Summary
Background
Disruption of circadian (daily) timekeeping enhances the risk of metabolic syndrome, obesity, and Type 2 diabetes. While clinical observations have suggested that insulin action is not constant throughout the 24 hour cycle, its magnitude and periodicity have not been assessed. Moreover, when circadian rhythmicity is absent or severely disrupted, it is not known whether insulin action will lock to the peak, nadir or mean of the normal periodicity of insulin action.
Results
We used hyperinsulinemic-euglycemic clamps to show a bona fide circadian rhythm of insulin action; mice are most resistant to insulin during their daily phase of relative inactivity. Moreover, clock-disrupted Bmal1-knockout mice are locked into the trough of insulin action and lack rhythmicity in insulin action and activity patterns. When rhythmicity is rescued in the Bmal1-knockout mice by expression of the paralogous gene Bmal2, insulin action and activity patterns are restored. When challenged with a high fat diet, arhythmic mice (either Bmal1-knockout mice or wild type mice made arhythmic by exposure to constant light) were obese prone. Adipose tissue explants obtained from high-fat fed mice have their own periodicity that was longer than animals on a chow fed diet.
Conclusions
This study provides rigorous documentation for a circadian rhythm of insulin action and demonstrates that disturbing the natural rhythmicity of insulin action will disrupt the rhythmic internal environment of insulin sensitive tissue, thereby predisposing the animals to insulin resistance and obesity.
Introduction
Many physiological processes display day-night rhythms, including feeding behavior, lipid/carbohydrate metabolism, and sleep. These daily oscillations are controlled by the circadian biological clock [1]. Circadian desynchrony in humans, a characteristic of shift work, jet lag, and/or sleep disruption, can have profound effects on both normal body weight regulation and glucose/lipid homoeostasis [2, 3, 4]. Improper circadian entrainment is associated with the onset of metabolic syndrome, obesity, and type 2 diabetes. Disruption of circadian rhythmicity may interact with other susceptibility factors to precipitate the disease state [2, 5, 6]. Many hormones that modulate insulin secretion, glucose homeostasis, and feeding are regulated cyclically by the circadian system, including orexin, leptin, glucagon, cortisol, growth hormone, catecholamines, and melatonin [6, 7]. Another linkage between clock-regulated sleep and insulin resistance/type 2 diabetes is the association between short sleep duration (due to e.g., insomnia and/or circadian disruption) and the risk of obesity and diabetes [8, 9].
In any given tissue, 3–10% of mRNA transcripts show circadian rhythmicity in vivo [10, 11], including those encoding transcriptional factors such as DBP, Rev-erbα, PPARγ, HLF and TEF [6, 12, 13]. These transcriptional factors then regulate downstream target genes involved in different biochemical pathways, including those relating to metabolism of glucose & lipids, synthesis of cholesterol, fatty acids & bile acids, and mitochondrial oxidative phosphorylation [11, 14, 15]. Therefore, polymorphisms of core clock genes or of clock-controlled hormone receptor genes that might influence the regulation of these metabolic pathways could have health consequences in humans. Indeed, polymorphisms of the clock genes Bmal1 (aka Arntl1 in humans) and Clock are associated with obesity, type 2 diabetes, and hypertension [16, 17]. Moreover, genome-wide association studies (GWAS) show an increased risk of type 2 diabetes associated with variants of the gene (mtnr1b) encoding a receptor for the clock-controlled hormone melatonin [18, 19].
