Abstract
Similar to bacteria, eukaryotic pathogens may also utilize common strategies of pathogenic secretion, as effector proteins from the oomycete Phytophthora infestans and virulence determinants from the human malaria parasite Plasmodium falciparum share a functionally equivalent host-cell targeting motif (RxLR-dEER in P. infestans and RxLxE/D/Q in P. falciparum). Here we summarize recent studies that reveal that the malarial motif may function differently than previously envisioned. Rather, binding the lipid phosphatidylinositol 3-phosphate [PI(3)P] is a critical step in accessing the host for both pathogens, but occurs in different locations. Nanomolar affinity for PI(3)P by these short amino acid motifs, suggests a new mechanism of phosphoinositide binding unexpectedly in secretory locations that has been exploited for virulence by diverse eukaryotic pathogens.
Keywords: malaria, oomycetes, host-targeting, phosphoinositides, secretion, pathogenesis
Eukaryotic pathogens target proteins into plant and human cells
Numerous microbes parasitize the intracellular environment of plants and mammals. To invade and survive in a host cell, prokaryotic and eukaryotic pathogens communicate across several membrane barriers and are able to modulate host endomembrane systems. Consequently, intracellular microbes develop strategies to modulate host membrane trafficking. Phosphoinositides (PIs) are key players in trafficking and signaling processes in eukaryotes that control membrane–cytoskeleton interactions and vesicle trafficking [1]. In particular, for endocytic trafficking, phosphatidylinositol-3-phosphate [PI(3)P] plays a crucial role in conferring identity to endosomes and regulating vesicle fusion on the cytoplasmic face of the membrane [2] and these properties may be exploited by pathogens to establish intracellular infection.
For some bacterial pathogens, manipulation of cytoplasmic PIs and membrane trafficking begins prior to becoming intracellular. Bacteria such as Salmonella, invade epithelial cells that are not phagocytic. They do so by actively injecting effectors into host cells, which induces phagocytosis in non-phagocytic cells [3]. These effectors are delivered through a bacterial secretion apparatus inserted into the host plasma membrane [4]. Once injected, these effectors modulate functions of small GTPases as well as cytoskeleton and membrane lipids in the cytoplasm of the host cell. The Salmonella effector protein SopB is a phosphatase whose activity recruits the host Rab5 and PI(3)P kinase Vps34 to the Salmonella-containing vacuole (SCV) [5]. This induces remodeling in the cytoplasmic actin network underneath the plasma membrane and facilitates bacterial engulfment by host cells (Figure 1, panel A). Subsequently, SopB-induced enrichment of PI(3)P also contributes to the maturation of the SCV. In contrast, intravacuolar Mycobacterium tuberculosis uses a phosphatase SapM (secreted acid phosphatase M) to decrease cytoplasmic PI(3)P levels on the vacuole and thus arrests maturation of the vacuole at the early endosome stage in which M. tuberculosis is able to replicate [6].
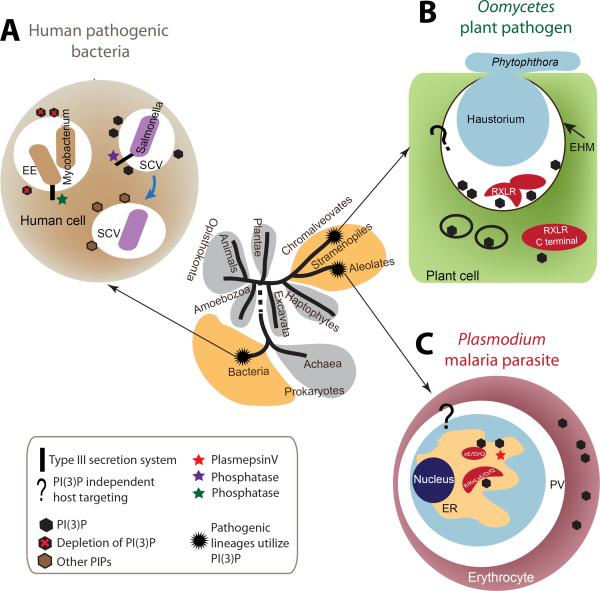
Figure 1. Phylogenetic position, modulation of PI(3)P and host targeting in different intracellular pathogens.
The tree of major life domains shown is based on the analysis of [55]. Pathogenic lineages that utilize PI(3)P for host targeting and host modulation are chromalveolates, alveolates and bacteria. The life domains to which they belong are colored yellow. Host targeting refers to the process by which intracellular pathogens deliver pathogen-produced enzymes, toxins and other proteins directly into host cytoplasm and membrane. Bacterial pathogens use a type III secretion system to inject effectors into host cells. Eukaryotic pathogens utilize endocytic pathways to deliver proteins. (a) Intracellular bacteria use type III effectors to modulate PI(3)P levels for survival and replication within vacuoles inside the host cell cytoplasm. Mycobacterium tuberculosis decreases PI(3)P levels to arrest endosome maturation, avoiding destruction by the host. Salmonella transiently increases PI(3)P levels to facilitate invasion and biogenesis of the vacuoles for intracellular survival. (b) Plant pathogenic oomycetes partly grow into plant cells with a structure called the haustorium. Host targeting utilizes PI(3)P at the extra haustorial membrane surrounding the haustorium to enable RxLR-dEER effector entry. How effectors are delivered across the extra haustorial membrane into the host cell is not known (and thus shown by a question mark), but once in the cytoplasm, their C-terminal domain(s) can bind cytoplasmic PI(3)P to modulate host physiology. (c) The malaria parasite Plasmodium survives in a membranous structure in erythrocytes. Host targeting starts at the parasite ER with binding of PI(3)P by a host targeting motif R/KxLxE/D/Q. Plasmepsin V is an aspartic protease that does not cleave KxL motifs. However it is detected in association with PI(3)P (see Figure 3) cleaves after the RxL to yield xE/D/Q, which does not bind PI(3)P but supports export by unknown mechanisms (depicted by question mark). In oomycetes, PI(3)P-independent host targeting pathways also exist. Vaid et al. [56] report the presence of PI(3)P in the infected erythrocyte. Abbreviations: EE, early endosome; SCV, Salmonella-containing vacuole; EHM, extra haustorial membrane; ER, endoplasmic reticulum; PV, parasitophorous vacuole.
