SUMMARY
Meiotic recombination, crucial for proper chromosome segregation and genome evolution, is initiated by programmed DNA double-strand breaks (DSBs) in yeasts and likely all sexually reproducing species. In fission yeast, DSBs occur up to hundreds of times more frequently at special sites, called hotspots, than in other regions of the genome. What distinguishes hotspots from cold regions is an unsolved problem, although transcription factors determine some hotspots. We report the discovery that three coiled-coil proteins – Rec25, Rec27, and Mug20 – bind essentially all hotspots with unprecedented specificity even without DSB formation. These small proteins are components of linear elements, are related to synaptonemal complex proteins, and are essential for nearly all DSBs at most hotspots. Our results indicate these hotspot determinants activate or stabilize the DSB-forming protein Rec12 (Spo11 homolog) rather than promote its binding to hotspots. We propose a new paradigm for hotspot determination and crossover control by linear element proteins.
INTRODUCTION
During meiosis, a defining feature of all sexually reproducing species, homologous chromosomes segregate from each other to convert diploid cells into haploid cells (eggs and sperm in animals, ovules and pollen in plants, or spores in fungi). Homolog segregation requires in most species a physical connection between them, which imparts tension when homologs begin to segregate properly to opposite poles at the first meiotic division. The physical connection, a crossover, arises by homologous recombination, which also reassorts genetic differences between homologs, thereby increasing genetic diversity important for evolution.
In the species most thoroughly studied at the molecular level, the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, meiotic recombination is initiated by DNA double-strand breaks (DSBs) formed by a topoisomerase II-like protein Spo11 (Rec12 in fission yeast) (Keeney, 2007). Because a Spo11 ortholog is found in all sexually reproducing species examined, DSBs likely initiate meiotic recombination in all species; indeed, Spo11-deficient mutants of worms, flies, mice, and plants are deficient in meiotic crossing-over. During DSB formation, a tyrosine residue in the active site of Spo11 becomes covalently linked to the DNA via a 5′ phosphodiester bond. Unlike topoisomerase II enzymes, however, Spo11 does not appear to be catalytic, and it absolutely requires several partner proteins for its action. Nine such partner proteins have been identified in budding yeast, and six in fission yeast. Several additional proteins, discussed below, strongly stimulate but are not absolutely essential for DSB formation.
Meiotic DSBs are not uniformly distributed across the genomes studied. Instead, there are preferred sites, called hotspots, of DSB formation. What determines hotspots has been a long-standing problem, one addressed here. In a few known cases, hotspots are determined, at least in part, by a simple DNA sequence bound by a transcription factor. At the HIS4 locus of S. cerevisiae, Bas1, Bas2, and Rap1 factors bind closely spaced sequences and increase DSB formation nearby (White et al., 1993). Elimination of Bas1 decreases DSB formation at eight other genomic sites but, curiously, also increases DSB formation at four others and has no significant effect at 58 other Bas1-binding sites (Mieczkowski et al., 2006). The ade6-M26 single base-pair mutation of S. pombe creates a binding site for the Atf1-Pcr1 transcription factor, which is essential for increased gene conversion conferred by the M26 hotspot (Kon et al., 1997). Elimination of Pcr1 reduces DSB formation at the M26 hotspot and at about a dozen selected sites with the DNA binding sequence (Steiner and Smith, 2005); however, only a minority of DSB hotspots are likely bound by Atf1 (Cromie et al., 2007; Eshaghi et al., 2010); unpublished data). Other transcription factors also activate recombination hotspots in S. pombe ade6 mutants containing their cognate binding sequences (Steiner et al., 2011). Because there are hundreds or thousands of DSB hotspots in S. pombe and S. cerevisiae, respectively, no single transcription factor seems responsible for most or all DSB hotspots. Collectively, transcription factors may account for hotspots (Wahls and Davidson, 2010; Pan et al., 2011), but too few data are available to allow a firm conclusion. Thus, although a few hotspots clearly are determined at least in part by sites bound by transcription factors, wide-spread protein determinants that bind to the hotspots have been unknown.
Overall chromatin structure also appears to strongly influence hotspot activity. In S. cerevisiae, DSB hotspots often occur in nucleosome-depleted regions (NDRs) (Pan et al., 2011). In S. pombe, hotspots often contain an NDR over a small fraction of the DSB region, but most NDRs are not near hotspots, indicating that NDRs are poor predictors (de Castro et al., 2011). Set1, a histone H3 Lys4 methyltransferase, is important for most but not all DSB formation at the majority of hotspots in S. cerevisiae, but set1Δ mutants have high spore viability (Borde et al., 2009); thus, this chromatin feature is not required for crossing-over. The Prdm9 H3 Lys4 methyltransferase is essential for recombination stimulation at hotspots in some mammals but apparently not others (e.g., Baudat et al., 2010; Munoz-Fuentes et al., 2011), and not all Prdm9 binding sites are hotspots (Wang et al., 2012). The role of histone methylation in DSB formation is currently unclear (Tischfield and Keeney, 2012). The failure of hotspots, including the well-defined M26 hotspot of S. pombe, to act when transplaced from their active locus may reflect long-distance effects of chromatin structure (Ponticelli and Smith, 1992). Furthermore, M26 hotspot activity depends in part on histone modifications and chromatin remodeling factors (Hirota et al., 2008). Additional proteins that bind to chromatin during meiosis to form the axial element precursors of the synaptonemal complex (SC), such as S. cerevisiae Red1 and Hop1 of S. cerevisiae, are also important for DSB formation (Keeney, 2007). Meiotic cohesins are required in some regions but not others for DSB formation in both S. pombe and S. cerevisiae (Ellermeier and Smith, 2005; Kugou et al., 2009). In none of these cases, however, are these chromatin modifying factors known to directly bind and activate hotspots with high specificity.
S. pombe lacks a full-fledged SC but has structures, called linear elements (LinEs), whose temporal appearance and morphology by electron microscopy of nuclear spreads are similar to those of the axial element precursors of the SC of S. cerevisiae (Loidl, 2006). LinEs may serve a role similar to that of the SC – to align homologs for proper recombination. Four components of LinEs have been identified: Rec10, Rec25, Rec27, and very recently Mug20 (Lorenz et al., 2004; Davis et al., 2008; Spirek et al., 2010; Estreicher et al., 2012). In the few cases tested, these proteins co-localize by light microscopy, and focus formation of one depends on the others, implying that they intimately interact, perhaps by forming a complex; indeed, Rec10 co-immunoprecipitates with the other three proteins, and in two-hybrid assays Rec10 interacts with Rec25 and Rec15, a Rec12 partner protein (Spirek et al., 2010; Miyoshi et al., 2012). rec10Δ mutants are nearly as defective as rec12Δ mutants in the formation of recombinants (Ellermeier and Smith, 2005), but rec25Δ and rec27Δ mutants retain significant, though reduced, levels of recombination (Davis et al., 2008), as do mutants lacking the meiosis-specific Rec8 or Rec11 subunits of sister chromatid cohesin and the non-null rec10-109 mutant (DeVeaux and Smith, 1994; Ellermeier and Smith, 2005). These phenotypes are understandable because LinE focus-formation and binding to chromosomes depend on meiotic cohesins, but cohesins form foci equally in the absence or presence of LinE proteins (Molnar et al., 1995, 2003; Lorenz et al., 2004; Davis et al., 2008; Miyoshi et al., 2012). These gene dependencies and physical interactions are summarized in the pathway for DSB formation in S. pombe shown in Figure 1: binding of Rec8 and Rec11 leads to assembly of LinEs (Rec10, Rec25, and Rec27), which in turn leads to the stabilization or activation of Rec12 and its partner proteins and the formation of DSBs (Davis et al., 2008).
Figure 1. Pathway of Meiotic DSB Formation and Repair in S. pombe.
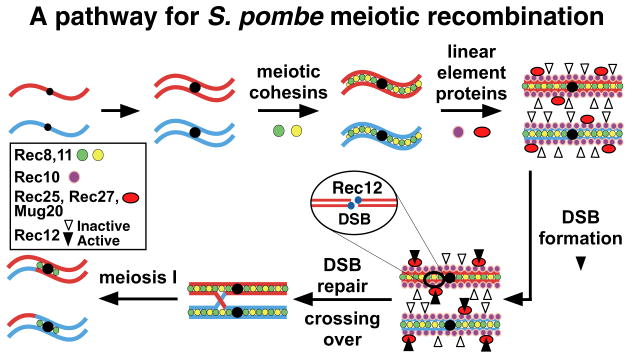
About the time of replication, loading of meiotic cohesin subunits Rec8 and Rec11 is followed by loading of LinE proteins Rec10, Rec25, Rec27, and Mug20 and loading or activation of Rec12 and its six partner proteins. Rec12 makes DSBs, becoming covalently linked to the DNA. Removal of Rec12 allows repair of the DSB with the sister chromatid or homolog. Repair with the homolog can form a crossover, which allows proper segregation of homologs at the first meiotic division. See also Figure S5.