Disruptions of the normal light/dark cycle in wild-type mice–analogous to shiftwork disruption in humans–can lead to increased body weight, hyperleptinemia and elevated insulin secretion [20, 21]. Altering the circadian timing of food intake stimulates weight gain in mice [22]. Conversely, feeding a high-fat diet can itself change the period of the circadian activity rhythm and alter the expression of core clock genes in mice in vivo [23]. Genetic mouse models have also elucidated the linkage between metabolism and the circadian system. For example, mice that are homozygous for a loss-of-function mutation in the circadian gene Clock overeat, become obese and develop hyperglycemia and dyslipidemia [24]. These Clock mutant mice develop the adipocyte hypertrophy and excessive accumulation of fat in the liver that are hallmarks of the metabolic syndrome. Regulation of the histone deacetylase Hdac3 by the clock-regulated transcriptional factor Rev-erbα directs a circadian rhythm of histone acetylation and gene expression that is required for normal hepatic lipid metabolism [25, 26]. Knockout of the three circadian Period genes in mice cause arhythmicity in behavior and increased weight gain on high-fat diets [27]. Similarly, Bmal1 knockout mice display arhythmic behavior in constant conditions, increased fat deposition, elevated triglycerides/free fatty acid levels, and disrupted insulin responsiveness [28, 29, 30, 31, 32].
Therefore, a preponderance of evidence supports a close relationship between clocks and metabolism. Consequently, manipulating biological timing could be used to develop non-invasive therapies for metabolic disorders. However, whether insulin action itself is rhythmic is unclear, nor has the impact of clock disruption upon insulin action been well characterized. Using a hyperinsulinemic-euglycemic clamp procedure that was developed at Vanderbilt to eliminate the need to handle, restrain, or stress mice [33, 34], we show herein that mice show a bona fide circadian rhythm of insulin action such that mice are most resistant to insulin during the phase of relative inactivity. Knockout of the Bmal1 gene leads to profound insulin resistance, which can be rescued by constitutive expression of the Bmal2 gene. In addition to insulin resistance and hyperglycemia, arhythmic mice exhibit metabolic phenotypes related to fat accumulation. By analysis of food intake and activity levels in rhythmic vs. arhythmic mice in light/dark and continuous light, these metabolic phenotypes are associated with disruption of rhythmic circadian behavior.
Results
Circadian rhythm of insulin action and its elimination in “clockless” mice
The hyperinsulinemic-euglycemic clamp, or insulin clamp, is widely considered to be the “gold standard” method for assessing insulin action in vivo. During an insulin clamp, hyperinsulinemia is achieved by constant insulin infusion. Euglycemia is maintained via a concomitant glucose infusion at a variable rate. The glucose infusion rate (GIR) is determined by measuring blood glucose at brief intervals throughout the experiment and adjusting the GIR accordingly to maintain constant blood glucose levels. We applied the insulin clamp methodology [33, 34] over the circadian cycle in conscious and unrestrained mice to assess most precisely circadian control over insulin action and glucose homeostasis in wild-type mice (WT C57/BL6) and in mice whose key circadian clock gene Bmal1 has been knocked out (B1ko) such that the circadian system is abolished or at least severely disrupted [35].
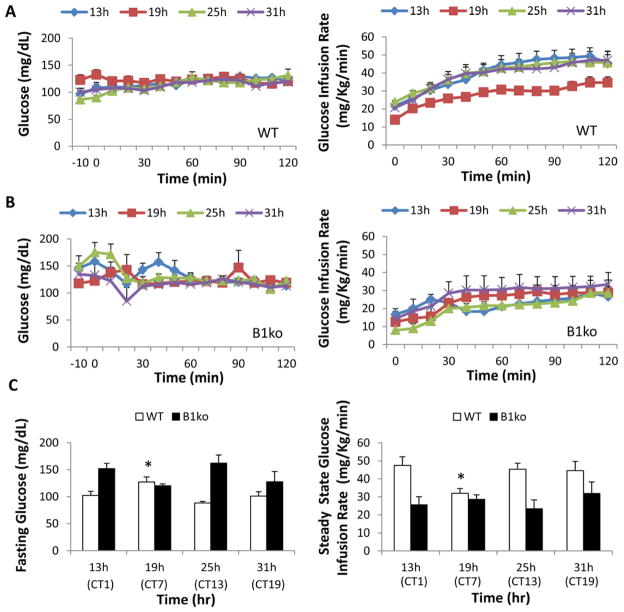
