.
Figure 4. Association of U1 and hnRNP A1 with the 3’ end of exon 4 is required for ESS1-and hnRNP L-dependent exon repression.
(A) In vitro splicing as in Figure 1B with the indicated substrates described in Figures 2 and 3. (B) In vitro splicing in the absence (−) or presence (60 ng) of purified, recombinant hnRNP L (GST-L). (C) RT-PCR analysis of minigene expressed RNA harvested from 293 cells transfected with the CD45 exon 4 minigene containing the wildtype (W) or mutant (Q, A, U as defined in Figures 2, 3) C-15 region (top). (D) RT-PCR analysis of minigene expressed RNA harvested from 293 cells transfected with the wildtype CD45 exon 4 minigene and nothing (−) or siRNAs to hnRNP L (L), hnRNP A1 (A1) or GFP as control. See also Figure S4.
Importantly, the hnRNP A1 and U1 interactions with exon 4 are also required for maximal repression in vivo. Specifically, the QC9 and A-to-C mutations reduce skipping of CD45 exon 4 from a minigene that we have previously shown to recapitulate all aspects of regulation of the endogenous gene (Figure 4C). Further consistent with the in vitro studies, knock-down of hnRNP A1 results in a reduction in exon 4 skipping from the minigene, although knockdown of hnRNP L has a more prominent effect consistent with hnRNP L functioning as the driving factor in repression (Figure 4D, S4D). We note that the U1Up mutation does not significantly increase repression either in vitro or in vivo (Figure 4A, C). While hnRNP L is sufficient for a significant amount of repression, this exon has also been shown to be positively influenced by several SR proteins (Motta-Mena et al., 2010). We conclude that this balance of enhancing and inhibitory influences on exon 4 (typical of regulated exons) enforces a maximum limit on how much hnRNP L can repress the exon.
The linker region of hnRNP L recruits hnRNP A1 and is required for exon repression
The simplest explanation for the ESS1-dependence of the hnRNP A1 and U1 association with the 3’ end of exon 4 is that hnRNP L promotes the recruitment of these components to otherwise weak binding sites. Consistent with this, addition of excess recombinant hnRNP L promotes association of hnRNP A1 with G-16 in the UV crosslinking assay (Figure 5A). Furthermore, epitope-tagged hnRNP L efficiently precipitates hnRNP A1 (Figure 5B). Interestingly, the ability of hnRNP L to co-precipitate hnRNP A1 depends on a proline-rich linker sequence within hnRNP L (Figure 5B,C). This co-precipitation is lost in the presence of RNase, suggesting that RNA stabilizes the association of these proteins (data not shown).
Figure 5. HnRNP L interacts with hnRNP A1 through linker domain and recruits hnRNP A1 to exon 4 to achieve repression.
(A) UV crosslinking as in Figure 3A in the absence (−) or presence of indicated amount of purified, recombinant GST-L. (B) Co-precipitation of hnRNP A1 with hnRNP L. Extract was made from cells transfected with Flag-MS2 control or Flag-MS2 hnRNP L constructs shown in panel C. Transfected protein was precipitated using anti-FLAG antibody, resolved on SDS-PAGE and analyzed for co-precipitation of hnRNP A1 by western blots with antibody to hnRNP A1 relative to Flag control. (C) Schematic of hnRNP L domains included in MS2 fusion proteins. FL contains the complete hnRNP L protein sequence fused downstream of the MS2 coat protein as used in Figure 5. ΔLink lacks the hnRNP L sequence between RRM 2 and 3. ΔR34 lacks all sequences C-terminal of the beginning of RRM3. (D) % inclusion of the wildtype MS2-exon 4 as in Figure S5A, comparing the activity of the indicated MS2-hnRNP L deletion mutants relative to full-length. Data are graphed as mean +/− SEM.