The phenotypes of mutants lacking cohesin subunits or LinE components suggest that these structures are required for most but not all DSB formation. To determine the genome-wide effects of the corresponding rec mutations, we have analyzed Rec12-DNA covalent complexes, a measure of DSBs, genome-wide by microarray hybridization. We have also localized each of these proteins along the genome by immunoprecipitation of chromatin crosslinked to each GFP-tagged protein followed by microarray hybridization (ChIP-chip). Our results show, unexpectedly, that Rec25, Rec27, and Mug20 are enriched with exceptionally high specificity at DSB hotspots and that Rec27 is required for formation of nearly all DSBs at hotspots. Furthermore, hotspot DNA is bound by Rec27 even without DSB formation, and mutants with altered Rec27 binding have correspondingly altered hotspot DSB formation. Rec27 and Mug20 respectively show similarity to C. elegans SYP-2, an SC component, and DDL-1, which interacts with SYP-2 (Colaiacovo et al., 2003; Simonis et al., 2009). Thus, these proteins, whose functions may be conserved for meiosis in other species, are the first highly specific determinants of essentially all meiotic DSBs.
RESULTS
Linear Element Proteins Rec25, Rec27, and Mug20 Interdependently Co-localize in Meiotic Nuclei
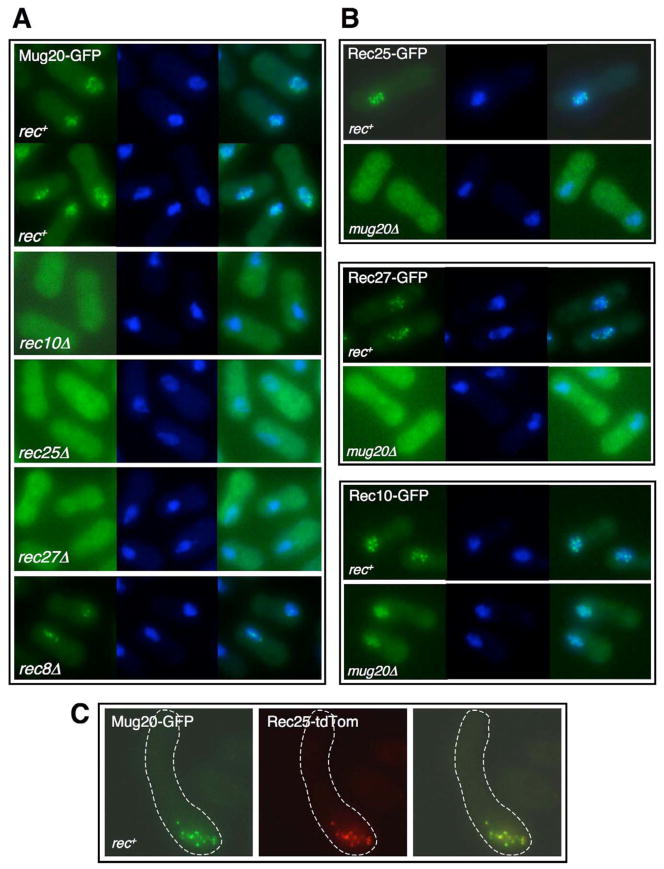
Mug20 was identified in a Rec10 immunoprecipitate by mass spectrometry (Spirek et al., 2010). This 17 kDa protein is induced early in meiosis, as are the 16 – 17 kDa proteins Rec25 and Rec27 (Davis et al., 2008; Estreicher et al., 2012). We analyzed by fluorescence microscopy cells bearing the mug20-GFP fusion gene and synchronously induced for meiosis in pat1-114 (Ts) diploid strains. [The encoded fusion protein is nearly fully active for recombination (Table S1).] In intact cells Mug20-GFP behaved much like Rec25-GFP and Rec27-GFP: all were visible from about 1.5 – 2 hr after meiotic induction, when cells were replicating DNA, to about 4 hr, when cells were beginning the first meiotic division; at 3.5 hr, when proteins and DSBs were analyzed below, all three proteins are prominent in most cells (Figures 2A, S1, S2, and S3; Davis et al., 2008). Mug20-GFP was exclusively in the nucleus and formed both fuzzy dot-like foci and linear structures. Nucleus-specific localization of Mug20-GFP was completely lost in the rec10Δ, rec25Δ, and rec27Δ mutant cells (Figures 2A and S4A and unpublished data for other induction periods). Conversely, in mug20Δ mutant cells foci of Rec25-GFP and Rec27-GFP were completely lost, and Rec10-GFP foci were less distinct (Figure 2B). Like the LinE components Rec25 and Rec27, Mug20-GFP formed only a limited number of discrete foci or elongated structures in rec8Δ mutants (Figure 2A); Rec8-GFP, however, formed normal grainy nuclear structures in mug20Δ cells (Figure S4B), as we previously showed for Rec8 in rec10Δ, rec25Δ, and rec27Δ mutants using nuclear spreading (Davis et al., 2008). Also like the LinE component Rec10 (Lorenz et al., 2004; Davis et al., 2008), Mug20-GFP formed abundant structures in nuclear spreads of control and rec12Δ mutant cells during meiotic prophase (Figure S4A and unpublished data). Furthermore, Rec25-dtTomato and Mug20-GFP co-localized during meiotic prophase in zygotic pat1+ meiosis (Figure 2C); co-localization was complete throughout pat1-114 prophase, as well (unpublished data). The strikingly similar behavior of Mug20 and other LinE components and the interdependence of the four proteins for distinct focus-formation strongly support Mug20 being a genuine LinE component and confirm that LinE formation depends on sister chromatid cohesins but not on DSB-formation, as shown in the pathway in Figure 1. Recently, Estreicher et al. (2012) also showed that Mug20, in nuclear spreads rather than in intact cells as used here, is a LinE-associated protein. We infer that the dots in intact cells, whether fixed or not, reflect the in vivo structures and that the proteins may become reorganized during nuclear spreading.
Figure 2. Mug20 Is a LinE Component Interacting with Rec25, Rec27, and Rec10.
In A and B, cells with the indicated GFP fusion protein and rec gene were synchronously induced for meiosis, fixed 3 hr later, stained with DAPI, and examined by fluorescence microscopy. Each set of three images shows the GFP protein (green; left), DAPI-stained DNA (blue; middle), and merge (right). In C live cells from zygotic meiosis were examined for Mug20-GFP (green; left), Rec25-tdTomato (red; middle), and both (right). See also Figures S1, S2, S3, and S4.
(A) Mug20 forms nuclear foci that depend on LinE components Rec10, Rec25, and Rec27 but only partially on the sister chromatid cohesin Rec8.
(B) Mug20 is required for nuclear focus-formation of Rec25 and Rec27 and, partially, of Rec10.
(C) Mug20 and Rec25 co-localize in live zygotic cells.
In spite of clear similarities, there are differences in the behavior of the four LinE components. Rec10-GFP remained primarily in the nucleus in the absence of Mug20, whereas Rec25-GFP, Rec27-GFP, and Mug20-GFP were evenly distributed throughout the cell in the absence of any other LinE component (Figures 2A and 2B; Davis et al., 2008), likely because Rec10 has a predicted nuclear localization sequence but the other proteins do not (Lorenz et al., 2004; unpublished data). Rec10-GFP, Rec25-GFP, and Rec27-GFP foci are sharper than those of Mug20-GFP, which appeared to make some thin lines as well as fuzzy dots (Figures 2A, S1, S2, and S3) (Davis et al., 2008). Although DSB-formation and recombination are essentially eliminated in rec10Δ mutants (Ellermeier and Smith, 2005) (see below), there is significant residual, region-specific recombination in rec25Δ and rec27Δ mutants (Davis et al., 2008) as well as in mug20Δ mutants (Table S1; Estreicher et al., 2012). We discuss the implications of these observations later.
