Abstract
Glutathione (GSH), the major intracellular antioxidant, protects against cancer development by detoxifying carcinogens and free radicals and strengthening the immune system. Recently, a GAG-trinucleotide repeat polymorphism in the 5’-untraslated region of the gene for the rate limiting enzyme for GSH biosynthesis, γ-glutamine cysteine ligase (GCL), was shown to be associated with lowered GCL activity and GSH levels in vitro and in vivo. We tested the hypothesis that this functional polymorphism in GCL is associated with the risk for lung and aerodigestive tract cancers. To this end, we conducted a case-control study that included 375 lung cancer cases, 200 aerodigestive tract cancer cases and 537 controls. GAG repeat genotype (4, 7, 8, 9 and 10 repeat alleles) was determined by capillary electrophoresis of PCR products from the repeat region of the GCL catalytic subunit (GCLC). Odds ratios were calculated by logistic regression and adjusted for risk factors including age, sex, body mass index and smoking history. The GAG-7/7 genotype was associated with a 1.9-fold increased risk of lung cancer and 2.6-fold increased risk of aerodigestive tract cancer compared to the wild type GAG-9/9 (p<0.05). Similarly, the GAG-7 allele was associated with an increased risk of lung cancer (OR=1.5, p=0.01) and aerodigestive tract cancer (OR=2.3, p<0.001) compared to subjects without GAG-7 allele. These findings suggest that glutathione synthesis affects the risk of lung and aerodigestive tract cancers, and further implicates a role for oxidative stress in the development of these cancers.
Keywords: γ-Glutamylcysteine ligase, Glutathione, GAG-repeat polymorphism, Lung cancer, Aerodigestive tract cancer
Introduction
Oxidative stress is thought to play a significant role in carcinogenesis, however, specific mechanisms remain poorly understood [1,2]. Glutathione (GSH) is the most abundant antioxidant in cells and tissues, and plays a primary role in protection against oxidative stress. GSH is also an important substrate for the conjugation and detoxification of known carcinogens such as the polycyclic aromatic hydrocarbons (PAHs) present in tobacco smoke [3,4]. In addition, GSH may protect against neoplasia through its role in the maintenance of immune function, as it regulates mitogenic response and lymphocyte proliferation [5]. Low levels of blood GSH have been associated with chronic diseases including cancer [6,7] and GSH administration has been shown to inhibit tumor initiation, promotion and progression in animal models [8]. While there has been a growing interest in the role of GSH and cancer inhibition, there have been few clinical studies of factors that regulate GSH levels or its biosynthesis.
All cells in the body synthesize GSH in a two-step ATP requiring process catalyzed by γ-glutamyl cysteine ligase (GCL) and GSH synthetase. GCL, the rate limiting enzyme, is a heterodimer of catalytic (GCLC) and modifier (GCLM) subunits. While several genetic polymorphisms in both GCLC and GCLM, have been identified [9–11], a GAG-trinucleotide repeat polymorphism in the 5’-untranslated region (UTR) of GCLC (rs3830798) may be of particular importance as it has been linked to altered GCL activity and GSH levels [12,13]. For this polymorphism, five different alleles have been identified (GAG-4, GAG-7, GAG-8, GAG-9 and GAG-10 containing 4, 7, 8, 9, and 10 GAG repeats, respectively). The GAG polymorphism has been associated with GSH levels in vitro and in vivo [12,14]. Luciferase reporter studies on the functional mechanism of the GAG repeat polymorphism show that the repeats affect gene expression through translation and that GCLC 5’-UTRs with 7 GAG repeats have lower luciferase activity compared to those with 8 or 9 repeats [13,15]. Individuals having the GAG-9/9 genotype had lower GSH levels and GCL activity than those having GAG-7/9 and GAG-7/7 genotypes. This polymorphism has also been associated with the risk for several diseases including schizophrenia [16], chronic beryllium disease [17] and diabetes [18]. Based upon the many important roles for GSH in protection against carcinogenesis, particularly for tobacco related cancers, we hypothesized that the GAG repeat polymorphism is associated with increased risk for tobacco and or oxidative stress related cancers. We tested this hypothesis by studying GAG repeat polymorphisms in a case control study of lung and aerodigestive tract cancers.
Methods
Recruitment of Study Subjects
We conducted a case-control study of lung and aerodigestive tract cancers at the H. Lee Moffitt Cancer Center (Tampa, Florida, USA) from 1999–2003. Results for other genetic variants from the lung cancer component have been previously described [19–21]. The current analysis was limited to white study subjects. Informed consent was obtained from all subjects as per the guidelines of the institutional review board. Cases were patients recruited within one year after diagnosis with histologically confirmed cancers of the lung and aerodigestive tract (gingiva, hard-palate, dorsal tongue, floor of mouth, inner lip, soft-palate, buccal mucosa, tongue, tonsil, oropharynx, larynx, and esophagus). Controls were randomly selected from the Moffitt Lifetime Cancer Screening Center. The Center screens healthy individuals for prostate-specific antigen testing, skin examinations, endoscopy, or mammography. A trained interviewer used a structured questionnaire to collect information on demographics, medical history, smoking and alcohol drinking habits. DNA was available for 375 lung cancer cases, 200 aerodigestive tract cancer cases and 537 controls.
Determination of GAG Genotype
Genomic DNA was extracted from exfoliated buccal mucosal cells collected from all the subjects as described previously [14]. Briefly, buccal mucosal cells were obtained by having the subjects wash their mouth with distilled water, brush their cheeks and gums with a soft tooth brush, and rinsing with 20 ml of saline. The rinse was centrifuged at 3000 × g for 10 min and genomic DNA was extracted from the cell pellet using phenol:chloroform:isoamylalcohol (25:24:1). GAG genotype was determined by PCR amplification of the GAG repeat region from genomic DNA followed by capillary electrophoresis. DNA containing the GAG repeat region was amplified in 96-well PCR plates using Qiagen multiplex PCR kits (Qiagen, Valencia, CA) in a reaction volume of 10 μL.