To more fully probe the nature of the interaction between hnRNP L and hnRNP A1 we turned to an MS2-tethering system in which the ESS1 of exon 4 minigene is replaced by a MS2 binding sequence and hnRNP L is expressed as an MS2 fusion protein (Figure S5). Importantly, repression of the MS2 hairpin-containing exon by MS2-hnRNP L is dependent on both the integrity of the 3’ end of exon 4 and on hnRNP A1, indicating that MS2-hnRNP L causes repression by the same mechanism as does hnRNP L bound to the ESS1 (Figure S5B,C). Moreover, the linker sequence that is required for recruitment of hnRNP A1 to RNA by hnRNP L is required for MS2-hnRNP L-mediated exon repression (Figure 5D). By contrast, deletion of other domains of hnRNP L that are not required for repression activity in the MS2-tethering assay, do not reduce co-precipitation of hnRNP A1 with hnRNP L (Figure 5B–D). Therefore, the ability of hnRNP L to interact with hnRNP A1 correlates with the ability of hnRNP L to promote exon repression, although we have not determined whether this interaction is direct or indirect. Given the cooperative binding of hnRNP A1 with the U1 snRNP at the 3’ end of exon 4 (Figure 3), the observed recruitment of hnRNP A1 by hnRNP L is sufficient to explain the requirement for hnRNP L/ESS1 for the C-15 crosslink of U1; although we cannot rule out additional direct contacts between hnRNP L and U1. In sum, we show that the ability of hnRNP L to recruit hnRNP A1, and the ability of hnRNP A1 and U1 snRNA to interact with the 3’ end of exon 4, are all required for repression of exon 4.
Interaction of U1 with the 3’ end of exon 4 inhibits exchange with U6 and NTC recruitment to regulate splicing
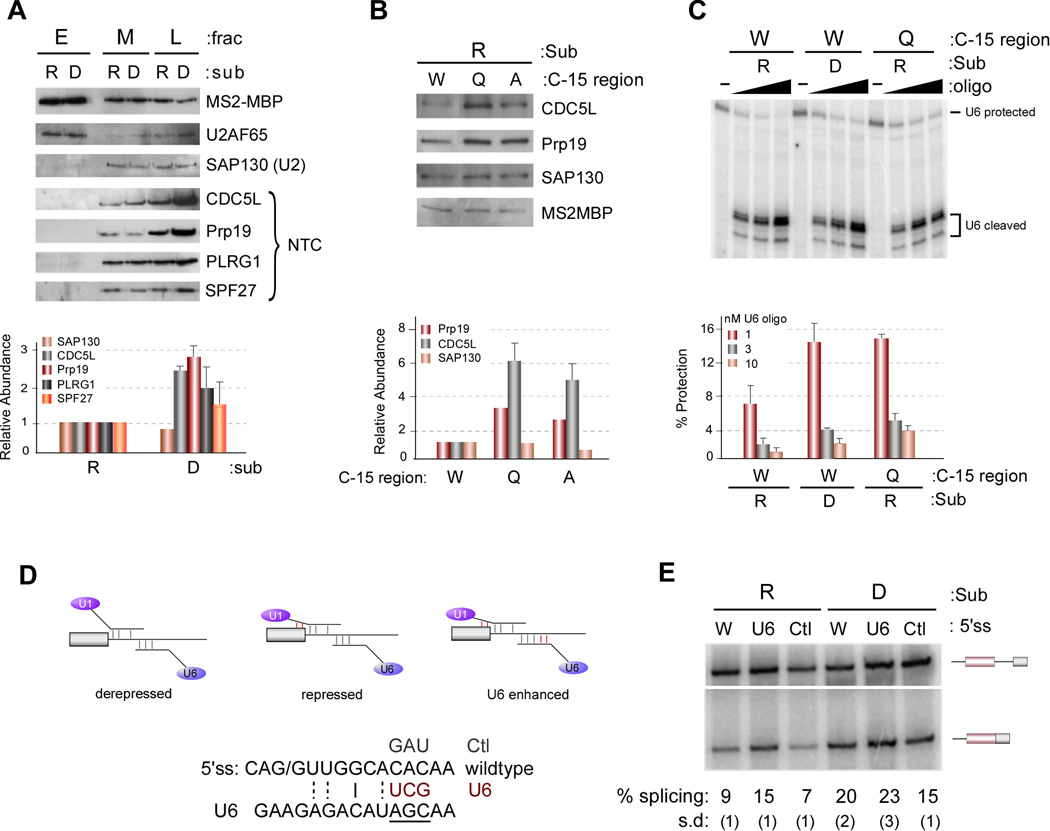
Previous studies in yeast have demonstrated that artificially hyperstabilizing the association of U1 with the 5’ss blocked the replacement of U1 by U6 at the 5’ss, thereby blocking subsequent spliceosome assembly and catalysis (Staley and Guthrie, 1999). To investigate if the hnRNP L-induced interaction of U1 snRNA with the 3’ end of exon 4 might likewise inhibit proper loading of U6 and subsequent spliceosome assembly, we isolated the hnRNP L-repressed and derepressed spliceosomes by pooling fractions from the glycerol gradient, followed by purification via the MS2-MBP tag (see Experimental Procedures). Analysis of the purified spliceosomes is consistent with a stall of the repressed substrate at an early precursor to B complex that has been described by others (Maroney et al., 2000; Roybal and Jurica, 2010; Schneider et al., 2010a; Schneider et al., 2010b). Specifically, the tri-snRNP is associated with the stalled spliceosome, but in a heparin-sensitive manner (Figure S6B and compare Figure S6A with Figure 1C); and there is a block in subsequent NTC recruitment and splicing (Figure 6A, S6C). Therefore, we conclude that ESS1 represses spliceosome assembly by blocking the transition from weak to strong association of the tri-snRNP, precluding further assembly of spliceosomal components.