Conservation of Coiled-Coil Domain Proteins Rec27 and Mug20 Among Species
Three LinE proteins (Rec25, Rec27, and Mug20) are rather small proteins with predicted coiled-coil domains also found in SC proteins and some DNA-binding transcription factors, although LinE and SC proteins lack an obvious DNA-binding motif. We compared the amino acid sequences of these three LinE proteins encoded by four Schizosaccharomyces species with the sequences of the small (~25 kDa) SC proteins SYP-2 and SYP-3 of four Caenorhabditis species and found remarkably conserved similarity among the Rec27 and SYP-2 proteins (Figure S5). The similarity encompasses the predicted coiled-coil domain as well as a region toward the N-terminus. Like Rec27, localization of SYP-2 on meiotic chromosomes strongly depends on Rec8 but not Spo11 (Rec12) (Colaiacovo et al., 2003). We also found similarity among Schizosaccharomyces Mug20 and Caenorhabditis DDL-1 proteins; in a two-hybrid assay DDL-1 interacts with SYP-2 (Simonis et al., 2009), suggesting that DDL-1 is associated with the SC. Although LinEs have been considered to be only distantly related to SCs (Loidl, 2006), these similarities suggest a more highly conserved function common to the two structures than previously realized.
Rec25, Rec27, and Mug20 Co-localize at DSB Hotspots
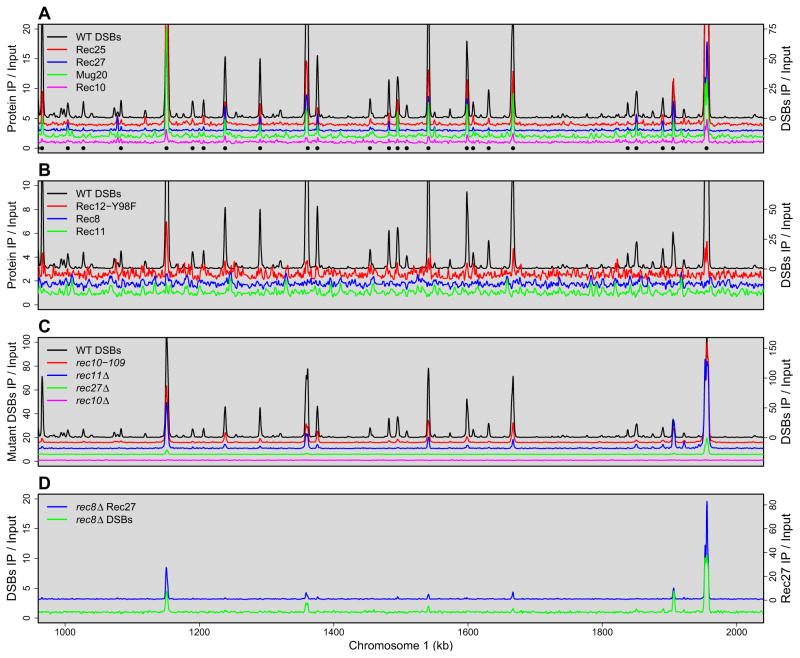
The preceding microscopic analyses suggested that Rec25, Rec27, and Mug20 bind chromosomes at the same or closely linked sites. To determine their localization at high resolution, we analyzed by ChIP-chip the localization of the GFP-tagged proteins described above. We found that all three proteins are highly enriched at certain sites across the genome. Figure 3A shows a representative 1 Mb interval of the 12.5 Mb genome; graphical representation of the data in Figures 3 and 6 for the whole genome are on the Lab Websites. In this 1 Mb interval there are 25 DSB hotspots, taken as sites at which >0.3% of the DNA is broken, as determined by the nearly linear relation, for 25 hotspots, between Rec12-DNA covalent linkages and DSBs assayed directly by Southern blots (Cromie et al., 2007); this DSB level is at the limit of detection by Southern blots. At some hotspots in the genome these linkages are >250 times the genome median, as in previous analyses (Cromie et al., 2007; Hyppa et al., 2008). Linkages were determined in rad50S mutants, in which DSBs are made with the same distribution as that in rad50+ but are not repaired and hence accumulate, allowing sensitive measures of DSBs (Hyppa et al., 2008). Between these hotspots, Rec12-DNA linkages do not rise significantly above the genome median, defining cold regions.
Figure 3. Rec25, Rec27, and Mug20 Bind DNA at Meiotic DSB Hotspots with High Preference, but Rec8, Rec10, Rec11, and Rec12 Bind with Little or No Preference.
DNA covalently linked to Rec12-FLAG (signifying DSBs; harvested at 5 hr, when DSBs are maximal) or DNA crosslinked to the indicated GFP fusion protein (harvested at 3.5 hr, when foci are prominent). was analyzed by microarray hybridization. Data are median-normalized, smoothed using an 11-probe window, and plotted with an offset for legibility. “Input” is whole cell extract. Complete genome data are on the Lab Websites. See also Figures S6 and S7.
(A) LinE proteins Rec25, Rec27, and Mug20 (plotted on left axis; offsets of 3, 2, and 1, respectively) bind preferentially to DSB hotspots (right axis), but LinE protein Rec10 (left axis) binds nearly uniformly except for modest preference at strong hotspots. Black circles beneath the traces indicate wild-type hotspots (see Supplemental Methods for peak-calling criteria).
(B) Sister cohesin subunits Rec8 and Rec11 (left axis; offsets of 1 and 0, respectively) bind nearly uniformly, as does the inactive Rec12-213 (Y98F) mutant protein (left axis; offset of 2).
(C) DSBs are nearly eliminated in rec10Δ and are significantly reduced at most hotspots in rec27Δ a nd rec11Δ null mutants and the rec10-109 missense mutant (left axis; offsets of 0, 5, 10, and 15, respectively).
(D) LinE protein Rec27 preferentially marks the DSB hotspots remaining in rec8Δ.
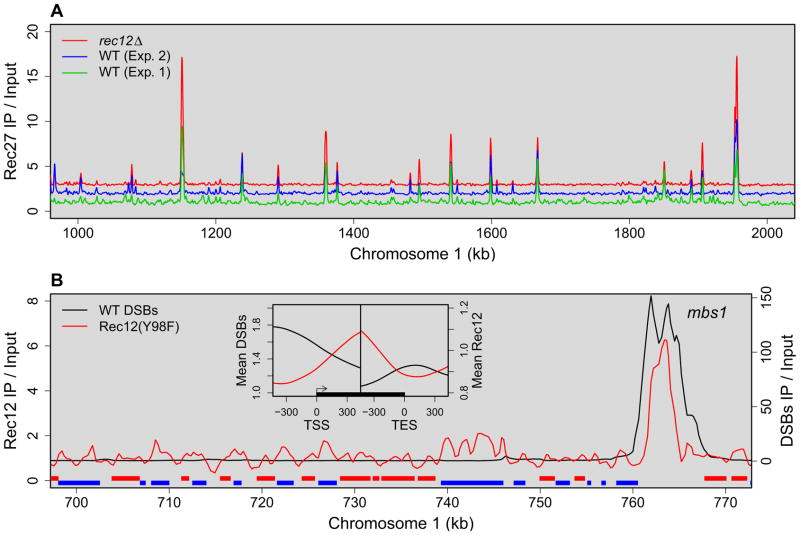
Figure 6. Rec27 Binds to DSB Hotspots in the Absence of DSB-formation by Rec12, Which Binds to DSB Hotspots with Only Modest Preference.
(A) Rec27 binding in rec12+ (Figure 3, experiment 2, offset of 1) or in rec12Δ (offset of 2; analyzed concurrently with rec12+ in experiment 2). r = 0.77 for single probes and 0.81 for 11-probe smoothing (Figure 4C and Table S2). Complete genome data are on the Lab Websites.
(B) Binding of inactive Rec12-213 (Y98F) as in Figure 3 is largely independent of Rec27 (Figure S8). Binding is higher in protein-coding genes (red and blue bars for upper- and lower-strand coding) and lower between genes. Inset: 4003 genes were aligned at their transcription start sites (TSS) and transcription end sites (TES) (Lantermann et al., 2010). Rec12-Y98F binding (red line) is higher in genes (black rectangle with arrow) than between genes, whereas mean DSBs (Rec12-DNA covalent linkages; black line) is higher between genes than in genes. Note recombinant frequency in ura1 (7 kb blue bar near 740 kb) is 19 times lower than genome average (Supplemental Information), although Rec12-Y98F binding is about twice the genome median. See also Figure S9.