Our protocol measures GIR during a hyperinsulinemic-euglycemic clamp at different phases of the circadian cycle in freely roaming, non-stressed mice whose circadian system is “free-running” in constant dim red light (Fig. S1). This protocol revealed a clear circadian rhythm of insulin action in WT mice (Fig. 1). In particular, mice are significantly more insulin resistant at hour 19 in constant dim red light as indicated by a lower GIR (Fig. 1A, 1C). This phase corresponds with the middle of their subjective day (Circadian Time 7, or “CT7,” see Fig. S1), when they are relatively inactive. Constant dim red light is perceived as darkness by the circadian system of mice [30, 36], so mice in constant dim red light express their endogenous “free-running” circadian patterns. Statistical analyses of the data depicted in Figure 1 addressed two questions for both the fasting glucose levels and GIR data sets: (i) are there significant differences among the phases of the WT or B1ko samples, and (ii) are the WT and B1ko groups different statistically? One-way ANOVA analysis of the phase data revealed statistically significant phase differences among the WT data for both fasting glucose levels (peak at CT7, p = 0.004) and for GIR (trough at CT7, p = 0.047). Therefore, there are circadian differences in fasting glucose levels and insulin action in WT mice such that CT7 is the phase that is different for both rhythms; arterial glucose was higher at the same phase (CT7) when the mice were less sensitive to insulin as measured using the hyperinsulinemic-euglycemic clamp (Fig. 1A, 1C)[37].
Fig. 1.
Hyperinsulinemic-euglycemic clamps on conscious, unrestrained wild-type (WT) and Bmal1 knockout (B1ko) mice at four circadian phases under constant red light. Arterial glucose levels (left panels) and glucose infusion rates (GIR, right panels) during hyperinsulinemic-euglycemic clamps for wild-type (A) and B1ko (B) mice are shown for four time points under constant red light (CT0 = subjective dawn; CT12 = subjective dusk). The light/dark and fasting protocol prior to the clamps is shown in Fig. S1.
(C, left panel) Arterial glucose levels in mice subjected to a 5-h fast (the average of -10 and 0 min prior to the start of clamps, left) during clamps for WT and B1ko mice (left panel, WT: open rectangle, B1ko: solid rectangle) at four time points. (C, right panel) Glucose infusion rates for the final 50 minutes (steady state of glucose levels) during the clamps at four circadian time points under constant red light (right panel, WT: open rectangle, B1ko: solid rectangle). The times/phases of the clamp measurements are plotted as both hours in red light and circadian time {CT}(clamp time should not be expressed in CT for B1ko mice since the clock appears to be abolished in these animals). Data are presented as mean ± SEM (WT: 6–8 mice/group, B1ko: 3–5 mice/group). Asterisks denote the phase of the WT data (CT7) that is significantly different (p < 0.05) from the other phases of the WT samples for both fasting glucose levels and GIRs as described in the text.
In contrast, in the Bmal1-knockout (B1ko) mice, one-way ANOVA analyses indicate no significant differences in the phase for either fasting glucose levels (p = 0.163) or GIR (p = 0.675), which is consistent with the interpretation of the B1ko strain as being “clockless.” Moreover, one-way ANOVA analyses indicate no significant differences between the CT7 values of WT with any of the values of the B1ko mice for GIR (p = 0.331) or fasting glucose levels (p = 0.116)(Fig. 1C). This means that the disruption of the circadian system in B1ko mice renders them relatively insensitive to insulin and locks them into a circadian phase of insulin action and glucose metabolism that is similar to that of WT mice in the inactive segment of their daily cycle (CT7). WT and B1ko are significantly different from each other; two-way ANOVA analyses of the WT and B1ko data together confirm that the data for these two strains are significantly different for both the fasting glucose levels (p < 0.0001) and for GIRs (p < 0.0001). Two-way ANOVA analysis of strain X phase interaction indicated significant difference for the fasting glucose data (p = 0.001), but not for the GIRs (p = 0.182).