Viruses are not known to utilize cellular PI(3)P, but utilize other PIs. Lipid enveloped viruses that harbor a lipid membrane bilayer derived from their host cell, such as human immunodeficiency virus-1 (HIV-1) and human T-lymphotropic virus-1 (HTLV-1), utilize PI(4,5)P2 and mono- and polyvalent PIs enriched on the inner leaflet of the plasma membrane (PM) including phosphatidylinositol (4)-phosphate [PI(4)P], phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] and phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3], respectively, to assemble and exit the cell [7, 8]. Further, PI(4)P is required for the replication of RNA viruses from the Picornaviridae (i.e., poliovirus, coxsackivirus, Aichi virus, and enterovirus 71) and Flaviviridae (i.e., hepatitis C virus) families [9] and inhibition of host PI 4-kinases may serve as a mechanism of panviral therapy [10].
In contrast, eukaryotic pathogens, such as the oomycete Phytophthorainfestans and the apicomplexan Plasmodium falciparum, appear to utilize PI(3)P at the host plasma membrane and the parasite endoplasmic reticulum (ER) lumen to respectively modulate endocytic and exocytotic trafficking pathways for both secretion and pathogenesis (Figure 1, panel B and C). P. infestans causes potato blight. Infection begins when spores germinate and develop into an infection structure called the appresorium to penetrate plant cells. Subsequently, the pathogen develops a structure called haustorium for nutrient acquisition and host immunity modulation. The haustorium is surrounded by an extra haustorial membrane (EHM) that separates the pathogen from the plant cytoplasm (Figure 2A). When blood stage malaria parasites invade human erythrocytes they too become enclosed within a parasitophorous vacuolar membrane (PVM) that forms during invasion and encloses the intracellular parasite during the asexual life cycle (Figure 2B). In P. falciparum, invasion is completed in minutes but intracellular development in the erythrocyte takes ~ 48 hours. Parasite progeny proliferate within, then rupture out of the PVM and the infected erythrocyte membrane to re-invade new erythrocytes and thus maintain blood stage infection which is responsible for all the symptoms and pathologies associated with malaria.
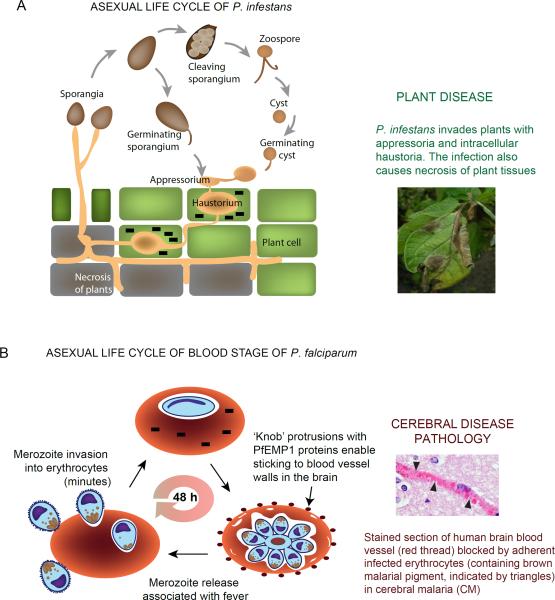
Figure 2. Life cycles and pathologies of Phytophthora and Plasmodium.
(a) P. infestans is able to initiate an infection with both sporangia and zoospores. The pathogen forms an appressorium and penetrates plant leaf cells. Subsequently, a haustorium is developed within host cytoplasm for nutrient acquisition and host immunity modulation. Effectors (black bricks) are trafficked into host cells from the haustoria. Later during infection, P. infestans causes necrosis in plant tissue and sporulates to form new sporangia. The photo of infected potato leaves with sporulating P. infestans is kindly provided by Klaas Bouwmeester and Francine Govers. (b) Asexual, P. falciparum blood stage infection begins when the extracellular merozoite invades the erythrocyte, a process which although complex, is completed in minutes. Effectors (black bricks) are exported across the vacuolar membrane that surrounds the intracellular parasite. Subsequent parasite development, maturation and emergence of merozoites takes ~48 h. In the second half of the asexual cycle, exported parasite effectors assemble in electron dense `knobs' that display the cell surface parasite protein PfEMP1. PfEMP1s are endothelial adhesins that enable infected erythrocytes to bind to the walls of blood vessels. In the brain this can lead to inflammation as well as occlusion of blood vessels (as seen in the adjacent tissue section) and the fatal, neurological pathology of cerebral malaria. The photo is kindly provided by Dan Millner.
Both P. infestans and P. falciparum employ a host targeting (HT) motif on secretory proteins, to deliver hundreds of effectors that must cross the haustorial/vacuolar membrane to enter the cytoplasm of host cells (Figure 2 A–B). Interestingly, their major host targeting motifs, RxLR-dEER from P. infestans and R/KxLxE/D/Q from P. falciparum, not only share protein sequence similarity, but also are functionally interchangeable in both binding PI(3)P [11] and in targeting effectors to the host [12, 13]. These motifs reflect the first well defined pathogenic secretion signals conserved across diverse eukaryotic pathogens. In this review, we will discuss how both oomycetes and Plasmodium target PIs but function in unexpected locations, namely the plasma membrane of the plant cell or the lumen of the parasite's ER rather than the cytoplasmic face of cellular membranes.
Eukaryotic pathogens use PI(3)P to target host cells
The utilization of PI(3)P by both P. infestans and P. falciparum provides remarkable evidence for the presence and function of PIs in secretory locations in eukaryotes. P. infestans and additional oomycete pathogens use the RxLR-dEER motif and additional sequences [14] for PI(3)P binding on the plant host plasma membrane for internalization. However, the motif RxLR in absence of dEER sequences recognizes a tyrosine-O-sulfated host membrane protein in the oomycete fish parasite Saprolegnia parasitica [15], indicating that in different proteins (as well as possibly in absence of the dEER sequences) the motif RxLR may facilitate binding to different host determinants, distinct from PI(3)P. Moreover in a P. infestans effector such as Avh5, both the RxLR-dEER sequence and the C terminal domain bind PI(3)P and the latter shows greater affinity for PI(3)P [16]. Differences in both specificity and amount of binding arise due to use of different methodologies as summarized in Box 1, which discusses best practices for measuring PI-binding and implementing appropriate controls.
P. falciparum secretory proteins with the R/KxLxE/D/Q motif bind PI(3)P in the ER of the parasite to initiate host targeting to the erythrocyte [11]. In the subsequent discussion, we refer to the RxLxE/D/Q motif as the RxL motif, and KxLxE/D/Q as KxL motif. In plasmodial proteins with the RxL motif, the HT signal is subsequently cleaved by an aspartic protease plasmepsin V (pV), which releases the signal anchor [17, 18]. KxL is not a substrate for pV [17]. KxL motifs are present in the major virulence adhesin family P. falciparum erythrocyte membrane protein 1 (PfEMP1, or Var), with 59 members that are central to disease in malaria [19]. The N terminal leaders of two P. infestans proteins, Avr3a and Nuk10, when expressed transgenically in P. falciparum, target reporters like green fluorescent protein (GFP) to the erythrocyte [13] dependent on their RxLR-dEER motifs and with efficiencies comparable to that shown for malarial leaders [20, 21]. Further, cell biological and biochemical analyses confirm that with Nuk10, its export is dependent on its RxLR motif binding to PI(3)P and independent of cleavage by pV [11]. However, it is unknown whether PI(3)P is detected in the P. infestans ER and/or whether export of effectors from Phytophthora to the haustorium are dependent on interactions of RxLR-dEER with PI(3)P in the pathogen's ER.