The distribution of each of the three LinE proteins (Rec25, Rec27, and Mug20) closely parallels that of DSBs. For example, Rec25 was enriched >2 times the genome median at 23 of the 25 DSB hotspots in the 1 Mb interval shown in Figure 3A, whereas between the hotspots Rec25 was found at essentially the genome-median level. Enrichments up to 80 times the genome median were found at some hotspots (Lab Websites). Rec27 and Mug20 showed similar enrichments, up to 24 and 28 times the genome median respectively (Figures 3A). The binding profiles of all three proteins closely parallels that of DSBs, determined either by ChIP-chip analysis of Rec12-DNA linkages or by high resolution Southern blots (Figures 3A and S6). We estimate from these data that the resolution (maximal genome distance by which two peaks could be offset but appear to be coincident) is <1 kb. Most hotspots, such as the well-studied mbs1 hotspot, are clusters of closely spaced sites of variable breakage; these DSB regions are up to 7 kb wide and on average are ~45 kb apart (Figures S6; Cromie et al., 2005, 2007). We consider the entire region of breakage one hotspot.
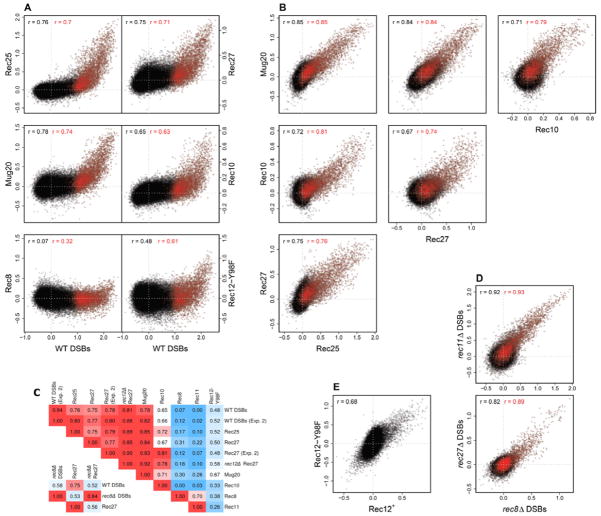
To quantify the correlations, we compared the enrichments of each of these proteins to those of the Rec12-DNA linkages at each of the ~44,000 genome positions (probes) represented on the microarray. Scatter plots showed that Rec25, Rec27, and Mug20 are highly enriched at DSB hotspots (Figure 4A; red data) but not in DSB cold regions. From these data, we calculated the Pearson correlation coefficient r. Without smoothing of the data (i.e., analysis of all individual probes), r = 0.76, 0.75, and 0.78 for Rec25, Rec27, and Mug20, respectively, vs. Rec12-DNA linkages (DSBs). These values rise only slightly to 0.80, 0.79, and 0.85, when smoothed over an 11-probe (~3 kb) window (Table S2), an indication of the high precision and resolution of these data. For comparison, we note that r for two independent Rec27 protein distributions is 0.77 (Figure 4C), which we take as the upper bound for identity of genome-wide features determined on microarrays. Thus, these data show that Rec25, Rec27, and Mug20 are enriched nearly exclusively, and exceptionally highly, at DSB hotspots. These proteins may also be present in cold regions, but if so their frequency is, conservatively, <10 % of that at strong hotspots.
Figure 4. Rec25, Rec27, and Mug20 Binding Is Highly Correlated with Genome-wide DSB Frequencies, but Rec8, Rec11, and Rec12 Binding Is Not.
(A and B) Scatter plots of the genome median-normalized data for the two parameters indicated on the axes. All data points (~44,000; in black) are plotted on a log10 scale (IP/input), but most are obscured by their high density. Red data are points within DSB hotspots. Pearson correlation coefficients (r) are for unsmoothed data. See also Table S2.
(C) Summary of r for unsmoothed data. Dark red indicates highest correlation, dark blue the lowest, and lighter values between these extremes.
(D) Scatter plots and r from two rec8Δ inductions.
(E) Scatter plot and r, as above, for Rec12-213 (Y98F) and Rec12+ proteins (Lab Websites). The Rec12+ data reflect both self-linkage (DSBs) and crosslinking (binding), but a positive correlation is still observed.
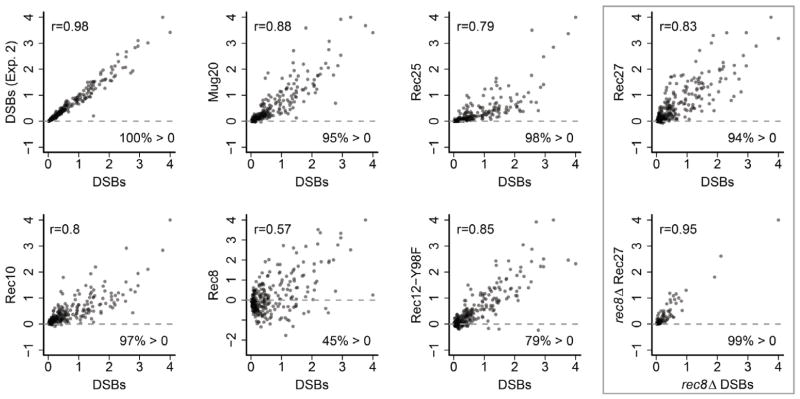
The correlation between protein binding and DSBs was also dramatic when we analyzed hotspots individually. For this analysis we integrated the values for DSB frequency and for protein binding across each of the 288 hotspots in the genome (those with >0.3% DSBs, as defined above). The data show that the DSB frequency is a linear function of the relative amount of protein bound (Figure 5); r = 0.79, 0.83, and 0.88 for DSBs vs. Rec25, Rec27, and Mug20, respectively. Thus, binding of these proteins determines not only the position of DSB hotspots, but accounts for the majority of the variation in breakage: the coefficient of determination R2 is 0.62 – 0.78. Similarly in rec8Δ, in which Rec27 binding is altered (Figure 3D), the few hotspots that remain show an exceptionally strong correlation (r = 0.95) between the amount of breakage and Rec27 bound (Figure 5).
Figure 5. Correlations between DSB Frequencies and Protein Abundances at DSB Hotspots Are Especially Strong.
Scatter plots and r for DSBs (integral above or below median of Rec12-DNA covalent linkages across hotspots) and the indicated protein similarly integrated. Points above the line indicate protein enrichment at that hotspot; points below imply depletion. Data are in arbitrary units on linear axes.
A corollary of these observations is that these three proteins should be highly colocalized on the genome, as suggested by the microscopy data (Figures 2. S1, S2, S3, and S4; Davis et al., 2008). The data in Figure 3A show that this is the case: Rec25, Rec27, and Mug20 are enriched at only a limited number of sites in the genome, and essentially always co-localize with each other. Scatter plots of these data confirm this conclusion. For example, there is a high, linear correlation between the abundance of Rec25 and Rec27, of Rec25 and Mug20, and of Rec27 and Mug20 at each probe (Figure 4B). Thus, these data confirm the colocalization implied by microscopy (Figures 2, S1, S2, and S3; Davis et al., 2008) and show that these three LinE proteins are strongly enriched exclusively at the same sites – hotspots of DSB formation.
Linear Element Proteins and Cohesins Are Required for DSB Formation at Most Hotspots
Rec25 and Rec27 are enriched specifically at DSB hotspots (Figures 3, 4, and 5) and are required for DSB-formation at the few loci previously tested by Southern blot hybridizations (Martin-Castellanos et al., 2005). To determine the extent of this requirement, we determined the genome-wide DSB distributions in mutants lacking Rec27. DSBs were essentially eliminated at more than 80% of the hotspots in the rec27Δ mutant (Figures 3C). Low-level DSBs were seen at some of the strongest hotspots, but even these DSBs were reduced or nearly eliminated.
Similar DSB patterns were seen in the absence of the meiosis-specific cohesin subunits Rec8 or Rec11. DSBs at most of the hotspots were nearly eliminated, and residual levels were seen at the same hotspots at which DSBs remained in rec27Δ (Figures 3C and 3D). There is a striking correlation between DSB frequency in rec27Δ or r ec11Δ and that in rec8Δ (Figures 4D and Table S2). Thus, DSBs at most hotspots depend on both Rec8 and Rec27, in accord with Rec8 being required for binding of Rec27 to most hotspots and the requirement of meiotic cohesins for LinE focus-formation and localization on DNA (Molnar et al., 1995, 2003; Lorenz et al., 2004; Davis et al., 2008; Miyoshi et al., 2012).