The reaction mixture consisted of 5 μL of 2x Qiagen mastermix, 0.25 μL each of 20 μM 6FAM-labeled forward (51-6FAM-CGGCTGAGTGTCCGTCTCGC-31) and unlabeled reverse primers (51-CCACTTGAGAACGTCCTTGTGCCGG-31; Applied Biosystems, Foster City, CA), 500 ng of genomic DNA, and distilled water to a final volume of 10 μL. Thermocycling parameters were as follows: initial activation at 95°C for 15 min followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 63°C for 45 sec and extension at 72°C for 30 sec; and a final extension at 72°C for 10 min. Amplification of the PCR product was confirmed by running 10% of randomly chosen samples per 96-well plate in 2% agarose gels.
The number of GAG repeats in the 5’ UTR region was derived based on the number of bp in the PCR products as determined by capillary electrophoresis of fluorescent PCR products in an ABI 3130 xl genetic analyzer (ABI, Foster City, CA). Depending on the number of GAG repeats, the length of the PCR products ranged from 211 bp (for 4 GAG repeats) to 229 bp (for 10 GAG repeats). Amplified PCR products (1 μl) were denatured by addition of 8.5 μl of deionized formamide. DNA ladder (0.5 μl, GeneScan 500 LIZ containing DNA fragments of various lengths ranging from 35 nucleotides to 500 nucleotides) was added and samples were incubated at 95°C for 2 min and immediately cooled on ice. Denatured products were subjected to capillary electrophoresis and the number of nucleotides (and therefore, the number of GAG repeats) was determined by analyzing electropherograms with Genemapper software (ABI, Foster city, CA). Genotypes from DNA of 20 individuals determined by this method were confirmed by alternate methods of direct sequencing and running P32 PCR products in 10% polyacrylamide gels as described previously [14] and found to be 100% concordant.
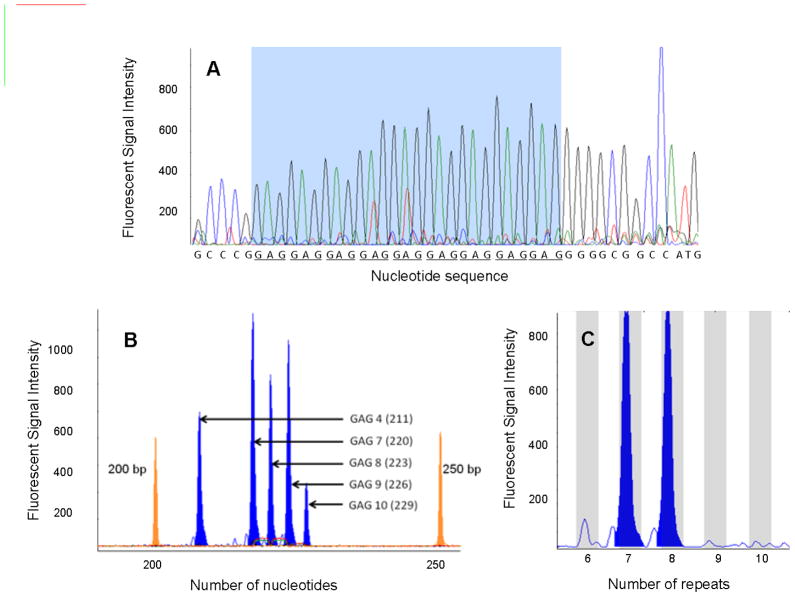
Several quality control measures were followed during the genotyping procedure. The genotypes of several actual samples as obtained by capillary electrophoresis were confirmed by direct sequencing (Figure 1A). An allelic ladder was constructed by pooling genomic DNA from several individuals with different genotypes (determined by direct sequencing) representing each of the five observed alleles (GAG-4, GAG-7, GAG-8, GAG-9 and GAG-10) and run as an assay control during capillary electrophoresis (Figure 1B). The number of GAG repeats for unknown samples were determined based on the elution times of DNA fragments from the Liz 500 internal DNA ladder (Figures 1B and 1C).
Figure 1. Determination of GAG repeat genotypes by capillary electrophoresis.
A) Several samples were selected at random and the resulting PCR products were sequenced to determine the number of GAG-repeats (representative sample result shown). The GAG repeats are underscored. B) Electropherogram of allelic ladder, made by pooling samples with different genotypes (determined by DNA sequencing) to represent all the five GAG alleles. This ladder was also was run on each plate as a control. The two peaks at 200bp and 250bp are the DNA fragments from LIZ 500 internal ladder. c) A representative electropherogram of a PCR product from a single individual shows alleles GAG-7 and GAG-8 (LIZ 500 not shown).
Statistical Analysis
Data were analyzed using SAS 9.2 (SAS Cary, NC). Hardy-Weinberg equilibrium (HWE) between the major allele and major genotype frequencies was calculated for each type of cancer and controls using the chi-square test for goodness of fit. Odds ratios for lung and aerodigestive tract cancers were calculated by unconditional logistic regression using the entire control series as the comparison group for both cancer sites. For lung cancer, unadjusted odds ratios and odds ratios adjusted for age, sex, body mass index and smoking status (former smokers, current smokers and never smokers) were calculated. For aerodigestive tract cancer, odds ratios were adjusted for age, sex, body mass index, smoking status (former smokers, current smokers and never smokers) and alcohol drinking. Odds ratios were calculated for men and women together and separately as well. Effect modification of smoking and histology for lung cancer and smoking, alcohol consumption, and site for aerodigestive tract cancer was assessed by including multiplicative interaction terms in the regression models. All tests were two-sided and considered significant if p≤0.05. Subgroup analysis for lung cancer was performed by histologic type of lung cancer, and by site of aerodigestive tract cancer.