The studies of Whisson et al. [22] definitively established that during infection export of a P. infestans effector like Avr3a from the pathogen to the plant cell was dependent on RxLR-dEER. Subsequent work revealed the presence of PI(3)P on host cells and its ability to bind P. infestans effectors, other oomycete RxLR effector proteins, and rust fungus effectors with RxLR-like motifs [23], when recombinant effectors were with cultured host cells in the absence of infection (Table 1). These studies suggest that PI(3)P-RxLR-dEER dependent uptake of effector proteins into endosomes occurs in the absence of additional pathogenic factors and link PI(3)P-dependent uptake to RxLR-dEER as well as possibly other host targeting signals present on Phytophthora proteins. However, the consequences of post-translational modification of effectors by the pathogens have not been assessed. Further, uptake of effectors by cell surface PI(3)P results in a concentration of effectors in the endosome rather than in the cytoplasm where they are normally delivered in infection. Thus, although it is likely that effectors bind to PI(3)P as the first step during uptake into plant cells, how effectors are concentrated and translocated across the endosomal membrane to the host cytosol is poorly understood. Prokaryotic and possibly eukaryotic pathogens actively insert effector translocation machinery into pathogenic vacuoles and it is likely that Phytophthora may similarly modify the extra-haustorial membrane (Figure 1).
Table 1.
Host targeting characteristics for PI(3)P binding and protease sensitivity of effectors in Plasmodium, oomycetes and fungi
| Species | Protein | Protein functiona | HT motif | Cellular localization for motif activityb | PI(3)P binding | Kd for PI(3)P binding | Cleavage by protease | Experimental methods for host targeting | Experiment al methods for PI(3)P bindingc | Refs |
|---|---|---|---|---|---|---|---|---|---|---|
| Plasmodium parasites | ||||||||||
| P. falciparum | PfHRPII | Unknown | RLLHE/RLLYE | Parasite ER | Yes | 36 nM | Yes | in vivo | RP/SPR/In cells | [11, 51] |
| P. falciparum | RIFIN | Large antigenic family | RTLSE | Parasite ER | Yes | 20 nM | Yes | in vivo | RP/SPR | [11] |
| P. falciparum | PfEMP1 | Cell surface adhesin | KDVLE | Parasite ER | Yes | 30 nM | nd | in vivo | RP/SPR | [11] |
| P. falciparum | STEVOR | Antigen family | RLLAQ | Parasite ER | Yes | 70 nM | Yes | in vivo | RP/SPR | [11, 20, 21] |
| P. falciparum | PfEMP1 | Cell surface adhesin | KELLD | Parasite ER | Yes | 90 nM | No | in vivo | RP/SPR | [11, 21] |
| P. falciparum | Hsp40 | Heat shock protein | RSLAE | Parasite ER | Yes | 110 nM | Yes | in vivo | RP/SPR | [11, 20] |
| Oomycetes | ||||||||||
| P. infestans | Nuk 10 | nd | RQLR..dEER | h | Yes | 28 nM | No | in vitro | RP/SPR | [11] |
| P. infestans | Avr3a | Suppress host immunity | RLLR..dEER | h | No | nd | No | in planta | RP/LS | [22, 25] |
| P. sojae | Avr1b | Suppress host immunity | RFLR..dEER | h | Yes | nd | nd | in planta | RP/LS | [12, 14, 25] |
| P. sojae | Avh5 | nd | RFLR..dEER | h | Yes | nd | nd | in vitro | RP/LS | [14] |
| P. sojae | Avh331 | nd | RSLR..dEER | h | Yes | nd | nd | in vitro | RP/LS | [14] |
| S. parasitica | SpHTP1 | nd | RHLR | Host plasma membrane | No | nd | nd | in vitro | RP/LS | [15] |
| Fungi | ||||||||||
| Melampsora lini | AvrL567 | nd | RFYR | h | Yes | nd | nd | in vitro | RP/LS | [14] |
| Fusarium oxysporum | Avr2 | nd | RIYER | h | Yes | nd | nd | in vitro | RP/LS | [14] |
| Puccinia | Ps87 | nd | KRLTG | h | nd | nd | nd | in vitro | RP | [23] |
| striiformis | ||||||||||
| Leptosphaer ia maculans | AvrLm6 | nd | RYWT | h | Yes | nd | nd | in vitro | RP | [14] |
nd, no data available.
h, host membrane surrounding the haustorium.
RP, in vitro recombinant protein; SPR, surface plasmon resonance; LS, lipid strips.
Detailed comparative analyses of P. infestans and P. falciparum host targeting signals
Binding to PI(3)P
Functional properties of P. infestans and P. falciparum host targeting signals have been defined in the context of N-terminal leader sequences of proteins, that when coupled to exogenous reporters such as GFP and RFP, enable their export to the host cell. Using this basic reporter design, Kale et al. [14] reported that PI(3)P lipid was present on plant and mammalian cell surfaces and bound by the P. infestans HT signal RxLR-dEER to enable effector proteins access to host endosomal compartments. They further showed that a P. falciparum host targeting signal (with a consensus motif of RxLxE/D/Q, also known as PEXEL, for plasmodial export element) bound PIs. However, since PI binding was determined by using lipid strips, the mechanism and quantitative affinity for PI(3)P were unknown (as elaborated further in Box 1). Bhattacharjee et al. [11] demonstrated that the HT Nuk10 signal (RQLR) of P. infestans showed a high affinity interaction for PI(3)P with a Kd of 28 nM. As shown in Table 1, this was comparable to the Kd's (20–35 nM) for PI(3)P with HT motifs of three P. falciparum proteins: PfHRPII (PF3D7_0831800; a protein implicated in heme polymerization with an HT motif RLLYE), a RIFIN (member of a large antigenic family, Rif, of unknown function; PF3D7_0401600.1; HT motif RTLSE), and PfEMP1 protein (PF3D7_1200400; HT motif KDVLE). PfEMP1 is a family of variant surface virulent adhesins that enable infected erythrocytes to bind endothelial cells. A second PfEMP1 (PF3D7_1240600) with a motif of KELLD closely related to KDVLE, showed an approximately threefold lower Kd of 90 nM for PI(3)P. Similarly a heat shock protein (Hsp40, PF3D7_0501100.1) with a RSLAE motif and a member of STEVO,R another antigenic family of unknown function (PF3D7_0101800), with a RLLAQ motif display Kd's of 110 nM and 70 nM, respectively. Thus, even a limited set of P. infestans and P. falciparum HT motifs display a fivefold range for Kd's varying from 20 to 100 nM. As can be inferred from Table 1, this is not easily explained by differences in sequences of the HT motif sequence alone. It is likely that PI(3)P binding is mediated by a domain, a critical component of which is contributed by the R/K residues of the HT signal. As elaborated later in this review, multiple cationic residues are expected to contribute to nanomolar affinity of PI(3)P recognition, hydrophobic residues are likely needed for membrane penetration and hydrogen bonding through consensus D/E and will likely distinguish specificity for PI(3)P relative to other PIs [24]. Requirement of a domain is also consistent with early data indicating that in a vacuolar translocation signal (VTS; of ~40 amino acids) of PfHRPII both the HT motif and sequences upstream are needed for optimal P. falciparum protein export to the erythrocyte [13].