A distinctly different pattern was seen in the rec10Δ mutant: DSBs were completely eliminated except for an almost invisible amount at an exceptionally strong hotspot near 4.0 Mb on the right end of chromosome 2 (Figures 3C). Southern blot analysis shows that the very low level DSBs at this site are meiosis-specific (Ellermeier and Smith, 2005). Thus, our ChIP-chip analysis has the power to detect even tiny amounts of DSBs. With this one exception, the DSB pattern in rec10Δ was indistinguishable from that in rec12Δ, which lacks the protein with the active site for DSB formation (Young et al., 2002). In contrast, significant levels of DSBs remained in the initial rec10 isolate, rec10-109, which harbors two closely spaced missense mutations and retains significant region-specific recombination (DeVeaux and Smith, 1994; Ellermeier and Smith, 2005): the DSB pattern closely resembles that in rec8Δ, rec11Δ, and rec27Δ mutants (Figures 3C and 3D). In the Discussion, we propose an explanation for the DSB patterns seen in these mutants.
Rec27 Binds Sites Poised To Be DSB Hotspots Even in the Absence of DSB Formation
Given the hotspot-enriched binding shown above and that LinE components are required to form DSBs (Ellermeier and Smith, 2005; Martin-Castellanos et al., 2005), we would expect these proteins to be present before DSB formation and, thus, even in the absence of DSB formation. To test this hypothesis, we determined the genome-wide distribution of Rec27 in the absence of Rec12. We found that the distributions were practically identical (Figures 4C and 6A; Table S2). The simplest interpretation of these data and the genome-wide requirement for Rec27 for most DSB formation is that Rec27 localizes to sites poised to be DSB hotspots before DSBs are formed and recruits one or more DSB-forming proteins to their sites of action or activates them after they bind or both.
The high correlation of LinE protein binding with both hotspot position and hotspot intensity predicts that altering the LinE binding profile should alter the DSB landscape. Rec8 is required for proper LinE formation (Molnar et al., 1995; Lorenz et al., 2004; Davis et al., 2008), and a rec8Δ mutant has co-coordinately altered DSB and Rec27 binding profiles (Figures 3D). Similarly, insertion of exogenous DNA (the bacterial kan drug-resistance determinant) at the rec8+ locus creates both a Rec27 binding site and a DSB hotspot (Figure S7); similar results were found with other insertions, such as FLAG (unpublished data). The generation of DSB hotspots at manipulated loci has previously been observed in S. pombe and S. cerevisiae, but the mechanism remains unresolved (e.g., Ponticelli and Smith, 1992; Borde et al., 1999; de Castro et al., 2011). Thus, sites poised for DSB formation, even unusual de novo sites, are predetermined by Rec27, indicating a mechanistic relationship between Rec27 binding and DSB formation.
Rec12 Binds DNA with Only Modest Preference for DSB Hotspots
The strong preference for DSB formation at hotspots could reflect either preferential binding of Rec12 at hotspots or its preferential activation there. To distinguish these possibilities, we determined the genome-wide binding profile of Rec12 with phenylalanine in place of the active site tyrosine. This protein, from the rec12-213 (Y98F) mutant, lacks a single oxygen atom necessary for wild-type Rec12 DSB formation and is as recombination-deficient as rec12Δ (Cervantes et al., 2000). Using this mutant protein eliminates self-linkage, which could be exceptionally strong at DSB hotspots and thereby obscure the true Rec12 binding profile. Rec12-213 (Y98F) binding has only low-level peaks above the genome median across most of the genome, although at exceptionally strong DSB hotspots it is clearly more abundant than the genome median (Figures 3B and 6B). A similar pattern was seen for DSB-proficient Rec12 crosslinked with formaldehyde (Figures 4E and S8; Ludin et al., 2008), indicating that Rec12 and Rec12-213 (Y98F) bind similarly.
The pattern of binding in the low-level regions, with multiple adjacent probes significantly above the genome median, implies that the low-level peaks in meiosis are not background “noise.” The distributions about these peaks are nearly identical for Rec12 and Rec12-213 (Y98F) (Figure S8A), again indicating that these proteins bind the same. Furthermore, Rec12-213 (Y98F) binds significantly above the genome median in genes but less than the genome median between genes, whereas DSB frequency has the opposite pattern (Figure 6B, inset). These differentials between genes and intergenic regions, both for binding and DSB formation, increase with increasing abundance of meiotic transcripts (Figure S9), suggesting that transcription can promote Rec12 binding and DSB-formation but in distinctly separate regions. As expected, r for DSBs vs. Rec12-213 (Y98F) binding is much lower than r for DSBs vs. Rec27 binding, for example (Figures 4A and C). If only individual hotspots are considered, however, r for DSBs vs. Rec12-213 (Y98F) binding is 0.85, about the same as r for DSBs vs. Rec25, Rec27, or Mug20 (Figure 5); with hotspots excluded it is −0.05. Thus, although Rec12 binds to hotspots in proportion to the amount of DSBs that will be formed, it also binds outside hotspots but nearly at random with respect to the amount of DSBs that will be formed. We account for this pattern in the Discussion.
Rec12 Binds to Some DNA Sites Independent of Rec27 without Forming DSBs
We noted ~20 loci at which both Rec12 and Rec12-213 (Y98F) bind significantly above the genome median, yet at which few if any DSBs are formed. Two such sites, denoted C and D in Figure S8B, are near two prominent DSB hotspots, denoted A and B. Although Rec12-213 (Y98F) binding (with formaldehyde crosslinking) is nearly equivalent at all four sites, DSBs (Rec12 self-linkages, without formaldehyde) are much more prominent at A and B than at C and D. These data show directly that Rec12 can bind without making DSBs and indicate that Rec12 is activated at some sites (hotspots) by another factor. One of these factors appears to be Rec27, since Rec27 is abundant at sites A and B but not at C and D (Lab Websites). Furthermore, in the absence of Rec27, binding of Rec12 is abundant at all four sites despite DSBs (self-linkage) being barely detectable at these sites in rec27Δ (Figure S8B). Thus, Rec12 can bind without Rec27 but makes DSBs at most hotspots only in its presence.
Meiotic Cohesin Subunits Rec8 and Rec11 and Linear Element Protein Rec10 Are Nearly Uniformly Distributed Across the Genome
Microscopic analyses show that most but not all LinE formation requires Rec8 and Rec11 (Figure 2A; Molnar et al., 1995, 2003; Lorenz et al., 2004; Davis et al., 2008). To determine if the hotspot-specific binding of Rec25, Rec27, and Mug20 reflects hotspot-specific binding of Rec8 and Rec11, we determined the genome-wide distributions by ChIP-chip of Rec8-GFP and Rec11-GFP, which are nearly fully active for DSB-formation and recombination (Table S1). Unexpectedly, these two proteins were distributed nearly uniformly across the genome, with no preferential enrichment or depletion at hotspots (Figures 3B). Scatter plots confirm this impression (Figures 4A and C). Considering hotspots individually, at roughly half of the hotspots each protein is below the genome median, as expected for uniform binding with some variation; in sharp contrast, at ≥94% of the hotspots the LinE proteins Rec25, Rec27, and Mug20 are enriched above the median (Figure 5). These data suggest no significant correlation between Rec8 and Rec11 binding and DSB-formation, a marked difference from the negative correlation of DSBs and Rec8 binding in S. cerevisiae (see Discussion) (Blat et al., 2002; Glynn et al., 2004; Panizza et al., 2011). [Ding et al. (2006) reported somewhat greater excursions in Rec8 binding density across the part of the genome they assayed; this difference may reflect the Rec8-HA tag or the use of haploids instead of diploids, as used here.]
We were further surprised by the distribution of Rec10, which by fluorescence microscopy appears to colocalize with Rec25, Rec27, and Mug20 (Davis et al., 2008; Estreicher et al., 2012). Rec10-GFP binds nearly uniformly across the genome, but with modest enrichment at many hotspots (Figure 3A). The enrichment at hotspots was generally <3 times the genome median, and the highest enrichment was 6-fold at an exceptionally strong hotspot on the right end of chromosome 1. r for Rec10 vs. DSBs is 0.65, slightly lower than that for Rec27 (0.77) (Figure 4C). Considering only enrichment at hotspots, r = 0.80 for Rec10 (Figure 5), and with hotspots excluded, r = 0.35. Thus, DSB frequency and Rec10 abundance are more highly correlated at DSB hotspots than in DSB-cold regions, similar to the distribution of Rec12-213 (Y98F) binding, indicating two chromosomal domains with respect to DSB-formation as we discuss below.