Genetic data was analyzed using two different approaches. Genotype modeling was conducted by comparing the genotypes of the 7, 8, and 9 alleles, using the GAG-9/9 genotype as the referent group. Since the prevalence of GAG-4 and GAG-10 alleles were less than 1%, meaningful odds ratios could not be calculated for these alleles. Allelic association models were used where all GAG genotypes were separated into groups based on the presence or absence of a single allele. For example, there were two possible genotype groups for GAG-7, one where at least one of the two alleles is a GAG-7 and the other where neither of the alleles is a GAG-7. Similarly two groups were considered for both alleles GAG-8 and GAG-9. This 2nd genetic model was considered to determine if any of the three major alleles were individually associated with cancer risk.
Results
Demographics and study subject characteristics are shown in Table 1. For cancer cases and controls, the mean ages were between 56 and 65 y. The majority of cases of lung and aerodigestive tract cancer were men (60–75%). As expected, the percentage of never smokers among lung cancer cases (11%) and aerodigestive tract cancer cases (24%) were lower than among controls (40%) (P<0.0001) and lifetime tobacco smoke exposure expressed in pack years was ~2-fold greater in cases than in controls (P<0.0001). Comorbidities significantly associated with cancer cases included chronic bronchitis (lung), emphysema (lung), heart disease (lung and aerodigestive tract), diabetes (lung and aerodigestive tract) and osteoarthritis (aerodigestive tract). For lung cancers, adenocarcinoma was the most common histologic type (37%), followed by squamous cell carcinoma (21%). For aerodigestive tract cancers, squamous cell carcinoma was the major histologic type (87%). The most common site for aerodigestive tract cancer was the tongue (24%), followed by the larynx (21%), pharynx (16%), esophagus (13%) and floor of mouth (6%).
Table 1.
Characteristics of study subjects
| Category | Lung Cancer | Aerodigestive tract Cancer | Controls | P-value Lung Cancer vs. Control | P-value Aerodigestive Tract Cancer vs. Control |
|---|---|---|---|---|---|
| Number of Subjects, n | 375 | 200 | 537 | ||
| Age (y) | <0.0001* | 0.57* | |||
| Mean ± SD | 64.3 ± 9.8 | 58.8 ± 11.4 | 58.3 ± 10.4 | ||
| Range | 38–85 | 25–85 | 28–83 | ||
| Sex, n (%) | |||||
| Male | 225 (60) | 151 (75) | 300 (56) | ||
| Female | 150 (40) | 49 (24) | 237 (44) | ||
| BMI (mean ± SD) | |||||
| Male | 27.6 (4.58) | 27.8 (5.34) | 27.9 (4.44) | 0.77* | 0.69* |
| Female | 25.7 (5.22) | 25.9 (6.60) | 26.8 (5.33) | 0.97* | 0.52* |
| Smoking Status, n (%) | <0.0001† | <0.0001† | |||
| Never | 43(11) | 49 (24) | 219 (40) | ||
| Former | 192 (51) | 70 (35) | 226 (42) | ||
| Current | 140 (37) | 80 (40) | 92 (17) | ||
| Pack Yrs, mean (SD) | 53 (38) | 47 (41) | 23(31) | <0.0001* | <0.0001* |
| Comorbidities, n (%) | |||||
| Asthma | 18 (5) | 20 (10) | 34 (6) | 0.33† | 0.09† |
| Chronic Bronchitis | 33 (9) | 8 (4) | 26 (5) | 0.017† | 0.63† |
| Diabetes | 41 (11) | 23 (12) | 32 (6) | 0.006† | 0.011† |
| Emphysema | 45 (12) | 12 (6) | 24 (4) | <0.0001† | 0.39† |
| Heart disease | 76 (20) | 27 (14) | 24 (4) | <0.0001† | <0.0001† |
| Rheumatoid arthritis | 12 (3) | 10 (5) | 20 (4) | 0.67† | 0.44† |
| Thyroid disease | 26 (7) | 14 (7) | 51 (9) | 0.17† | 0.29† |
| Asbestosis | 5 (1) | 1 (0.5) | 4 (0.7) | 0.38† | 0.72† |
| Epilepsy | 4 (1) | 1 (0.5) | 5 (0.9) | 0.84† | 0.56† |
| High BP | 128 (34) | 62 (31) | 157 (29) | 0.12† | 0.64† |
| Osteoarthritis | 35 (9) | 10 (5) | 66 (12) | 0.16† | 0.004† |
| Stroke | 19 (5) | 12 (6) | 16 (3) | 0.11† | 0.056† |
| Histology, n (%) | |||||
| Squamous cell carcinoma | 82 (21) | 126 (87) | NA | ||
| Small cell carcinoma | 34 (9) | 0 | NA | ||
| Adenocarcinoma | 134 (37) | 18 (12) | NA | ||
| Large cell carcinoma | 24 (6) | 0 | NA | ||
| Non-smallcell carcinoma | 60 (16) | 0 | NA | ||
| Others | 13 (3) | 0 | NA | ||
| Site, n (%) | |||||
| Esophagus | NA | 24 (12) | NA | ||
| Tongue | NA | 48 (24) | NA | ||
| Floor of the mouth | NA | 12 (6) | NA | ||
| Pharynx | NA | 31 (16) | NA | ||
| Larynx | NA | 42 (21) | NA | ||
| Other | NA | 42 (21) | NA | ||
t-Text
χ2-Test
A representative electropherogram obtained from capillary electrophoresis for determining GAG repeat genotype is shown in Figure 1C. A total of five alleles GAG-4, GAG-7, GAG-8, GAG-9, and GAG-10 were identified. Genotyping was repeated for 5% of the samples from the study and the repeated genotypes were 100% concordant. In both cases and controls, GAG-7 was the most frequent allele (control, 58%; lung cancer, 61%; aerodigestive tract cancer, 62%), followed by GAG-9 (control, 27%; lung cancer, 24%; aerodigestive tract cancer, 22%) and GAG-8 (control, 13%; lung cancer, 12%; aerodigestive tract cancer, 15%). GAG-4 and GAG-10 occurred less frequently in both cases and controls (0–0.6% for GAG-4 and 0–2.3% for GAG-10). These GAG allele frequencies are similar to those observed in previous studies [12,14,22]. When genotypes were examined, GAG-7/7 occurred most frequently genotype, followed by GAG-7/9, GAG-7/8, GAG-8/9, GAG-9/9 and GAG-8/8 in both cases and controls (Table 2). Allele and genotype frequencies were in HWE for cancer cases (p>0.05) but not controls (p=0.01).