Figure 6. Association of U1 and hnRNP A1 with the 3’ end of exon 4 represses U1/U6 exchange and recruitment of NTC.
(A) Western blots (top) and quantification (bottom) of proteins from MS2-MBP purified complexes from M and L fractions (Figure S6A, +ATP) or E fraction (Figure S6A, −ATP). Quantification here and in following panels is averaged from three experiments and data are represented as mean +/− SEM. (B) Western blot (top) and quantification (bottom) of splicing complexes purified from L fractions as in panel A. (C) Splicing complexes purified by MS2-MBP were treated with varying concentrations of an oligo complementary to the ACAGA box of U6 to induce RNase H cleavage. Cleavage of U6 snRNA was detected by primer extension (top) and quantified by phosphorimager (bottom). (D) Schematic of the U6 enhanced experiment and U6 enhanced (U6) and control (Ctl) mutations made in the wildtype substrate (W). (E) In vitro splicing as in Figure 1B with the substrates indicated in panel (D).
To confirm the significance of the U1 snRNA and hnRNP A1 interaction in this pre-B complex block, we first used recruitment of the NTC as a convenient read-out of B complex. Consistent with the prediction that extended U1 snRNA and hnRNP A1 interactions with exon 4 inhibit progression to B complex, mutations in the 3’ end of exon 4 that disrupt the association of hnRNP A1 and U1 exhibit an increased association of the NTC relative to wildtype (Figure 6B). Similarly, addition of anti-3’E4 to the spliceosome purification assay substantially increases recruitment of the NTC components to the ESS-containing substrate, resulting in a level of NTC binding similar to that observed in the absence of the ESS1 (Figure S6D).
To more directly assess the state of tri-snRNP recruitment we utilized an oligo complementary to the ACAGAGA box of U6, to interrogate the accessibility of this sequence to oligo-directed RNAse H cleavage. The ACAGAGA box of the U6 snRNA is unpaired (thus subject to RNAse H cleavage) in the free U6 snRNA or when paired to U4 as part of the incoming tri-snRNP. By contrast, when the U6 snRNA replaces U1 at the 5’ splice site the ACAGAGA region makes RNA contacts with the substrate RNA and thus should be more protected from RNAse H digestion. Consistent with an ESS1-dependent block in U1–U6 exchange we indeed observe less U6 protection in the repressed versus derepressed complexes at all concentrations of oligo tested (Figure 6C). Importantly, the ability of U1 to make contact with the 3’ end of exon 4 is required for this differential U6 accessibility, as the QC9 mutation also increases the relative protection of U6, indicative of increased association of U6 with the 5’ splice site (Figure 6C).