DISCUSSION
It has long been recognized that meiotic recombination does not occur at uniform frequency across the genome; rather, there are hotspots of recombination – sites at which recombination occurs at higher-than-average frequency – and intervening cold regions (Keeney, 2007). But what determines hotspots has been largely elusive except for a few particular sites activated by certain transcription factors and a more wide-spread effect of chromatin structure (see Introduction). Here, we identify three coiled-coil proteins, Rec25, Rec27, and Mug20, likely acting as a complex, that bind to and, at least for Rec27, activate nearly all DSB hotspots across the genome of the fission yeast S. pombe. These proteins are components of linear elements and are related to the SC proteins of other species (Loidl, 2006; Figure S5). This feature provides the basis for an additional level of control, discussed below, for formation of crossovers, the crucial connection between homologs that allows their successful segregation in meiosis.
Rec25, Rec27, and Mug20 Bind Hotspots with High Specificity and Are Hotspot Determinants
Microarray-based assays for binding of these three proteins to DNA show directly that they are enriched with unprecedented specificity at DSB hotspots, with an enrichment up to 80 times the genome median (Figures 3A). Quantitative analysis shows a linear relation between the frequency of DSB formation at a hotspot and the frequency of protein binding at that hotspot (Figure 5). Elimination of Rec25 or Rec27 protein strongly reduces or eliminates DSB formation at hotspots (Figure 3C; Martin-Castellanos et al., 2005); to our knowledge, Mug20 has not been similarly tested. Thus, these proteins determine both the position and the activity of nearly all hotspots across the genome and can be considered essential components of meiotic DSB hotspots. As predicted by this conclusion, when exogenous DNA was inserted into the chromosome, it created both a hotspot for Rec27 binding and a hotspot for DSB formation (Figure S7). Furthermore, deleting rec8 coordinately reduces DSB formation and Rec27 binding, leaving a DSB landscape that mirrors the residual Rec27 binding profile (Figures 3D and 5).
Previous reports have shown that certain transcription factors also bind to and activate hotspots. But the factor examined most thoroughly, Bas1 of S. cerevisiae, activates only a few of the thousands of hotspots across the genome, and the M26 binding sequence for Atf1-Pcr1 is a poor predictor of DSB hotspots in S. pombe (Steiner and Smith, 2005; Mieczkowski et al., 2006). Other factors, including chromatin remodeling and histone modifications, have more wide-spread effects (Hirota et al., 2008; Borde et al., 2009; de Castro et al., 2011; Pan et al., 2011), but it is not clear that these modifications act directly (as opposed to altering replication or gene expression and thereby having indirect effects on recombination), nor is it clear that they are hotspot-specific. Indeed, most such factors are poor predictors of hotspots (Tischfield and Keeney, 2012). Rec25, Rec27, or Mug20 detectably bind at 86% of all DSB hotspots (97% of the hottest two-thirds of sites, or about 200 hotspots), and they are enriched nearly exclusively at hotspots, making their binding the best predictor for hotspot position in any species reported to date.
Meiosis-specific Cohesin Subunits Rec8 and Rec11 and Linear Element Protein Rec10 Bind Chromosomes with Little Site Specificity
Meiotic cohesins are required for LinE formation (Figure 2; Molnar et al., 1995, 2003; Lorenz et al., 2004; Davis et al., 2008; Estreicher et al., 2012), and Rec8 and Rec11 make discrete foci in chromosome spreads or live cells (e.g., Ding et al., 2006; Davis et al., 2008); however, they bind essentially uniformly across the genome (Figures 3B, 4A and 4C). We suppose that this uniformity reflects a limited amount of these proteins (to account for their punctate foci in individual cells) that binds with nearly equal probability at any point along the DNA (to account for the uniform binding to DNA in the population of cells). If so, on an individual chromosome a limited number of nearly random sites may be bound by these meiosis-specific cohesins.
Rec10, the protein that defined linear elements seen by light microscopy (Lorenz et al., 2004), also binds along the chromosomes nearly uniformly, although it binds somewhat more frequently to DSB hotspots (Figure 3A). Miyoshi et al. (2012) recently reported a similar profile during haploid meiosis: Rec10-FLAG is modestly enriched at hotspots with more uniform binding elsewhere. The relative magnitude of Rec10’s binding to hotspots is not, however, as great as that of Rec25, Rec27, or Mug20: the maximal enrichment of Rec10 at a hotspot is 6 times the genome median, whereas the maximal enrichments for Rec25, Rec27, and Mug20 are 80, 24, and 28 times the genome median, respectively (Figures 3A). The absolute amount of Rec10 at a hotspot may be as high as that of Rec25, for example, but if so the level of Rec10 between hotspots would seem to be higher than that of the other proteins. We infer that Rec10, like Rec8 and Rec11, binds nearly uniformly across the genome but, unlike Rec8 and Rec11, with additional enrichment at hotspots.
Rec10, Rec25, Rec27, and Mug20 Stabilize or Activate Rec12 To Make DSBs, Rather Than Recruiting Rec12 to DSB Hotspots
The nearly uniform binding of Rec12, as the DSB-inactive Y98F mutant, along chromosomes strongly contrasts with the much higher specificity of DSB formation at hotspots (Figures 3B, 6, and S8). In addition Rec12 binds to many points along the chromosome where it makes few if any DSBs (Figure S8). We propose that Rec12 is stabilized or activated to a high level by Rec25, Rec27, and Mug20, specifically at hotspots, and is activated at a low level by Rec10 alone, to make low-level DSBs between hotspots (i.e., in DSB-cold regions). Since Rec10 is also required for Rec25, Rec27, and Mug20 focus-formation (Figures 2A and S4; Davis et al., 2008) and Rec27, and perhaps Rec25 and Mug20 as well, is required for most DSBs at hotspots (Figures 3C; Martin-Castellanos et al., 2005), this view predicts, as observed, that Rec10 is essential for virtually all DSBs across the genome (Figures 3C; Ellermeier and Smith, 2005).
Our conclusion that Rec12 binds to both DSB-cold regions and DSB hotspots but is activated at DSB hotspots by Rec25, Rec27, and Mug20 is supported by the distinct pattern of global Rec12 binding and DSB formation along the chromosome. In DSB cold regions, low-level DSBs are more frequent between genes, but Rec12-binding is more frequent within genes (Figure 6B inset). This preferential loading in genes correlates with transcriptional activity and is strongly influenced by the transcript start and stop sites (Figures 6B and S9). Furthermore, in the ura1 gene recombination is about 20 times lower than the genome mean, but Rec12-binding is about twice the genome median (Supplemental Information). This inverse relation is most readily explained by a requirement for Rec12 to be activated after it has bound DNA (i.e., loading is not sufficient for breakage). At DSB hotspots, this activation depends on Rec25, Rec27, and Mug20: the level of DSBs is proportional to the amount of each of these three proteins bound (Figure 5). DSB hotspots thus depend on the strong localization of Rec25, Rec27, and Mug20 to hotspots. Our proposal also explains the observation that only ~10% of the total Rec12 is covalently linked to DNA (Milman et al., 2009). The majority of bound Rec12 may have another role, such as chromosome segregation at the second meiotic division (Sharif et al., 2002). In S. cerevisiae and mice, there also appears to be a large excess of the Rec12 ortholog, Spo11, which may play a role independent of DSB formation (Keeney, 2007; Bellani et al., 2010).
Rec12 may be activated for DSB formation by the binding of one or more of its partner proteins, dependent on one or more of the LinE proteins. Formation of nuclear foci by two Rec12 partner proteins, Rec7 and Rec24, depends on Rec10 (Lorenz et al., 2006; Bonfils et al., 2011). One of these partner proteins, such as Rec15, which interacts with Rec10 in a two-hybrid assay (Miyoshi et al., 2012), may be rate-limiting for DSB formation and more abundant at hotspots than in DSB-cold regions. Rec10 may be the crucial link between the “early” proteins (cohesins and other LinE proteins) and the “late” proteins (Rec12 and its partners) for DSB formation, with additional activation at hotspots by Rec25, Rec27, and Mug20 (Figure 1).