Table 2.
Distribution of GAG repeat genotypes and lung and aerodigestive tract cancer risk
| GAG Genotype | Controls n (%) | Lung Cancer Cases* | Aerodigestive Tract Cancer Cases† | ||
|---|---|---|---|---|---|
|
| |||||
| n (%) | OR (95% CI) | n (%) | OR (95% CI) | ||
| GAG-9/9 | 52 (9.6) | 20 (5.7) | 1.00 (reference) | 9 (4.5) | 1.00 (reference) |
| GAG-7/7 | 190 (35.3) | 133 (38.3) | 1.87 (1.00–3.51) | 79 (39.5) | 2.56 (1.16–5.63) |
| GAG-7/8 | 79 (14.7) | 45 (12.9) | 1.68 (0.83–3.42) | 35 (17. 5) | 2.60 (1.10–6.12) |
| GAG-7/9 | 155 (28.8) | 102 (29.3) | 1.67 (0.88–3.16) | 55 (27.5) | 1.94 (0.87–4.35) |
| GAG-8/8 | 15 (2.7) | 6 (1.7) | 1.16 (0.36–3.73) | 5 (2.5) | 1.83 (0.50–6.68) |
| GAG-8/9 | 32 (5.9) | 23 (6.6) | 2.23 (0.96–5.16) | 16 (8.0) | 3.09 (1.16–8.22) |
| GAG-4/4 | 0 | 2 (0.5) | ‡ | 0 | ‡ |
| GAG-7/10 | 9 (1.6) | 12 (3.4) | ‡ | 0 | ‡ |
| GAG-8/10 | 1 (0.1) | 1 (0.3) | ‡ | 0 | ‡ |
| GAG-9/10 | 2 (0.3) | 3 (0.8) | ‡ | 0 | ‡ |
| GAG-4/9 | 0 | 0 | ‡ | 1(0.5) | ‡ |
| GAG-4/7 | 0 | 0 | ‡ | 0 | ‡ |
| GAG-10/10 | 2(0.3) | 0 | ‡ | 0 | ‡ |
OR adjusted for age, sex, body mass index and smoking history (categorical).
OR adjusted for age, sex, body mass index, smoking history (categorical) and the amount of alcohol intake.
Minor genotypes not included in statistical model.
The results of multivariate logistic regression models and GAG genotypes are shown in Table 2. In initial analysis, unadjusted ORs were significant for GAG-7/7 (OR=1.80, 95% CI=1.03–3.16) for lung cancer and for GAG-7/7 (OR=2.40, 95% CI=1.13–5.09), GAG-7/8 (OR= 2.56, 95% CI=1.14–5.76) and GAG-8/9 (OR=2.89, 95% CI=1.14–7.30) for aerodigestive tract cancer. Adjusted odds ratios resulting from multivariate models controlling for age, sex, BMI, and smoking history for lung cancer and age, sex BMI, smoking history and alcohol consumption for aerodigestive tract cancers for the major genotypes were estimated (Table 2). Due to low frequency (<3%), odds ratios could not be obtained for genotypes having alleles GAG-4 and GAG-10. Among the six most frequent genotypes (GAG-7/7, GAG-7/8, GAG-7/9, GAG-8/8, GAG-8/9 and GAG-9/9), the GAG-7/7 genotype was associated with increased risk for both lung cancer (OR=1.87, p=0.04) and aerodigestive tract cancer (OR=2.56, p=0.03) compared to the GAG-9/9 genotype. Higher risk was also observed for aerodigestive tract cancer in individuals with the GAG 7/8 genotype (OR=2.60) and GAG-8/9 (OR=3.09) genotypes (P<0.05). When examined separately by sex, no significant associations with risk were observed except for GAG-7/8 which was associated with lung cancer in women (OR=3.21, 95% CI=1.01–10.22, p=0.04) (data not shown).
The analysis of the association of lung cancer with GAG-7/7 genotype stratified by specific histologic type showed a significant association for adenocarcinoma (OR=2.91, P<0.05) compared to GAG-9/9 (Supplementary Material, Table 1). While no significant associations were observed for other histologic cell types, these analyses were limited due to the relatively low numbers of cases. Likewise, examination of the association between GAG genotype and aerodigestive tract cancer by site did not provide conclusive results due to the relatively low number of cases for each individual site (Supplementary Material, Table 2).
Allele specific effects on risk for cancer were also observed (Table 3). Individuals with at least one GAG-7 allele were at greater risk for lung cancer (OR=1.52, p=0.01) and aerodigestive tract cancer (OR=2.40, p=0.0001) compared to those not having any GAG-7 alleles. No differences were observed when comparing subjects with all genotypes having at least one GAG-8 allele versus subjects without a GAG-8 allele, or when comparing subjects with all genotypes having at least one GAG-9 allele versus subjects without a GAG-9 allele.
Table 3.