Additional host targeting properties
Host targeting is a complex process, with different transport properties ascribed to peptidic regions of effectors known to catalyze export to the erythrocyte. A summary of transport properties for oomycete and Plasmodium protein domains is provided in Figure 3. Emerging evidence from Phytophthora suggest that in addition to RxLR-dEER, the C-terminal domain of an effector protein can also bind PI(3)P and thus access the extra-haustorial membrane [25]. Indeed for the effector of Avr1b in Phytophthora sojae, the C-terminal domain shows a greater affinity for PI(3)P compared to the N-terminal RxLR-dEER [25]. The four positively charged K residues on the first alpha helix from the N-terminus of the effector domain are important for PI(3)P binding activity. Mutation of any of these residues abolishes the binding activity of PI(3)P. These data are also consistent with the idea that PI(3)P binding is the property of a domain.
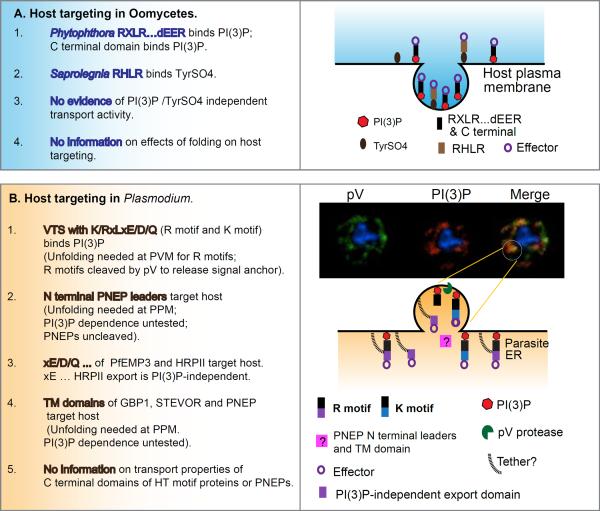
Figure 3. Summary of transport properties of (a) oomycete and (b) Plasmodium protein domains.
(a) In Phytophthora, leaders containing RxLR- dEER and/or C terminal domains (black bars) of effector proteins (purple circles) binds the lipid PI(3)P (red dots) at the host plasma membrane resulting in their endosomal internalization. In Saprolegnia, leaders containing RHLR (brown bars) binds tyrosine sulfate (TyrSO4; grey dot, a post translational modification of host proteins) to also undergo endosomal internalization. Presently, there is no evidence of host targeting properties independent of binding PI(3)P or TyrSO4 or protein folding, such as seen in Plasmodium. (b) In Plasmodium, leaders known as vacuolar translocation sequences (VTS) containing a host targeting motif KxLxE/D/Q (black/blue bar) or RxLxE/D/Q (black/purple bar) bind PI(3)P (red dots) in the ER lumen. Micrographs in the right hand side top panel show distribution of the lipid PI(3)P in the ER lumen (red) and a parasite protease plasmepsin V (pV; green) colocalized (yellow/orange) in membrane domains in the ER. This suggests a model (shown immediately below) for a PI(3)P enriched vesicle (containing pV) emerging from the ER. In this regard, PI(3)P-dependent exit from the ER is topologically equivalent to PI(3)P-dependent endosomal uptake at the plasma membrane. Proteins with RxLxE/D/Q may be cleaved by the protease pV in the emerging PI(3)P vesicle, (which is reinforced by the finding that the specificity of PI(3)P binding mimics that of pV mediated cleavage). The xE/D/Q fragments of PfEMP3 and PfHRPII (purple rectangle) also have intrinsic export activity suggesting a second mechanism of inclusion and retention of soluble proteins (indicated by black tethers). N terminal leaders lacking K/RxL domains called PNEPs (pink rectangles) appear to share a common export mechanism whose dependence on PI(3)P has not been tested (and thus indicated by a question mark). Unlike Phytophthora, C-terminal domains of plasmodial effectors have not yet been shown to have intrinsic PI(3)P binding or export activity and parasite host targeting is sensitive to protein folding.
C-terminal regions of malarial proteins have not been tested for PI(3)P binding. Gruring et al. [26] have recently shown that short leader sequences from PNEPs (or PEXEL negative exported proteins) which lack intrinsic activity to target soluble reporters, are capable of exporting membrane chimeras when used in conjunction with transmembrane domains derived from PNEP proteins [26]. Dependence of this process on PI(3)P was not investigated. It is important to do so because PI(3)P binding is not expected to be restricted to KxL or RxL motif, but rather supported by diverse structural moieties across pathogenic and mammalian proteins (Figures 4 and 5). When the PNEP leaders were substituted by sequences downstream from HT motifs of STEVORs and glycophorin binding protein 130 (GBP130, another parasite protein of unknown function) and erythrocyte membrane protein 3 (EMP3; targets the erythrocyte skeleton), export of the membrane chimeras was preserved. However, the sequences in EMP3 show intrinsic erythrocyte-export activity with soluble reporters [11] even when the PfEMP1 signal anchor is engineered to be cleaved by signal peptidase. Indeed this observation was key to recognizing that the aspartic protease pV does not target export as previously proposed [17] but rather releases the signal anchor from the ER membrane. Sequences downstream of the HT motif in GBP130 and STEVOR have not yet been tested, but those in HRPII also show intrinsic export activity with soluble reporters [27]. This activity is independent of PI(3)P and less efficient than the dominant HT PI(3)P-dependent export when both signals function in an intact VTS [28]. Finally, the distribution of the aspartic protease pV corresponds to that of PI(3)P in the ER lumen (see Figure 3 and [28]). However, PfEMP1 proteins which are the major malarial virulence determinants, contain an HT motif and bind PI(3)P, are not substrates of pV. Thus despite the attraction of a `global' mechanism [29, 26] of export to the erythrocyte, further studies are required to establish whether PNEPs and HT containing proteins are similarly exported at a mechanistic level.