Extending the basepairing between U6 and the substrate has previously been shown to suppress the splicing defect caused by hyperstabilizing the U1:5’ss interaction (Staley and Guthrie, 1999), suggesting that the exchange of U6 for U1 is controlled by competing binding energy. If the hnRNP L-dependent alteration of U1 is indeed causing a similar energetic block to U6 recruitment as in the synthetic hyperstabilization experiments, we would predict that increasing the U6 basepairing potential would likewise alleviate repression and increase splicing (Figure 6D). Remarkably, using mutations similar to the previous study, we indeed observe that increasing the basepairing of U6 with the 5’ ss region increases the efficiency of splicing of exon 4 (Figure 6D,E). By contrast, control mutations have no effect on exon 4 splicing and increasing the basepairing potential of U6 has no effect on the derepressed construct (Figure 6D,E). Taken together, we conclude that hnRNP L/ESS1 blocks splicing of CD45 exon 4 by trapping the 5’ss-bound U1 snRNP in a stabilized conformation that inhibits proper association of U6 with the 5’ss, thus blocking subsequent snRNP rearrangement, recruitment of the NTC, and catalysis (Figure 7A).
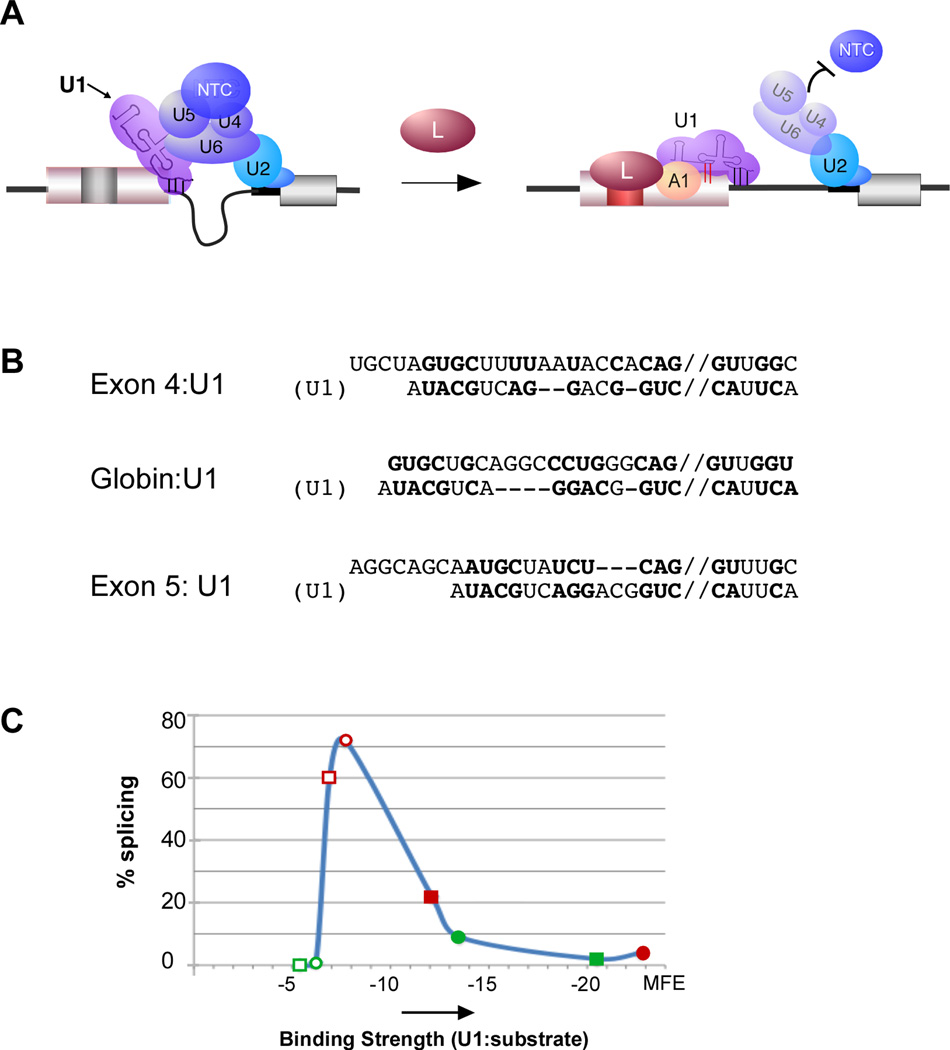
Figure 7. Model for hnRNP A1 and U1 recruitment by hnRNP L is sufficient to predict regulatory effect of hnRNP L on multiple exons.