Allele specific effects of GAG genotype on risk for lung and aerodigestive tract cancers.
| GAG Allele | GAG Allele | Controls n (%) | Lung Cancer Cases* | Aerodigestive tract Cancer Cases† | ||
|---|---|---|---|---|---|---|
|
| ||||||
| n (%) | OR (95% CI) | n (%) | OR (95% CI) | |||
| GAG-7 | No GAG-7 | 104 (19.4) | 55 (15.8) | 1.00 (reference) | 31 (15.5) | 1.00 (reference) |
| At least one GAG-7 | 433 (80.6) | 292 (84.1) | 1.52 (1.09–2.12) | 169 (84.5) | 2.40 (1.53–3.77) | |
| GAG-8 | No GAG-8 | 410 (76.3) | 272 (78.4) | 1.00 (reference) | 144 (72.0) | 1.00 (reference) |
| At least one GAG-8 | 127(23.6) | 75 (21.6) | 1.07 (0.75–1.54) | 56 (28.0) | 1.44 (0.96–2.16) | |
| GAG-9 | No GAG-9 | 296 (55.1) | 199 (57.3) | 1.00 (reference) | 119 (59.5) | 1.00 (reference) |
| At least one GAG-9 | 241 (44.9) | 148 (42.6) | 0.94 (0.70–1.27) | 81 (40.5) | 1.00 (0.71–1.42) | |
OR adjusted for age, sex, body mass index and smoking history (categorical).
OR adjusted for age, sex, body mass index, smoking history (categorical) and the amount of alcohol intake.
The association of GAG repeat genotype with risk for lung and aerodigestive tract cancers stratified by smoking status was examined (Table 4). We observed evidence of effect modification of smoking status on the associations between GAG genotype and both lung cancer and aerodigestive tract cancer (Pinteraction<0.05). For lung cancer significant associations were observed for GAG-7/7 (OR=2.53, P=0.01), GAG-7/8 (OR=2.20, P=0.05), GAG-7/9 (OR=2.12, P=0.04) and GAG-8/9 (OR=2.81, P=0.04) among ever-smokers but not in never-smokers. For aerodigestive tract cancers, significant associations were observed for GAG-7/7 (OR=3.72, P=0.01), GAG-7/8 (OR=3.82, P=0.02), GAG-7/9 (OR=2.92, P=0.04) and GAG-8/9 (OR=5.71, P=0.01) among ever-smokers but not in never-smokers.
Table 4.
Distribution of GAG repeat genotypes among lung and aerodigestive tract cancer patients by smoking status
| Smoking Status | GAG Genotype | Controls n (%) | Lung Cancer | Aerodigestive Tract Cancer Cases | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Cases n (%) | OR* (95% CI) | n (%) | OR† (95% CI) | |||
| Ever Smokers | GAG-9/9 | 31 (10.0) | 16 (5.4) | 1.00 (reference) | 5 (3.3) | 1.00 (reference) |
| GAG-7/7 | 112 (36.0) | 125 (42.1) | 2.52 (1.25–5.07) | 60 (39.7) | 3.72 (1.32–10.5) | |
| GAG-7/8 | 41 (13.2) | 39 (13.1) | 2.20 (1.00–4.88) | 25 (16.6) | 3.82 (1.26–11.6) | |
| GAG-7/9 | 102 (32.8) | 93 (31.3) | 2.12 (1.04–4.31) | 43 (28.5) | 2.92 (1.02–8.33) | |
| GAG-8/8 | 10 (3.2) | 5 (1.7) | 1.34 (0.37–4.96) | 4 (2.6) | 2.05 (0.43–9.69) | |
| GAG-8/9 | 15 (4.8) | 19 (6.4) | 2.81 (1.07–7.40) | 14 (9.3) | 5.71 (1.62–20.0) | |
| Never Smokers | GAG-9/9 | 21 (9.8) | 4 (12.1) | 1.00 (reference) | 4 (8.2) | 1.00 (reference) |
| GAG-7/7 | 81 (37.7) | 9 (27.2) | 0.55 (0.15–2.05) | 20 (40.8) | 1.56 (0.46–5.31) | |
| GAG-7/8 | 38 (17.7) | 6 (18.2) | 0.66 (0.16–2.76) | 10 (20.4) | 1.69 (0.45–6.40) | |
| GAG-7/9 | 53 (24.6) | 9 (27.3) | 0.75 (1.99–2.85) | 12 (24.5) | 1.23 (0.34–4.45) | |
| GAG-8/8 | 5 (2.3) | 1 (3.0) | 0.71 (0.06–8.38) | 1 (2.0) | 1.08 (0.09–13.0) | |
| GAG-8/9 | 17 (7.9) | 4 (12.1) | 1.09 (0.22–5.34) | 2 (4.1) | 0.85 (0.13–5.55) | |
OR adjusted for age, sex, body mass index and smoking history (categorical).
OR adjusted for age, sex, body mass index, smoking history (categorical) and the amount of alcohol intake.
The association of GAG genotype with aerodigestive tract cancer risk stratified by alcohol consumption was examined (Table 5). We observed evidence of effect modification of alcohol consumption on the associations between GAG genotype and aerodigestive tract cancer (Pinteraction<0.05).
Table 5.
Odds Ratios for aerodigestive tract cancer and GAG genotype by alcohol consumption
| Drinking Status | GAG Genotype
|
||||||
|---|---|---|---|---|---|---|---|
| GAG-9/9 | GAG-7/7 | GAG-7/8 | GAG-7/9 | GAG-8/8 | GAG-8/9 | ||
| Occasional/non drinker (< 1 drink/day) | No. of Cases: | 5 | 20 | 12 | 12 | 1 | 3 |
| OR (95% CI) †: | 1.00 (ref.) | 0.91 (0.27–3.10) | 1.28 (0.34–4.78) | 0.63 (0.17–2.32) | 0.98 (0.08–11.7) | 0.53 (0.08–3.51) | |
| Light drinker (1–2 drinks/day) | No. of Cases: | 3 | 14 | 4 | 13 | 0 | 2 |
| OR (95% CI) †: | 1.00 (ref.) | 2.64 (0.54–12.9) | 1.53 (0.25–9.54) | 2.67 (0.54–13.3) | - | 2.19 (0.26–18.6) | |
| Heavy drinker (>2 drinks/day) | No. of Cases: | 3 | 47 | 19 | 36 | 4 | 12 |
| OR (95% CI) †: | 1.00 (ref.) | 5.79 (1.47–22.8) | 6.39 (1.43–26.4) | 4.03 (1.00–16.1) | 5.22 (0.77–35.5) | 13.1 (2.40–72.2) | |
Abbreviations: 95% CI, 95% confidence interval; OR, odds ratio.