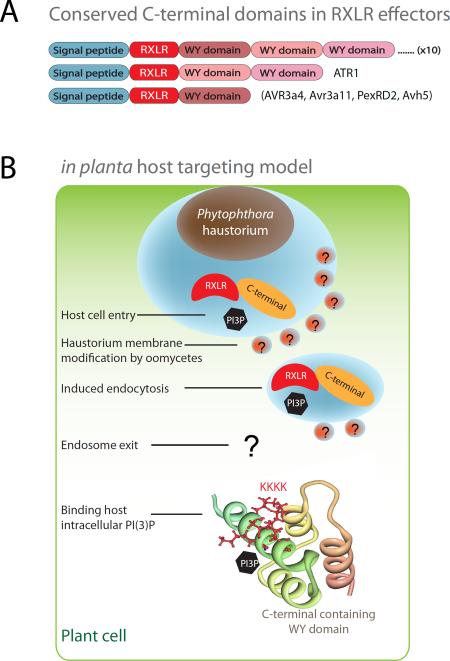
Figure 4. Conserved RxLR-dEER effector structure fold and different sites for PI(3)P binding.
(a) RxLR effector domain architecture. RxLR effectors have an N-terminal secretory leader, a host targeting RxLR motif and conserved C-terminal domains that are often arranged into tandem repeats. (b) Host targeting model and involvement of PI(3)P at different cellular sites. The N-terminal RxLR motifs play an important role in host cell entry at host plasma membrane; and in vitro recombinant proteins of Avr1b bind PI(3)P. The C-terminal region has a conserved effector fold and is able to suppress host defense responses. The positive patches in the C-terminal domain of Avr1b and Avr3a are important for PI(3)P and PI(4)P binding. The Avr1b C-terminal structure is used in the illustration. Several important conceptual steps such as modification of membrane by pathogen are postulated and indicated with question marks.
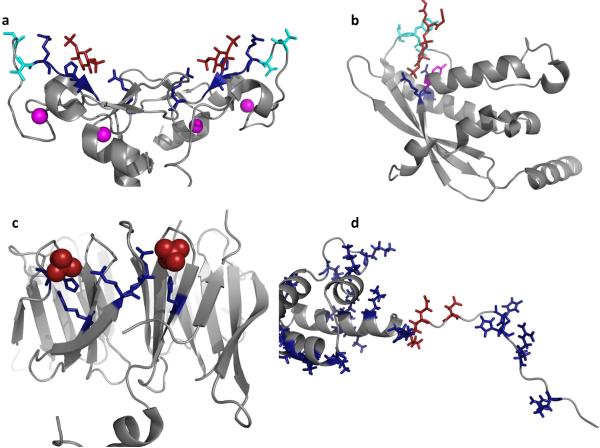
Figure 5. Mechanisms and models of PI(3)P binding by FYVE, PX, PROPPIN, RxLR and WY domains.
PI(3)P binding has been attributed to at least three distinct eukaryotic domains including FYVE, PX, and PROPPINS while pathogens utilize the RxLR or RxLxE/D/Q motif and/or WY domains. (a) The homodimeric structure of the EEA1 FYVE domain is shown (PDB ID: 1JOC), which binds PI(3)P (shown in red) in a sideways fashion where both electrostatic and H-bonding with PI(3)P can be optimized while achieving hydrophobic interactions through the turret loops [39]. Cationic residues that coordinate the PI(3)P headgroup are shown in blue, hydrophobic residues that penetrate into the hydrocarbon portion of the membrane are highlighted in cyan, and the zinc ions coordinated by the FYVE domains for structural stability are shown in magenta. (b) The p40phox PX domain (PDB ID: 1H6H) X-ray structure was solved bound to PI(3)P. Here, two critical arginine residues (shown in blue) mediate the recognition of PI(3)P, while a tyrosine residue (magenta) forms the bottom of the PI(3)P binding pocket. The PI(3)P binding pocket is deeper than that observed for the FYVE domain and harbors two loop regions that have been shown to facilitate nonspecific electrostatic interactions and membrane penetration (hydrophobic residues in cyan) [57]. (c) The yeast PROPPIN structure from Hsv2 (Homologous with SVP1), which is a key autophagic sensor protein, was solved bound to two sulfate ions (PDB ID: 4EXV). PROPPINs are PI(3)P sensors necessary for autophagy and are conserved from yeast to humans. The PROPPIN structure is a β-propeller composed of seven blades where two PI(3)P binding sites (sulfate ions in red and coordinating residues in blue) were discovered on blades five and six. In a similar manner to the FYVE and PX domains, a hydrophobic loop between blades five and six promotes membrane penetration of Hsv2, which is essential to act in concert with PI(3)P binding for membrane recognition and full autophagic function in yeast. (d) Shown is the N-terminal unstructured region and WY domain of Avr1b from P. sojae, which was modeled and generated using the available P. infestans Avr3a structure (PDB ID: 2LC2) [25]. The N-terminus harbors the RxLR motif (Arg residues in blue on the left) as well as number of other cationic (blue) and anionic (red) residues that may be critical tophosphoinositide binding. The WY domain of Avr1b is shown to highlight the cationic nature of this module (Arg, Lys and His shown in blue). In contrast to FYVE, PX, and PROPPINs a distinct phosphoinositide binding pocket or region is not obvious and nonspecific electrostatic association may occur based upon lipid charge.
There is as yet no evidence of HT-independent transport associated with P. infestans RxLR-dEER leaders. Indeed if the leader of P. infestans Nuk10 is expressed in blood stage malaria parasites with a mutated RxLR motif, export to the erythrocyte is abrogated, rather than reduced as seen with the P. falciparum VTS and HT motifs [11]. Sequences downstream of Avr3a also appear to have no intrinsic export activity [13, 26]. In P. infestans, another group of HT effectors are the crinklers, which possess a FLAK motif in the N-terminal end of the sequences [30]. It remains unknown whether crinklers use PI(3)P dependent or independent mechanisms to target the host.