(A) Schematic of model of hnRNP L-mediated repression of CD45 exon 4 through recruitment of hnRNP A1 and extended basepairing of U1 snRNA. (B) Predicted lowest free-energy extended basepairing of U1 snRNA with exons regulated by hnRNP L (CD45 exon 4, CD45 exon 5 and human β-globin exon 1). Pairing shown as predicted by RNAcofold (http://rna.tbi.univie.ac.at/cgi-bin/RNAcofold.cgi). Bold nucleotides represent basepairing. Double slash indicates 5’ splice site. (C) Binding strength of U1 basepairing in normal (−3 to +6) register (open shapes) or in predicted hnRNP L-induced extended conformation as in panel B (closed shapes) of two hnRNP L-repressed (red) and two enhanced (green) exons graphed against experimentally determined rate of exon inclusion. Details of minimal free energy (MFE) calculations and other data used for graph are shown in Figure S7B.
Finally, an immediate question from these data is whether extended pairing of U1 snRNA is unique to exon 4 or may play a more general role in splicing regulation. In previous studies, we have shown that the ESS1 is portable and recruitment of hnRNP L through the ESS1 represses at least two other chimeric test exons. Moreover, we recently reported that exon-bound hnRNP L can function as an enhancer in cases in which the 5’ splice site is especially weak. Importantly, in each of the cases we have observed hnRNP L function, the 3’ end of the exon contains a UGCU sequence (similar to the sequence surrounding the C-15 crosslink) within 20 nts upstream of the 5’ splice site, as well as an upstream AG dinucleotide which could serve as a binding site for hnRNP A1 (Figure 7B, S7A).
We therefore calculated the predicted minimal free energy (MFE) of binding of the U1 snRNA with the 3’ end of each of the above exons and 5’ splice site, making the assumption that U1 associates in the canonical fashion with nucleotides −3 to +6 (relative to the 5’ splice site) in the absence of hnRNP L, but in an extended fashion in the presence of hnRNP L (Figure 7B, S7B). Strikingly, we find Gaussian-like relationship between the predicted MFE of U1 binding and the in vivo splicing of these exons that can account for the observed hnRNP L activity (Figure 7C, S7B). In all cases the presence of hnRNP L is predicted to shift the MFE to tighter binding (opened to filled shapes), but the functional result of this shift depends on the starting MFE. Specifically, for 5’ splice sites in which basepairing of U1 snRNA is “optimal” without hnRNP L (open red shapes), the extended pairing results in repression (filled red shapes). By contrast, if the binding of U1 is too weak in the absence of hnRNP L to observe any splicing (high MFE, open green shapes), the extended pairing brings the MFE to a range at which splicing is at least detectable (filled green shapes). An implication of this analysis is that maximal splicing occurs in an intermediate window of U1 binding energy, whereas excessively high or low binding energy results in exon repression. This “goldilocks effect” is entirely consistent the equilibrium model put forth by others (Smith et al., 2008), the U1 hyperstabilization studies in yeast (Staley and Guthrie, 1999) and data from our earlier work (Motta-Mena et al., 2010).