Localization of Rec25, Rec27, and Mug20 to DSB Hotspots
Unlike the protein determinants, the DNA determinants of most DSB hotspots remain unclear. Sequence comparisons, such as that by MEME (http://www.meme.sdsc.edu), do not reveal an obvious consensus sequence for hotspots, although polypurine stretches on one strand have a limited correlation with hotspots (Cromie et al., 2007). Non-coding RNAs (ncRNAs) are correlated with hotspots (Wahls et al., 2008), but this correlation may simply reflect the higher-than-average density of ncRNA genes in large intergenic regions and not be directly causative. Since Rec10 binds to hotspots with modest preference (Figures 3A, 4, and 5) and since formation of nuclear foci by Rec25, Rec27, and Mug20 depends on Rec10 (Figures 2A and S4A; Davis et al., 2008; Estreicher et al., 2012), we infer that this protein complex has the intrinsic ability to bind hotspots; their coiled-coil structure suggests they may act like certain transcription factors with extensive coiled-coil domains. The hotspot specificity may reside within one of these proteins but be effective only when in the putative complex.
This proposal is concordant with the chromosome interval-dependent reduction of DSB formation and recombination in rec25Δ, rec27Δ, mug20Δ, and rec10-109 mutants. In the mutants tested in this set, DSBs are strongly reduced at most hotspots (Figures 3C; Davis et al., 2008), and recombinant frequencies are reduced in some intervals by factors of >100 but in other intervals by factors of <3 or even not significantly in rec10-109 (Table S1; DeVeaux and Smith, 1994; Davis et al., 2008; Estreicher et al., 2012). Although these differentials were initially described as “region specific,” our data suggest that they are “site specific,” since no large region of the mutant genomes retains all hotspots present in wild type (Lab Websites). We note, however, that DSB hotspots are nearly eliminated in these mutants on chromosome 3, on which the largest reductions in recombination are observed (Table S1; DeVeaux and Smith, 1994; Ellermeier and Smith, 2005; Davis et al., 2008; Estreicher et al., 2012).
We infer that the residual recombination in these four mutants reflects mostly non-hotspot DSBs with some contribution from residual DSBs at hotspots (see below). The rec10-109 missense mutant protein may have diminished ability to bind Rec25, Rec27, or Mug20 (or their complex) but retained the ability to activate Rec12 for DSB formation in DSB-cold regions; this hypothesized feature would account for the rec10-109 phenotype being similar to that of rec25Δ, rec27Δ, and mug20Δ mutants.
Rare DSB Hotspots Partially Independent of Cohesins and LinE Proteins
In null mutants lacking any one of these proteins other than Rec10, we discovered that DSBs still occur at some hotspots, roughly 10% of the total, although the frequency of DSBs at these sites is reduced (Figure 3; Ellermeier and Smith, 2005; Davis et al., 2008). rec10Δ lacks essentially all DSBs (Figures 3 and S5). Therefore, Rec10 can activate Rec12 at these few hotspots without the other proteins. What distinguishes these hotspots from the majority remains unknown, but it may reflect the meiotic transcription pattern: many of the residual hotspots are next to genes with large meiosis-specific 5′ UTRs (unpublished data). Alternatively, Rec10 may bind these sites in a manner that allows DSB formation at reduced level without cohesins, Rec25, Rec27, or Mug20.
Relation to Higher Order Chromatin Structure and Meiotic Recombination in Other Species
Our data contrast sharply with related observations in S. cerevisiae, the only other species in which meiotic DSBs have been directly determined genome-wide. To our knowledge, no other proteins have been shown to define DSB hotspots genome-wide with the high enrichment shown by Rec25, Rec27, and Mug20. Other S. cerevisiae proteins, notably including Rec8, preferentially bind to DSB-cold regions, although the degree of anti-correlation of Rec8 binding and DSBs is much less [R2 = 0.068 (Glynn et al., 2004), 0.036 (Panizza et al., 2011), or 0.14 (Pan et al., 2011)] than the degree of correlation of Rec27 binding and DSBs (R2 = 0.59; Figure 4). The binding of other S. cerevisiae axial element proteins also weakly anti-correlates with DSBs: for Red1 and Hop1 R2 = 0.068 and 0.04, respectively [our analysis of the protein data of Panizza et al. (2011) and the DSB data of Bühler et al. (2007)]. Thus, although the binding of Rec27 (positively) accounts for about 60% of the DSB distribution, the binding of S. cerevisiae Rec8, Red1, and Hop1 (negatively) accounts for only about 10 % of the DSB distribution.
In S. cerevisiae the anti-correlation of DSBs and binding of Scc1, the mitotic paralog of Rec8, and Red1 led Blat et al. (2002) to propose that DSBs form in chromatin loops but are repaired when that site is on the axis, since plant and animal DSB-repair protein Rad51 foci are on the axis. Panizza et al. (2011) and Miyoshi et al. (2012) observed that Spo11 (Rec12) partner proteins bind to the axis and interpreted their data in the same framework – DSB sites in the loops are brought to the axis before DSB formation and subsequent repair. Our data provide a different paradigm: the DSB-activating LinE (axial) proteins Rec25, Rec27, and Mug20 bind directly at or near the sites where DSBs are later made, perhaps by activating Rec12 bound nearby. That meiotic chromosome dynamics are different in these two yeasts is also illustrated by the microscopic lines (axial elements) of Rec8 in S. cerevisiae but dozens of dots of Rec8 in S. pombe (e.g., Ding et al., 2006; Davis et al., 2008). Thus, the mechanism by which Rec8, for example, promotes DSB formation apparently differs in the two yeasts.
Role of Hotspot-binding Proteins in Crossover Control
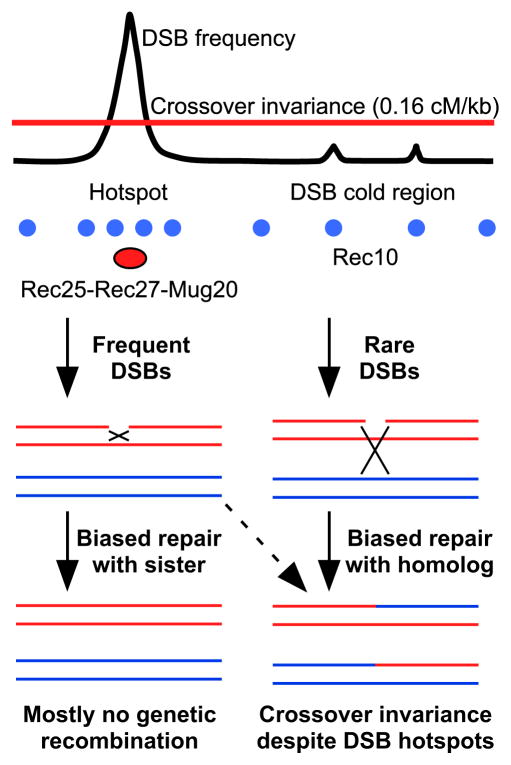
Crossover formation is carefully controlled, presumably to ensure proper homolog segregation at the first meiotic division (Figure 1), and numerous aspects of crossover control have been described (Phadnis et al., 2011). In S. pombe crossovers are much more evenly distributed across the genome than are DSBs, a feature called crossover invariance (Figure 7; Hyppa and Smith, 2010). At hotspots DSBs are repaired predominantly with the sister chromatid, which cannot yield a crossover, whereas in cold regions DSBs appear to be repaired primarily or exclusively with the homolog and yield a crossover in about 80% of repair events (Cromie et al., 2005; Hyppa and Smith, 2010). The mechanism of this partner choice for DSB repair presumably reflects some feature of the chromosome before DSB formation, since otherwise we suppose that, once formed, a DSB at one site is like a DSB at any other site.
Figure 7. Proposal for Crossover Control by Rec25-Rec27-Mug20.
Rec10 (blue balls) binds across chromosomes, enriched at DSB hotspots. Rec25-Rec27-Mug20 complex (red oval) binds hotspots and activates Rec12 to make high-frequency DSBs and biases DSB repair toward the sister, giving a low crossover:DSB ratio. In DSB cold regions repair is biased toward the homolog, giving a high crossover:DSB ratio. The result is a nearly uniform distribution of crossovers across the genome (crossover invariance; Hyppa and Smith, 2010).