Odds ratios were adjusted for age, sex, body mass index and smoking.
Significant associations for GAG-7/7 (OR=5.79, P=0.006), GAG-7/8 (OR=6.39, P=0.01), GAG-7/9 (OR=4.03, P=0.05) and GAG-8/9 (OR=13.1, P=0.003) were observed for heavy drinkers (>2 drinks/day), but not for occasional/non-drinkers (<1 drink/day) or light drinkers (1–2 drinks/day).
Discussion
Low GSH levels have been implicated in increasing risk for several diseases related to oxidative stress including cancer [7,23]. In this study we investigated if a functional trinucleotide repeat polymorphism in the gene for the rate-limiting enzyme for GSH biosynthesis can affect the risk for the major tobacco related cancers. Results from this study suggest that the GAG repeat polymorphism in the 5’ UTR of GCLC, which is linked to decreased production of GSH, is associated with increased risk for lung and aerodigestive tract cancers. Among the six most frequent GAG genotypes (GAG-7/7, GAG-7/8, GAG-7/9, GAG-8/8, GAG-8/9 and GAG-9/9), individuals with genotype GAG-7/7 were at approximately 2-fold higher risk for lung and aerodigestive tract cancers compared to those with genotype GAG-9/9. Overall, these findings support a critical role for the GSH biosynthetic pathway in the development of cancers which are linked to tobacco smoke. Protection from smoking related cancers by GSH could be due to its ability to fight oxidative stress induced by tobacco smoke, or alternatively due to its role in the detoxification of tobacco smoke derived carcinogens (e.g. polycyclic aromatic hydrocarbons) and toxins.
The association between the GAG-7/7 and GAG-7/8 genotypes and higher risk for lung cancer and aerodigestive tract cancer may be due to a lower capacity for GSH production and possibly lower GSH levels in target tissues (lung and tissues of the aerodigestive tract) compared to those with GAG-9/9 genotype. These results are consistent with findings from previous functional studies where GAG-7 constructs were associated with decreases in protein expression compared to GAG-8 and GAG-9 [13]. While this is the first report of an association between the GAG polymorphism and cancer risk, in a previous study comparing survival rates for lung cancer, patients with the GAG-7/7 genotype had lower 1-year survival rates (40%) than those having any other genotypes (100%) [24]. Findings from previous epidemiological studies for other diseases, such as cystic fibrosis [25] and chronic obstructive pulmonary disease [26], also showed associations of the GAG polymorphism with disease risk. In cystic fibrosis patients, the GAG-7/7 genotype was associated with decreased lung function [25]. Association of the GAG polymorphism with various lung related diseases including lung cancer highlights the importance of GSH levels in protecting the lung tissue. GSH levels are typically high in lung [27] and environmental exposures such as tobacco smoking can cause depletion of lung GSH, thus leading to oxidative damage [28,29].
It is likely that the GAG repeat polymorphism is having a direct effect on GCLC expression and hence GSH levels in target tissues. Previous in vitro studies demonstrated that GAG repeat number affects luciferase expression through translation with GAG-7 being associated with decreased expression compared to GAG-8 or GAG-9 [13]. In addition to the GAG repeat, a C/T single nucleotide polymorphism has been observed in the promoter of GCLC with the mutant T allele having reduced promoter activity compared to the wild type C allele [10]. It has been suggested that the C allele is in linkage disequilibrium with the GAG-8 and GAG-9 [26]. Thus, it is possible that both of these polymorphisms are acting together to impact GSH biosynthesis. In fact, an interaction between these two polymorphisms has been observed in vitro [15]. Given the association of this polymorphism with cancer and other diseases, it might be important to consider this polymorphism in genome wide association studies, which are usually related to SNPs but not trinucleotide repeat polymorphisms.
Given the important role of GSH in protection against oxidative stress, it is likely that the interaction of the GAG polymorphism with other antioxidant genes and environmental exposures may affect risk for oxidative stress related diseases. Indeed, functional polymorphisms have been identified in other critical antioxidant genes including superoxide dismutase, GSH peroxidases, GSH-S-transferases etc [24,30]. The GAG polymorphism is also likely to be affected by environmental factors such as smoking, alcohol consumption, occupational exposures etc. In this study, the effects of the GAG polymorphism on lung cancer risk were most readily apparent in smokers but not in non-smokers for both lung and aerodigestive tract cancers. In addition, the association of GAG genotype with aerodigestive tract cancer was predominantly apparent in heavy drinkers compared to moderate or non-drinkers. A similar effect of smoking was also found on the association of the GAG repeat polymorphism and measures of lung function in healthy adults [26]. Further studies aimed at identifying potential gene/gene and gene/environment interactions will aid in our understanding of the roles of oxidative stress and GSH in cancer development.
Our study has some limitations. We could not calculate odds ratios for rare genotypes (containing GAG-4 and GAG-10 alleles) due to the very low numbers of individuals with these genotypes. However, we have adequate power (0.8) for detecting differences among the major genotypes (GAG-7/7, GAG-7/8, GAG-7/9, GAG-8/8, GAG-8/9 and GAG-9/9). While the genotype frequencies observed in the present study were not found to be different from our previous study [14] it was noted that the allele and genotype frequencies in control subjects were not in HWE (P=0.01). This is not necessarily unexpected because unlike single nucleotide polymorphisms, trinucleotide repeat polymorphisms are often found to be not in HWE due to the pattern of inheritance (DNA polymerase slippage and frequent unequal crossing over) [31]. In one study designed to examine the inheritance patterns of trinucleotide repeats, only 4.6% of polymorphisms were in HWE [32]. Lastly, we recognize that the findings need to be replicated to accept the hypothesis that the GCL variants contribute to cancer risk. Although genetic association findings are often not reproducible, studies indicate that false negatives from underpowered replication studies are the major reasons for lack of consistency rather than false positives [33]. Our sample size is relatively small but significant results were observed suggesting some confidence in the finding. Still, confirmation of these results is needed to determine if the association is valid.