Discussion
Emerging studies of the spliceosome indicate that conformational dynamics play a critical role in maintaining splicing fidelity and transitions between mutually-exclusive RNA interactions both within the snRNAs and between snRNAs and the substrate often serve as check-points in assembly (Smith et al., 2008; Wahl et al., 2009). Remarkably, we demonstrate here that such spliceosomal dynamics are also naturally usurped by regulatory factors to induce alternative splicing. Specifically, we find that hnRNP L, together with hnRNP A1, effectively holds the U1 snRNA in an extended basepaired conformation that precludes the normal exchange of U1 for U6 at the 5’ splice site.
As shown in Figure 2, we propose that the interaction between the U1 snRNA and the 3’ end of exon 4 involves unwinding of the short intramolecular helix H within the U1 snRNA to form the intermolecular interactions with the exon. This putative conformation is consistent with all of our data and with previous reports demonstrating the ability of exogenous RNAs to make intermolecular basepairs with residues of helix H (Abad et al., 2008; Kaida et al. 2010). However, we cannot fully exclude other possible modes of interaction between U1 snRNA and exon 4. Indeed, a direct implication of our results is that any method of stalling the normal transition check-points in spliceosome assembly is likely to result in alternative splicing.
The simplest model for how hnRNP L-induced U1 interaction with exon 4 blocks U6 association is that increasing the binding energy of U1 snRNA to the substrate favors binding of the 5’ splice site to U1 relative to U6. This mechanism is consistent with previous studies in yeast (Staley and Guthrie, 1999) and with the fact the U6 hyperstabilization can overcome hnRNP L-induced repression (Figure 6E). A more recent study has questioned the relevance of the hyperstabilization model in mammals, however, basepairing of U1 snRNA in that study was not extended beyond the canonical −3 position of the exon and strength of pairing was measured by hydrogen bonds (Freund et al., 2005). By contrast, the hnRNP L-induced interactions between U1 and exon 4 extend to at least position −15 and we find hydrogen bond number to be poorly predictive of splicing efficiency (Figure S7). Instead, we find that the predicted minimal free energy of basepairing of U1 snRNA and the substrate is sufficient to account for previously observed regulatory effects of hnRNP L including our finding that hnRNP L promotes E complex formation on weak exons (Motta-Mena et al., 2010) and blocks stable recruitment of U6 snRNA and the NTC on stronger exons (Figure 6, 7). However, we cannot exclude the possibility that conformational changes in the U1 snRNP are also induced by its interaction with the exon and contribute to hnRNP L-mediated exon repression. For example, it is possible that opening of helix H has allosteric consequences within the snRNP, resulting in conformational changes or disruption of protein-RNA interactions within the snRNP that preclude recruitment of the U6 snRNP.
An additional layer of complexity in the mechanism of exon 4 repression is that binding of hnRNP A1 is required at the 3’ end of the exon, in addition to basepairing with U1 snRNA, in order to achieve regulation by hnRNP L. Interestingly, one of the first activities ascribed to hnRNP A1 was that of an RNA chaperone. Specifically, hnRNP A1 was shown to have both helix destabilizing and RNA annealing activities, such that hnRNP A1 promotes interconversion of RNA basepairing interactions (Kumar and Wilson, 1990; Pontius and Berg, 1990; Portman and Dreyfuss, 1994). However, no specific substrate of hnRNP A1 RNA chaperone activity has yet been identified in the spliceosome.
Recent work from the group of Tim Nilsen demonstrated that the presence of a high-affinity binding site for hnRNP A1 7–18 nuclelotides upstream of a weak 5’ splice site alters the footprint of the U1 snRNP and reduces splicing efficiency in the presence of competing splice sites (Yu et al., 2008). In these studies hnRNP A1 was shown to bind together with the U1 snRNP on the region around the 5’ splice site (Yu et al., 2008). This data, together with the annealing activity described above, suggests that direct interactions between A1 and the U1 snRNP might aid in the remodeling of the U1:substrate interaction. Notably, hnRNP A1 was proposed to play a general role in the silencing of pseudo splice sites, as high affinity A1 binding sites were found to be enriched immediately upstream of pseudo 5’ splice sites (Yu et al., 2008). Consistently, global analysis of hnRNP A1 binding and function on bona fide exons has revealed that this protein typically binds within the terminal 50 nucleotides of exons it represses (Huelga et al., 2012). Such biased localization of hnRNP A1 suggests that fostering extended interactions of the 5’splice site bound U1 snRNP with neighboring exonic sequences may be a widespread mode of hnRNP A1 action. Our data here further suggest that the presence of hnRNP L can induce hnRNP A1 to evoke its effect on U1 snRNP association in a “regulatable” manner so as to induce alternative use of bona-fide splice sites based on the occupancy of hnRNP L.