As elaborated in Figure 3 multiple export properties may be associated with peptidic domains of effectors in both Phytophthora and malaria parasites, and additional investigation is required to determine their related hierarchy and/or independence. It should be emphasized that since functional domains involved in protein-lipid interactions are larger than the HT motif, the design of constructs and understanding of the intrinsic export functions of flanking regions requires careful attention. For instance in P. infestans effectors the dEER sequences have not been found to have any intrinsic export activity, but they are needed in conjunction with RxLR to detect host targeting [13]. In addition, sequences upstream of the HT motif of HRPII are needed (in conjunction with RLLYE) for targeting the erythrocyte [13]. Finally, HT motifs and their downstream sequences appear inherently unstructured and thus unexpected bends (as well as folding) influences export [26, 31]. Thus careful biophysical measurements such as SPR or NMR are recommended for measuring lipid binding over more rapid but less discriminating lipid strips (see also Box 1).
Sites of action
A major difference between P. infestans and plasmodial HT signals and their binding to PI(3)P are their subcellular sites of action. P. infestans secretes proteins that bind at PI(3)P on the host cell surface, which provides the first step for effectors to access the host endosome ([14]; Figure 1). In contrast, the mature erythrocyte has no PI(3)P and thus merozoite proteins secreted during invasion do not bind PI(3)P on host erythrocytes. Further, HT signals with the RxL motif initially bind PI(3)P in the parasite ER but lose this property when they are subsequently cleaved in the ER (Figure 1). Hence RxL motif proteins when exported to the erythrocyte likely do not show affinity for PI(3)P there. Thus, at the end of the intraerythrocytic cycle, when the infected cell lyses and RxL-motif proteins are shed in plasma, they lack the HT motif and PI(3)P binding function.
Common fold for intracellular binding of PI(3)P in oomycete effectors
Emerging structural analysis is starting to provide insights into mechanisms of PI(3)P binding in diverse proteins. Protein structures for a total of five oomycetes effectors have been recently obtained (Figure 4), i.e., Avr3a4 and Avr3a11 (Phytophthora capsici), PexRD2 (P. infestans), Avh5 (P. sojae), and ATR1 (Hyaloperonospora arabidopsidis) [32, 33, 16]. Phytophthora RxLR-dEER effectors form large rapidly evolving superfamilies, and many effectors contain conserved sequence motifs (W, Y, and L) in their C-terminal domains arranged into tandem repeats [34]. Structural analysis of the four effectors uncovered a common fold conserved across oomycetes species. The conserved unit is termed WY-domain, and comprises a minimum three helices per domain [35].
Structural and functional analyses of the Phytophthora effector Avr3a shows a conserved, positively charged surface patch that binds PI(3)Ps on the first alpha helix of the conserved effector structure fold in the host cytoplasm [25]. Once delivered into plant cells, Avr3a is able to suppress host programmed cell death by stabilizing and inhibiting the host E3 ubiquitin ligase CMPG1. Mutations in the PI(3)P-binding patch are able to diminish the Avr3a effect of stabilization of CMPG1 in vivo [25]. PI(3)P-binding ability is therefore delineated to the C-terminus of the effector and shown to be involved in host defense suppression. It is possible that this activity is important for cytosolic effector action and also contributes to the PI(3)P recognition at the EHM between the pathogen and host cell (Figure 1). A dual recognition mechanism of PI(3)P has been shown by the RxLR effector Avh5 from P. sojae [16]. Structural analysis with NMR and a number of lipid binding assays revealed the C-terminal helical region bound PI(3)P while the RxLR motif also bound PI(3)P but played a more minor role; however, both PI(3)P binding sites mediated entry independently into the host cell. This type of mechanism of lipid recognition by Avh5 is reminiscent of mammalian lipid binding proteins, which are often coincidence detectors for membrane lipids harboring more than one lipid binding site for a lipid headgroup [36].
Structural studies of three mammalian PI(3)P binding proteins and domains (EEA1-FYVE, PX domains and PROPPINS) provide significant insights on how a diverse set of modular domains bind PI(3)P specifically with nanomolar affinity (Figure 5). The X-ray structure of EEA1-FYVE suggests that the FYVE domain stereo-specifically recognizes the PI(3)P head group [37], as a homodimer and the PI(3)P-bound FYVE may have an `angled' orientation with respect to the membrane surface, which allows hydrophobic residues in the turret loop to penetrate the membrane (Figure 5a) [38]. Due to the presence of cationic residues, the turret loop region of FYVE domains is surrounded by highly positive electrostatic potential, which drives the initial membrane association [39]. Subsequently, specific PI(3)P binding induces the interfacial penetration of hydrophobic and aromatic residues in the turret loop [39]. Further, PI(3)P binding is not a consequence of but a prerequisite for the interfacial penetration of turret loop residues. Thus PI(3)P binding serves as an electrostatic switch that greatly reduces the positive potential surrounding the turret loop and thereby promotes the interfacial penetration of hydrophobic and aromatic turret loop residues [39, 40].
The PX domain is a structural module (Figure 5b) composed of 100–140 amino acids [41, 42] shown to interact with different PIs via conserved basic residues [43]. PX domains exhibit broad PI specificity akin to PH domains, but many, including those of Vam7p [44], sorting nexin 3 [45], and p40phox [46], specifically interact with PI(3)P in vitro and target proteins to early endosomes in the cell. The crystal structure of the p40phox–PI(3)P complex illustrates how the domain also achieves stereospecific recognition of PI(3)P [46]. Non-specific electrostatic interactions between the cationic molecular surface of the PX domain and the anionic membrane are followed by specific PI binding and subsequent interfacial penetration of hydrophobic and aromatic residues [47, 48]. The CISK PX domain might also be involved in domain homodimerization in addition to PI-induced interfacial penetration [49]. Finally, the first crystal structure of PROPPINs (Figure 5c), which are key autophagic PI(3)P sensors conserved from yeast to humans, delineated a β-propeller structure harboring seven blades and two binding sites for PI(3)P [50]. A hydrophobic loop adjacent to the PI(3)P binding sites acts in tandem with PI(3)P to promote high membrane affinity and full autophagic function in yeast. Pathogenic effectors identified to date are unique in PI(3)P binding specificity. However a well-defined pocket has not yet been determined (Figure 5d) nor is the molecular basis of PI(3)P coordination known, which should undoubtedly provide H-bonding and electrostatic interactions with the 1- and 3-phosphate of PI(3)P if these effectors recognize PI(3)P over other PIs. Instead, Avh5 has been shown to bind PI(3)P using Lys62, Lys64, and Lys65 in helix 2 of the WY domain as well as the RxLR motif [16] in contrast to lysine residues present in helix 1 of the WY domain of Avr3a [25]. Thus, these effectors apparently use a cationic patch in different locations in the WY domain or RxLR region to recognize the PI(3)P headgroup. This type of specificity and PI(3)P binding mode is unique among PI(3)P binding proteins identified to date. As summarized earlier, FYVE and PX domains and the PROPPIN family typically insert into the hydrocarbon core of the membrane where aliphatic and aromatic amino acids mediate this step, which is induced by specific PI(3)P binding. The RxLR-dEER and K/RxLxE/D/Q motifs as well as WY domain are rich in aliphatic and aromatic amino acids, which may additionally play a role in increasing PI(3)P dependent membrane affinity and the ability of pathogenic effectors to enter the host cell or exit the ER through vesicular transport.