Findings from this study suggest that enhancement of GSH levels may be an important target for chemoprevention. In a case control study on oral cancer, an association between increased dietary intake of GSH from fruits and vegetables and a decreased risk for cancer was observed (OR=0.5) [34]. In another oral cancer case control study, we observed that high GSH levels in blood were associated with decreased cancer risk [35]. Indeed, numerous chemoprevention agents are known to act through induction of GSH levels as well as through up-regulation of GSH utilizing enzymes including GSH-S-transferases and GSH-peroxidases [36–38]. Further development of agents which specifically target GSH biosynthetic and recycling pathways as well as the transport of GSH and its precursors represents an important area for future research. Altogether these findings suggest that variation in GSH biosynthetic capacity is associated with differences in the risk for tobacco-related cancers. Susceptibility to other GSH-related diseases such as COPD, neurodegenerative diseases as well as drug and xenobiotic toxicities might also be affected by the GAG polymorphism.
Supplementary Material
Acknowledgments
Funding agencies: This study was supported in part by Public Health Service grants P01-CA68384 (P.I., Lazarus; Richie, Project Leader) and R01-DE13158 (Lazarus) from the National Institutes of Health.
We thank Dr. Carla Gallagher for her help with genotyping assay. Fragment analyses were conducted in the Molecular Genetics/DNA Sequencing Core at the Penn State University College of Medicine.
Abbreviations
- GSH
Glutathione
- GCL
γ-glutamine cysteine ligase
- GCLC
catalytic subunit of γ-glutamine cysteine ligase
- GCLM
modifier subunit of γ-glutamine cysteine ligase
- UTR
untranslated region
- HWE
Hardy-Weinberg equilibrium
References
- 1.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chemico-biological interactions. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS letters. 1995;358(1):1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 3.Coles B, Ketterer B. The role of glutathione and glutathione transferases in chemical carcinogenesis. Critical reviews in biochemistry and molecular biology. 1990;25(1):47–70. doi: 10.3109/10409239009090605. [DOI] [PubMed] [Google Scholar]
- 4.Ketterer B. Protective role of glutathione and glutathione transferases in mutagenesis and carcinogenesis. Mutation research. 1988;202(2):343–361. doi: 10.1016/0027-5107(88)90197-2. [DOI] [PubMed] [Google Scholar]
- 5.Hamilos DL, Wedner HJ. The role of glutathione in lymphocyte activation. I. Comparison of inhibitory effects of buthionine sulfoximine and 2-cyclohexene-1-one by nuclear size transformation. J Immunol. 1985;135(4):2740–2747. [PubMed] [Google Scholar]
- 6.Lang CA, Mills BJ, Mastropaolo W, Liu MC. Blood glutathione decreases in chronic diseases. The Journal of laboratory and clinical medicine. 2000;135(5):402–405. doi: 10.1067/mlc.2000.105977. [DOI] [PubMed] [Google Scholar]
- 7.Lang CA, Naryshkin S, Schneider DL, Mills BJ, Lindeman RD. Low blood glutathione levels in healthy aging adults. The Journal of laboratory and clinical medicine. 1992;120(5):720–725. [PubMed] [Google Scholar]
- 8.Trickler D, Shklar G, Schwartz J. Inhibition of oral carcinogenesis by glutathione. Nutrition and cancer. 1993;20(2):139–144. doi: 10.1080/01635589309514280. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, Ebbert JO, Sun Z, Weinshilboum RM. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol. 2006;24(11):1761–1769. doi: 10.1200/JCO.2005.02.7110. [DOI] [PubMed] [Google Scholar]
- 10.Koide S, Kugiyama K, Sugiyama S, et al. Association of polymorphism in glutamate-cysteine ligase catalytic subunit gene with coronary vasomotor dysfunction and myocardial infarction. Journal of the American College of Cardiology. 2003;41(4):539–545. doi: 10.1016/s0735-1097(02)02866-8. [DOI] [PubMed] [Google Scholar]
- 11.Custodio HM, Broberg K, Wennberg M, et al. Polymorphisms in glutathione-related genes affect methylmercury retention. Archives of environmental health. 2004;59(11):588–595. doi: 10.1080/00039890409603438. [DOI] [PubMed] [Google Scholar]
- 12.Walsh AC, Feulner JA, Reilly A. Evidence for functionally significant polymorphism of human glutamate cysteine ligase catalytic subunit: association with glutathione levels and drug resistance in the National Cancer Institute tumor cell line panel. Toxicol Sci. 2001;61(2):218–223. doi: 10.1093/toxsci/61.2.218. [DOI] [PubMed] [Google Scholar]
- 13.Nichenametla SN, Lazarus P, Richie JP., Jr A GAG trinucleotide-repeat polymorphism in the gene for glutathione biosynthetic enzyme, GCLC, affects gene expression through translation. Faseb J. 25(7):2180–2187. doi: 10.1096/fj.10-174011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichenametla SN, Ellison I, Calcagnotto A, Lazarus P, Muscat JE, Richie JP., Jr Functional significance of the GAG trinucleotide-repeat polymorphism in the gene for the catalytic subunit of gamma-glutamylcysteine ligase. Free radical biology & medicine. 2008;45(5):645–650. doi: 10.1016/j.freeradbiomed.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butticaz C, Gysin R, Cuenod M, Do KQ. Interaction of GAG trinucleotide repeat and C-129T polymorphisms impairs expression of the glutamate-cysteine ligase catalytic subunit gene. Free radical biology & medicine. 50(5):617–623. doi: 10.1016/j.freeradbiomed.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Gysin R, Kraftsik R, Sandell J, et al. Impaired glutathione synthesis in schizophrenia: convergent genetic and functional evidence. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(42):16621–16626. doi: 10.1073/pnas.0706778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekris LM, Viernes HM, Farin FM, Maier LA, Kavanagh TJ, Takaro TK. Chronic beryllium disease and glutathione biosynthesis genes. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine. 2006;48(6):599–606. doi: 10.1097/01.jom.0000201845.02369.ba. [DOI] [PubMed] [Google Scholar]