Significantly, we note that we do detect some weak binding of hnRNP A1 and the U1 snRNA to the 3’ end of CD45 exon 4 even in the absence of hnRNP L (e.g. in the Derepressed constructs). This weak binding suggests that hnRNP L is not absolutely required for the extended contacts between the spliceosome and the substrate, but rather promotes interactions that exon 4 is inherently poised to make. This raises the intriguing possibility that at least a subset of exons may have evolved to contain sequences inherently capable of forming interactions with snRNAs beyond those which are considered essential (e.g. the contacts at the 5’ splice site and branchpoint sequence). Such sequences may in some cases promote exon inclusion whereas in others, such as the CD45 exons, may predispose the exon to regulation. We also note that the regulation of U1 binding to exon 4 does not account for all of the repression observed. Specifically, the intron upstream of exon 4 is not required for altered contacts at the 3’ end of the exon, but is required for maximal repression (House and Lynch, 2006). We have preliminary data that this upstream intron contributes an independent level of repression that functions in conjunction with the hnRNP L-induced mechanisms described here. While outside the scope of this study, such a multi-layered mechanism of repression is consistent with the splicing of exon 4 being under extensive combinatorial control (Motta-Mena et al., 2011).
In sum, our demonstration here that hnRNP L blocks inclusion of CD45 exon 4 by altering the interaction of U1 snRNA with the substrate highlights the finely-balanced nature of spliceosome assembly and the potential for relatively subtle perturbations in snRNA interactions to dramatically alter the assembly pathway. We suggest that induction of extended U1 pairing with substrate may be responsible for the regulation of several exons for which aberrant U1 interactions have been observed; including the above mentioned studies with hnRNP A1 (Huelga et al., 2012; Yu et al., 2008) as well as regulation of P-element splicing by PSI (Labourier et al., 2001). Interestingly, blocking U1 interactions with other spliceosomal components has also been demonstrated as a mechanism of exon repression (Sharma et al., 2011), suggesting that appropriate binding of U1 within the spliceosome may broadly serve as a sensitive fidelity check-point in spliceosome assembly. Whether there are instances of alternative splicing occurring through extended interactions of other snRNPs remains to be shown; however, our data here strongly suggests that any interaction which functions as a check-point in assembly is likely also used as a point of control of alternative splicing in nature. Indeed, our findings here provide a compelling example of how the complexity inherent to the spliceosome is of benefit to higher organisms by offering a vast repertoire of regulatory opportunities.
Experimental Procedures
In vitro splicing and spliceosome assembly
Repressed (R), derepressed (D) RNA and the RNA substrates containing various mutations (Q, A and U) at the 3’ end of exon 4 were transcribed from PCR products that contain a T7 promoter sequence. Construction of the templates is described in Supplemental Methods. In vitro splicing reactions and spliceosome assembly were carried out as described previously (House and Lynch, 2006), with some modifications as described in Supplemental Methods.
Isolation and analysis of splicing complexes
Gradient separation of early (E), middle (M), and late (L) fractions (Figure 1C, S6A) is described in Supplemental Methods. The RNA recovered from the purified complexes was separated on 8% urea-PAGE gel followed by silver-staining (Bio-rad) or analyzed by phosphorimager. The proteins from ~0.02 pmole of purified complexes were separated by SDS-PAGE and analyzed by western blot. The antibodies against hnRNP L, hnRNP A1, Prp19, CDC5L and SAP130 were all from Abcam; anti-PLRG and anti-SPF27 were from Bethyl Laboratories; U2AF65 and MBP antibodies were from Sigma.