Concluding remarks and future directions
A major question raised by studies related to PI(3)P and host cell targeting in Plasmodium and Phytophthora is how an essential signaling molecule appears in unexpected cellular locations. Malaria parasites show PI(3)P in the ER lumen and host cells of oomycetes reveal PI(3)P in the outer leaflet of plasma membrane. Neither cellular location has been previously associated with notable levels of PI(3)P. All known enzymes for PI(3)P production are cytoplasmic and there are no well-defined mechanisms to account for the PIs in the ER or outer leaflet of the plasma membrane. Thus, questions emerge on how PI(3)P is generated and transported to the outer leaflet of the plasma membrane and to the ER lumen? How are their recycling and steady state maintenance performed? Are there other PI(3)P interacting proteins encoded by the parasites that assist with these processes?
A second set of questions arise on why binding of PI(3)P by a pathogenic effector induces endosomal budding at the plasma membrane or is associated with exit from the ER? What additional factors are required at the plasma membrane or in the ER? Are mechanisms related despite the action in different cellular locations? Expression of EEA1 and p40phox mammalian proteins that bind PI(3)P in P. falciparum ER does not result in their exit from the ER [11]. To date, the majority of lipid-binding proteins discovered are coincidence detectors [36] where they bind with high affinity and selectivity to two lipids [such as PI(3)P and PS] or to one lipid and a membrane physical property such as curvature [such as PI(3)P in highly curved membranes]. In addition, two PI(3)P binding sites may be present to increase avidity as observed for PROPPINs, the FYVE homodimer, or Avh5. Thus, PI(3)P binding by pathogenic effectors may be an important determinant of endosomal PM budding or exit from the ER. However, effector interactions with other lipids or membrane physical properties could also exist (Box 1), which may provide spatial and temporal cues in pathogenic export to the host.
Highlights
Eukaryotic pathogens use common strategies to target virulence proteins into host cells.
Binding the lipid PI(3)P is a critical step in virulence-related trafficking.
The high PI(3)P affinity of effectors from pathogens defines them as novel lipid interacting proteins.
Box 1. Methods and conditions of success for monitoring lipid-protein interactions.
Cellular membranes are composed of hundreds if not thousands of different lipid species in bilayers that compartmentalize a cell or pathogen with different organelle structure and function. Peripheral proteins, which transiently interact with one side of the bilayer usually, have high affinity and specificity for a key target lipid, which can mediate protein function and elicit communication across membrane bilayers. Lipid binding specificity among peripheral proteins can vary from anionic glycerophospholipids such as phosphatidlyserine and the phosphoinositides to sphingolipids including ceramide and ceramide-1-phosphate. Moreover, many peripheral proteins are now appreciated to be coincidence detectors where they harbor two lipid-binding sites that can provide exquisite regulation of protein localization to discrete membrane sites enriched in both lipids. Thus, to investigate lipid-binding specificity and membrane affinity for newly characterized proteins it is important that lipid-binding studies are carefully and accurately performed.
Methodologies can differ in their sensitivity and reliability
To this end vesicle sedimentation assays and surface plasmon resonance (SPR) with large unilamellar vesicles have robustly served to delineate lipid binding selectivity and membrane binding affinity, respectively. Lipid strips composed of different types of lipids blotted on nitrocellulose have been traditionally useful for probing lipid specificity such as which phosphoinositide a protein selectively binds, however, they are not able to provide quantitative information and their surfaces are not well characterized. This has been a contentious issue for the RxLR leaders where PI(3)P binding has been attributed to the RxLR motif [14], the WY domain [15, 25], or both [16] and also in one case denatured protein binding to PI(3)P [15]. For some of these studies lipid strips were the primary source of data collection. The behaviors and orientations of phosphoinositides spotted on nitrocellulose have not been rigorously investigated and reproducibility from experiment to experiment can be an issue. Assays that incorporate the key lipid into an environment that recapitulates the native environment of the protein binding can play a much more effective quantitative and predictive role of membrane binding mechanisms. For instance, the majority of peripheral proteins identified to date undergo hydrophobic or electrostatic interactions with the membrane surface in addition to phosphoinositide headgroup binding while others have a second binding site for an essential membrane lipid.
Lipid strips cannot provide such an environment for detecting these interactions unlike vesicles, which can be modulated to include a variety of lipid species to provide a surface more akin to a membrane bilayer. To further demonstrate that lipid blots may not accurately assess PI(3)P binding, mutation of the RxLR motif in Avh5 did not show an appreciable change in PI(3)P binding on lipid strips but had ~30% reduction in binding in lipid vesicle binding assays [16]. This may help explain some of the differences in PI(3)P binding analyzed earlier for Avr3a mutants where only lipid strips were used [25]. Lipid strips also may not be the best approach for studying some of these effectors that have two binding sites for PI(3)P. For instance, it is possible the two binding sites may compete for PI(3)P presented on the nitrocellulose making it difficult to quantify the role or affinity for either site for PI(3)P. Mutagenesis of the RxLR or WY domain in this case could just promote more binding of the other motif to PI(3)P or other PIs on the nitrocellulose membrane. Further, because these assays are usually performed at micromolar concentrations in the same vessel it is difficult to discern PI specificity for those effectors that demonstrate binding to two or more PIs without employment of a secondary assay (sedimentation or SPR).
Appropriate controls are important for phosphoinositide binding
It is also wise to use positive and negative controls for lipid surfaces whether it be lipid strips, SPR assays, or vesicle sedimentation assays to demonstrate the viability and reproducibility of the surface employed. For instance, if PI(3)P selectivity is shown for a newly identified pathogen effector one could use the PX domain of p40phox as a positive control for PI(3)P and the PH domain of PLCδ1 as negative control (PI(4,5)P2 specificity). This type of approach allows one to make quantitative and confident conclusions regarding lipid-binding specificity and affinity.