- 18.Bekris LM, Shephard C, Janer M, et al. Glutamate cysteine ligase catalytic subunit promoter polymorphisms and associations with type 1 diabetes age-at-onset and GAD65 autoantibody levels. Exp Clin Endocrinol Diabetes. 2007;115(4):221–228. doi: 10.1055/s-2007-970574. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher CJ, Muscat JE, Hicks AN, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(4):823–828. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher CJ, Ahn K, Knipe AL, et al. Association between haplotypes of manganese superoxide dismutase (SOD2), smoking, and lung cancer risk. Free radical biology & medicine. 2009;46(1):20–24. doi: 10.1016/j.freeradbiomed.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones NR, Spratt TE, Berg AS, Muscat JE, Lazarus P, Gallagher CJ. Association studies of excision repair cross-complementation group 1 (ERCC1) haplotypes with lung and head and neck cancer risk in a Caucasian population. Cancer epidemiology. 35(2):175–181. doi: 10.1016/j.canep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willis AS, Freeman ML, Summar SR, et al. Ethnic diversity in a critical gene responsible for glutathione synthesis. Free radical biology & medicine. 2003;34(1):72–76. doi: 10.1016/s0891-5849(02)01178-4. [DOI] [PubMed] [Google Scholar]
- 23.Townsend DM, Tew KD, Tapiero H. The importance of glutathione in human disease. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2003;57(3–4):145–155. doi: 10.1016/s0753-3322(03)00043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang P, Yokomizo A, Tazelaar HD, et al. Genetic determinants of lung cancer short-term survival: the role of glutathione-related genes. Lung cancer (Amsterdam, Netherlands) 2002;35(3):221–229. doi: 10.1016/s0169-5002(01)00426-3. [DOI] [PubMed] [Google Scholar]
- 25.McKone EF, Shao J, Frangolias DD, et al. Variants in the glutamate-cysteine-ligase gene are associated with cystic fibrosis lung disease. American journal of respiratory and critical care medicine. 2006;174(4):415–419. doi: 10.1164/rccm.200508-1281OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siedlinski M, Postma DS, van Diemen CC, Blokstra A, Smit HA, Boezen HM. Lung function loss, smoking, vitamin C intake, and polymorphisms of the glutamate-cysteine ligase genes. American journal of respiratory and critical care medicine. 2008;178(1):13–19. doi: 10.1164/rccm.200711-1749OC. [DOI] [PubMed] [Google Scholar]
- 27.Kleinman WA, Richie JP., Jr Determination of thiols and disulfides using high-performance liquid chromatography with electrochemical detection. Journal of chromatography. 1995;672(1):73–80. doi: 10.1016/0378-4347(94)00194-a. [DOI] [PubMed] [Google Scholar]
- 28.Cantin AM, North SL, Hubbard RC, Crystal RG. Normal alveolar epithelial lining fluid contains high levels of glutathione. J Appl Physiol. 1987;63(1):152–157. doi: 10.1152/jappl.1987.63.1.152. [DOI] [PubMed] [Google Scholar]
- 29.Deneke SM, Lynch BA, Fanburg BL. Transient depletion of lung glutathione by diethylmaleate enhances oxygen toxicity. J Appl Physiol. 1985;58(2):571–574. doi: 10.1152/jappl.1985.58.2.571. [DOI] [PubMed] [Google Scholar]
- 30.Yang P, Bamlet WR, Ebbert JO, Taylor WR, de Andrade M. Glutathione pathway genes and lung cancer risk in young and old populations. Carcinogenesis. 2004;25(10):1935–1944. doi: 10.1093/carcin/bgh203. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein DBSC. Microsatellites: evolution and applications. Oxford: Oxford University Press; 1999. [Google Scholar]
- 32.Calafell F, Shuster A, Speed WC, Kidd JR, Kidd KK. Short tandem repeat polymorphism evolution in humans. Eur J Hum Genet. 1998;6(1):38–49. doi: 10.1038/sj.ejhg.5200151. [DOI] [PubMed] [Google Scholar]
- 33.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature genetics. 2003;33(2):177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 34.Flagg EW, Coates RJ, Jones DP, et al. Dietary glutathione intake and the risk of oral and pharyngeal cancer. American journal of epidemiology. 1994;139(5):453–465. doi: 10.1093/oxfordjournals.aje.a117028. [DOI] [PubMed] [Google Scholar]
- 35.Richie JP, Jr, Kleinman W, Marina P, Abraham P, Wynder EL, Muscat JE. Blood iron, glutathione, and micronutrient levels and the risk of oral cancer. Nutrition and cancer. 2008;60(4):474–482. doi: 10.1080/01635580801956477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wattenberg LW. Inhibition of carcinogenesis by minor dietary constituents. Cancer research. 1992;52(7 Suppl):2085s–2091s. [PubMed] [Google Scholar]
- 37.De Flora S, Bennicelli C, Camoirano A, et al. In vivo effects of N-acetylcysteine on glutathione metabolism and on the biotransformation of carcinogenic and/or mutagenic compounds. Carcinogenesis. 1985;6(12):1735–1745. doi: 10.1093/carcin/6.12.1735. [DOI] [PubMed] [Google Scholar]
- 38.Richie JP, Jr, Kleinman W, Desai DH, et al. The organoselenium compound 1,4-phenylenebis(methylene)selenocyanate inhibits 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced tumorgenesis and enhances glutathione-related antioxidant levels in A/J mouse lung. Chemico-biological interactions. 2006;161(2):93–103. doi: 10.1016/j.cbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.