Partner choice may reflect the presence of Rec25, Rec27, and Mug20 almost exclusively at DSB hotspots (Figure 3A), effectively establishing domains of differential repair, since no other chromatin-associated pre-DSB proteins are known to distinguish the majority of hotspots from non-hotspots. The following observations support this hypothesis. In rec8Δ mutants residual DSB frequencies are proportional to the enrichment of bound Rec27 (Figure 5), suggesting that these DSBs are Rec27-dependent, but residual recombination is not Rec25-dependent [rec25Δ and rec27Δ single and double mutants are indistinguishable, suggesting that Rec25 and Rec27 act together (Davis et al., 2008)]. Thus, the Rec27-dependent DSBs in rec8Δ mutants apparently do not give rise to crossovers, perhaps because they are repaired by interaction with the sister chromatid (Hyppa and Smith, 2010). Dmc1 strand exchange protein is not required for DSB repair at strong hotspots and plays a larger role in recombination in DSB-cold regions than at DSB hotspots (Hyppa and Smith, 2010). Rec25, Rec27, and Mug20 may therefore prevent Dmc1 from acting at hotspots. Rad51, a paralog of Dmc1, acts both in DSB-cold regions and at hotspots (Hyppa and Smith, 2010) and, in this view, is immune to inhibition by Rec25, Rec27, and Mug20. Perhaps Dmc1 has an intrinsic preference for repair with the homolog when it can act. Regardless of these considerations, the requirement for Rec25, Rec27, and Mug20 for DSB formation at hotspots and for most crossovers implicates these proteins in crossover control by determining both the spatial position and the break frequency of DSB hotspots and perhaps their mode of repair as well.
EXPERIMENTAL PROCEDURES
S. pombe strains and culture conditions
S. pombe strains, genotypes, and sources of alleles are listed in Table S3. Diploid pat1-114 strains were thermally induced for meiosis and analyzed for DNA content by flow cytometry as described by Cervantes et al. (2000). Meiotic crosses were conducted and analyzed as described by Young et al. (2002).
Fluorescence microscopy
Diploid pat1-114 cells induced for meiosis were examined for a protein fused at its C-terminus to GFP as described by Davis et al. (2008). Cells were fixed sequentially with 100 % methanol and 100% acetone at the indicated time after induction of meiosis. Signals were similar to those in unfixed cells, except for Mug20-GFP, which gave more intense but less clearly defined structures in live cells. Details of these methods and those for nuclear spreads are in Supplemental Information.
Genome-wide DSB frequency and protein localization
DSB frequencies across the genome were determined by hybridization of DNA PCR-amplified from Rec12-DNA covalent linkages in diploid pat1-114 rad50S cells at 5 hr after being induced for meiosis (Cromie et al., 2007). Proteins, as GFP- or FLAG-fusions, were assayed across the genome by crosslinking proteins and DNA with formaldehyde 3.5 hr after meiotic induction of pat1-114 cells. Details are in Supplemental Information.
Supplementary Material
Highlights.
DSB hotspots for meiotic recombination are specified by largely unknown mechanisms
Linear element proteins, required for most DSBs, are highly enriched at hotspots
These proteins, first known global hotspot determinants, bind without DSB formation
Hotspot determination by these proteins provides a new paradigm for crossover control
Acknowledgments
We are especially grateful to Anna Estreicher and Josef Loidl for strains with mug20 mutations and unpublished information. We thank Emily Higuchi and Nishka Mittal for data in Table S1; Randy Hyppa and Naina Phadnis for strains; Luisa Bustamante-Jaramillo, Daniel Gómez-Sánchez, Jeff Delrow and Gareth Cromie for assistance; Monica Colaiácovo for fruitful discussions; and Sue Amundsen, Michael Lichten, Naina Phadnis, Walt Steiner, and Sarah Zanders for helpful comments on the manuscript. Supported by NIH grants GM031693 and GM032194 to GRS and grant FEDER-BFU2010-14954 from the Spanish Ministry of Science and Innovation to CMC. SG-V was supported by the JAE-Tech CSIC program. The Instituto de Biología Funcional y Genómica acknowledges support from “Ramón Areces Foundation.”
Footnotes
ACCESSION NUMBERS
Tiling data in this study have been deposited in the NCBI Gene Expression Omnibus (GSE43122) and are available in graphic form at http://labs.fhcrc.org/gsmith/Supplemental_Data/Fowler_MolCell__2013_Supplemental_Figures_2013.pdf.
Supplemental Information includes Extended Experimental Procedures, a supplemental calculation, 9 figures, and three tables, and can be found with this article online at ….
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani MA, Boateng KA, McLeod D, Camerini-Otero RD. The expression profile of the major mouse SPO11 isoforms indicates that SPO11β introduces double strand breaks and suggests that SPO11α has an additional role in prophase in both spermatocytes and oocytes. Mol Cell Biol. 2010;30:4391–4403. doi: 10.1128/MCB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Bonfils S, Rozalen AE, Smith GR, Moreno S, Martin-Castellanos C. Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J Cell Sci. 2011;124:1328–1338. doi: 10.1242/jcs.079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Wu TC, Lichten M. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HE, Farah JA, Grewal SHIS, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Rozalén AE, Moreno S, Smith GR, Martin-Castellanos C. Rec25 and Rec27, novel components of meiotic linear elements, link cohesin to DNA breakage and recombination in fission yeast. Curr Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Soriano I, Marin L, Serrano R, Quintales L, Antequera F. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 2011;31:124–137. doi: 10.1038/emboj.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M, Lee JH, Zhu L, Poon SY, Li J, Cho KH, Chu Z, Karuturi RK, Liu J. Genomic binding profiling of the fission yeast stress-activated MAPK Sty1 and the bZIP transcriptional activator Atf1 in response to H2O2. PLoS One. 2010;5:e11620. doi: 10.1371/journal.pone.0011620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estreicher A, Lorenz A, Loidl J. Mug20, a novel protein associated with linear elements in fission yeast meiosis. Curr Genet. 2012;58:119–127. doi: 10.1007/s00294-012-0369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Mizuno K, Shibata T, Ohta K. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol Biol Cell. 2008;19:1162–1173. doi: 10.1091/mbc.E07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Cromie GA, Smith GR. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 2008;4:e1000267. doi: 10.1371/journal.pgen.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Egel R, Lankenau D-H, editors. Recombination and meiosis: crossing-over and disjunction. Berlin: Springer-Verlag; 2007. pp. 81–123. [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:13756–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada T, Ito M, Sasanuma H, Katou Y, Itoh T, Matsumoto K, Shibata T, Shirahige K, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell. 2009;13:3064–3076. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lab Websites. "http://labs.fhcrc.org/gsmith/Supplemental_Data/Fowler_MolCell__2013_Supplemental_Figures_2013.pdf".
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova FE, Eisenhaber F, McFarlane RJ, Loidl J. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci. 2004;117:3343–3351. doi: 10.1242/jcs.01203. [DOI] [PubMed] [Google Scholar]
- Ludin K, Mata J, Watt S, Lehmann E, Bahler J, Kohli J. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma. 2008;117:431–444. doi: 10.1007/s00412-008-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol. 2005;22:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Gerton JL, Lieb JD, Petes TD. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1014–1027. doi: 10.1128/MCB.26.3.1014-1027.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, Oda A, Yamada T, Hirota K, Masai H, Ohta K. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell. 2012;47:722–733. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccaromyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci. 2003;116:1719–1731. doi: 10.1242/jcs.00387. [DOI] [PubMed] [Google Scholar]
- Munoz-Fuentes V, Di Rienzo A, Vila C. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. 2011;6:e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Hyppa RW, Smith GR. New and old ways to control meiotic recombination. Trends Genet. 2011;27:411–421. doi: 10.1016/j.tig.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc Natl Acad Sci USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif WD, Glick GC, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1–14. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Meth. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirek M, Estreicher A, Csaszar E, Wells JL, McFarlane RJ, Watts FZ, Loidl J. SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe. Chromosoma. 2010;119:59–72. doi: 10.1007/s00412-009-0241-5. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Davidow PA, Bagshaw AT. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics. 2011;187:385–396. doi: 10.1534/genetics.110.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol Cell Biol. 2005;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield SE, Keeney S. Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle. 2012;11:1496–1503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Davidson MK. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Siegel ER, Davidson MK. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLoS One. 2008;3:e2887. doi: 10.1371/journal.pone.0002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.