Membrane physical properties such as curvature and charge matter
Recently, membrane physical properties such as charge or curvature have also been shown to play a role in membrane recruitment of peripheral proteins[53]. In addition, some peripheral proteins induce changes to membrane structure (positive or negative curvature changes) upon membrane binding [54]. While testing membrane curvature dependency is not the first set of experiments one pursues it is important to be mindful that some lipid-binding proteins may bind to highly curved membranes more effectively than flat membranes while others themselves may induce changes to membrane structure upon lipid binding. Thus, if binding is not observed to one type of membrane curvature this may not rule out lipid binding affinity or selectivity for a particular protein. The membrane interface, which restricts proteins on the interface to 2-dimensions can promote avidity and protein oligomerization and could be another mechanism of PI(3)P recognition by these motifs. In closing, when performing lipid binding studies of previously uncharacterized proteins it is recommended to utilize at least two different lipid binding assays with appropriate controls to confirm lipid selectivity and to strive for an assay that is sensitive enough to delineate changes in affinity for point mutations if the origin of lipid binding is going to be investigated.
Acknowledgements
This work was funded by the National Institute of Health grants HL069630, AI039071, HL078826 (K.H) and AI081077 (R.V.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 3.Dai S, et al. Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic. 2007;8:1365–1374. doi: 10.1111/j.1600-0854.2007.00613.x. [DOI] [PubMed] [Google Scholar]
- 4.Galan JE. Salmonella interactions with host cells: Type III secretion at work. Ann. Rev. Cell Dev. Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Mallo GV, et al. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J Cell Biol. 2008;182:741–752. doi: 10.1083/jcb.200804131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V, et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol. 2006;8:719–727. doi: 10.1111/j.1462-5822.2006.00705.x. [DOI] [PubMed] [Google Scholar]
- 7.Chukkapalli V, et al. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. Journal of virology. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inlora J, et al. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. Journal of virology. 85:3802–3810. doi: 10.1128/JVI.02383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altan-Bonnet N, Balla T. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem Sci. 2012;37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharjee S, et al. Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell. 2012;148:201–212. doi: 10.1016/j.cell.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou D, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. The Plant cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharjee S, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kale SD, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Wawra S, et al. Host-targeting protein 1 (SpHtp1) from the oomycete Saprolegnia parasitica translocates specifically into fish cells in a tyrosine-O-sulphate-dependent manner. Proc Natl Acad Sci U S A. 2012;109:2096–2101. doi: 10.1073/pnas.1113775109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun F, et al. Structural basis for interactions of the Phytophthora sojae RxLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Mol Plant Microbe Interact. 2012 doi: 10.1094/MPMI-07-12-0184-R. http://dx.doi.org/10.1094/MPMI-07-12-0184-R. [DOI] [PubMed]
- 17.Boddey JA, et al. An aspartyl protease directs malaria effector proteins to the host cell. Nature. 2010;463:627–631. doi: 10.1038/nature08728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo I, et al. Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature. 2010;463:632–636. doi: 10.1038/nature08726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherf A, et al. Antigenic variation in Plasmodium falciparum. Annual review of microbiology. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 20.Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 21.Marti M, et al. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 22.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 23.Gu B, et al. Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLoS One. 2011;6:e27217. doi: 10.1371/journal.pone.0027217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nature chemical biology. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaeno T, et al. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci U S A. 2011;108:14682–14687. doi: 10.1073/pnas.1106002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruring C, et al. Uncovering common principles in protein export of malaria parasites. Cell Host Microbe. 2012;12:717–729. doi: 10.1016/j.chom.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharjee S, et al. PI(3)P-independent and -dependent pathways function together in a vacuolar translocation sequence to target malarial proteins to the host erythrocyte. Mol Biochem Parasitol. 2012;185:106–113. doi: 10.1016/j.molbiopara.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharjee S, et al. PI(3)P-independent and -dependent pathways function together in a vacuolar translocation sequence to target malarial proteins to the host erythrocyte. Mol Biochem Parasitol. 2012;185:106–113. doi: 10.1016/j.molbiopara.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg DE. Plasmodium protein export at higher PEXEL resolution. Cell Host Microbe. 2012;12:609–610. doi: 10.1016/j.chom.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Schornack S, et al. Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci U S A. 2010;107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehde N, et al. Protein unfolding is an essential requirement for transport across the parasitophorous vacuolar membrane of Plasmodium falciparum. Mol Microbiol. 2009;71:613–628. doi: 10.1111/j.1365-2958.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- 32.Boutemy LS, et al. Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J Biol Chem. 2011;286:35834–35842. doi: 10.1074/jbc.M111.262303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou S, et al. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci U S A. 2011;108:13323–13328. doi: 10.1073/pnas.1109791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang RH, et al. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Win J, et al. Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 2012;8:e1002400. doi: 10.1371/journal.ppat.1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott JL, et al. Emerging methodologies to investigate lipid-protein interactions. Integr Biol (Camb) 2012;4:247–258. doi: 10.1039/c2ib00143h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Molecular cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 38.Yokogawa M, et al. NMR analyses of the interaction between the EEA1 FYVE domain and phosphoinositide embedded in lipid bilayer. J Biol Chem. 2012;287:34936–34945. doi: 10.1074/jbc.M112.398255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahelin RV, et al. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 40.Diraviyam K, et al. Computer modeling of the membrane interaction of FYVE domains. Journal of molecular biology. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 41.Ponting CP. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- 43.Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Lee SA, et al. Molecular mechanism of membrane docking by the Vam7p PX domain. J Biol Chem. 2006;281:37091–37101. doi: 10.1074/jbc.M608610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, et al. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 46.Bravo J, et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Molecular cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 47.Stahelin RV, et al. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J Biol Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 48.Malkova S, et al. Orientation and penetration depth of monolayer-bound p40phox-PX. Biochemistry. 2006;45:13566–13575. doi: 10.1021/bi061133l. [DOI] [PubMed] [Google Scholar]
- 49.Xing Y, et al. Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX) J Biol Chem. 2004;279:30662–30669. doi: 10.1074/jbc.M404107200. [DOI] [PubMed] [Google Scholar]
- 50.Baskaran S, et al. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Molecular cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang HH, et al. N-terminal processing of proteins exported by malaria parasites. Mol Biochem Parasitol. 2008;160:107–115. doi: 10.1016/j.molbiopara.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Przyborski JM, Lanzer M. Protein transport and trafficking in Plasmodium falciparum-infected erythrocytes. Parasitology. 2005;130:373–388. doi: 10.1017/s0031182004006729. [DOI] [PubMed] [Google Scholar]
- 53.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Baumgart T, et al. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annual review of physical chemistry. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker G, et al. Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology. 2011;138:1638–1663. doi: 10.1017/S0031182010001708. [DOI] [PubMed] [Google Scholar]
- 56.Vaid A, et al. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood. 2010;115:2500–2507. doi: 10.1182/blood-2009-08-238972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stahelin RV, et al. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J Biol Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]