Psoralen and UV crosslinking
After assembly of splicing reactions for 30 min at 30°C or gradient fractionation of splicing reactions, the reactions or collected M fractions were placed on ice. AMT-poralen was added to a final concentration of 24 µg/µl and the reactions were irradiated with 365 nm light for 10 min on ice (Sharma et al., 2005). The R2 primer is complementary to nucleotide 56–76 of the downstream intron. UV crosslinking was done as previously described (Rothrock et al., 2005) using RNA substrates containing a single 32P-label, constructed as described (Sharma et al., 2011).
Oligonucleotide-directed RNase H cleavage
Depletion of snRNAs from JSL1 nuclear extract by RNase H, and analysis by primer extension, was done as previously described (House and Lynch, 2006). Probing accessibility of U6 snRNA in the purified complexes was done with oligo U6f (Konforti and Konarska, 1994). “U1internal” oligo targets residues 7–21 and 113–127 of the U1 snRNA.
Cell Culture, Transfections, and RNAi
All cell-based assays were done in HEK293 cells using Lipfectamine 2000 (Invitrogen) for transfection. The CD2 and CD24 minigenes, as well as the Flag-MS2-hnRNP L expression construct were described previously (Motta-Mena et al., 2010). SiRNA knock-down was done using the Dharmacon SmartPool against hnRNP L or A1. RNA was isolated from cells using RNA-Bee (Tel-Test), and semi-quantitative RT-PCR conducted as described previously (Rothrock et al., 2003). Construction of the Flag-MS2-hnRNP L deletion constructs and co-immunoprecipitation with hnRNP A1 are described in Supplemental Methods.
Supplementary Material
Acknowledgements
We wish to thank Melissa Moore, and Charles Query for helpful discussions on this project and Yoseph Barash, Tim Nilsen and Jon Staley for critical reading of the manuscript. This work was supported by R01 GM067719 to K.W.L. and a AHA Postdoctoral Fellowship to G.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. NAR. 2008;36:2338–2352. doi: 10.1093/nar/gkn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ, Schaal H. Extended base pair complementarity between U1 snRNA and the 5' splice site does not inhibit splicing in higher eukaryotes, but rather increases 5' splice site recognition. NAR. 2005;33:5112–5119. doi: 10.1093/nar/gki824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Molecular Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM. U4/U5/U6 snRNP recognizes the 5' splice site in the absence of U2 snRNP. Genes Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- Kumar A, Wilson SH. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990;29:10717–10722. doi: 10.1021/bi00500a001. [DOI] [PubMed] [Google Scholar]
- Labourier E, Adams MD, Rio DC. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Molecular Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A. A CD45 polymorphism associated with Muliple Sclerosis disrupts an exonic splicng silencer. J Biol Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. Functional recognition of 5' splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Molecular Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette J-F, Revil T, Chabot B. hnRNP Proteins and Splicing Control (Landes Biosciences) 2007 doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Molecular Cell. 2010;29:223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Smith SA, Mallory MJ, Jackson J, Wang J, Lynch KW. A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs) J Biol Chem. 2011;286:20043–20053. doi: 10.1074/jbc.M111.218727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius BW, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci U S A. 1990;87:8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Molecular Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. NAR. 2010;38:6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Hsiao HH, Will CL, Giet R, Urlaub H, Luhrmann R. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat Struct Mol Biol. 2010a;17:216–221. doi: 10.1038/nsmb.1718. [DOI] [PubMed] [Google Scholar]
- Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Luhrmann R. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Molecular Cell. 2010b;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5' splice site-dependent assembly of U2AF and the prespliceosomal E complex. Molecular Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH, Black DL. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Molecular Cell. 2011;41:579–588. doi: 10.1016/j.molcel.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. "Nought may endure but mutability": spliceosome dynamics and the regulation of splicing. Molecular Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. An RNA switch at the 5' splice site requires ATP and the DEAD box protein Prp28p. Molecular Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J Biol Chem. 2005;280:38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5' splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci U S A. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.