Expression of Bmal2 rescues the insulin resistance and signaling deficits of B1ko
The circadian clock mechanism in mammals is composed of autoregulatory transcription & translation feedback loops of central clock gene expression. These central clock genes include the positive transcriptional factors BMAL1/BMAL2 and CLOCK/NPAS2, and the feedback repressors PER1/2/3 and CRY1/2 [38, 39, 40, 41]. BMAL1 or BMAL2 form heterodimers with CLOCK or NPAS2 and bind to E-boxes in the promoters of a large number of target genes [30, 42, 43], including their own negative regulators Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2). Knockout of the gene encoding BMAL1 (B1ko strain) abolishes the circadian feedback loop as well as the rhythms of gene expression [35]. In a previous study, we reported that expression of Bmal2 from a constitutively expressed promoter (hEF1a promoter) rescues the clock phenotypes of B1ko mice, including rhythmic locomotor activity and rhythmic oxygen consumption [30]. We concluded from those data and the original Bmal1 knockout report [35] that Bmal1 and Bmal2 form a circadian paralogous pair that is functionally redundant, but that in the mouse, Bmal2 is regulated by Bmal1 such that knockout of Bmal1 alone results in a functional knockdown of Bmal2.
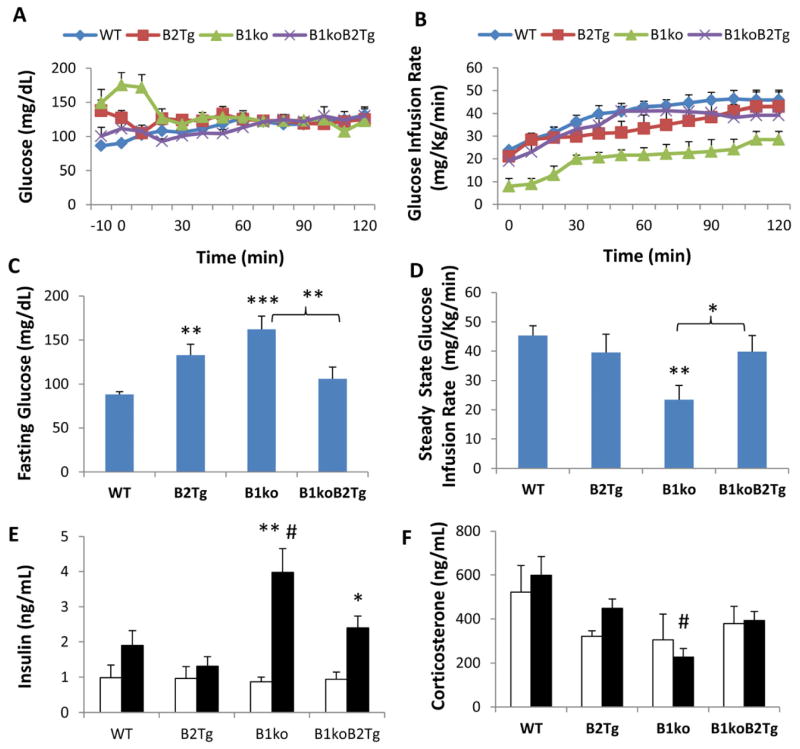
We find that transgenic expression of Bmal2 (the B2Tg mouse) not only restores rhythmicity, it also rescues the metabolic phenotypes of B1ko that are depicted in Fig. 1. Specifically, the insulin resistance and high fasting arterial glucose levels are both restored to wild-type levels in the B1ko mouse when Bmal2 is expressed from a constitutively expressed promoter (the B1ko/B2Tg mouse), as shown in Fig. 2. This rescue is also obvious from the arterial insulin levels during the clamp; the high insulin levels and low glucose infusion rate exhibited by the B1ko mice at the end of the clamp are eliminated in the B1ko/B2Tg mice (Fig. 2E). It is not clear why basal insulin levels are not high in B1ko mice (Fig. 2E), given their resistance to insulin action (Fig. 2B, 2D). A likely explanation is that beta cell function is compromised in B1ko mice resulting in inappropriately low insulin concentrations for the prevailing arterial glucose, as has been shown to be true by Marcheva et al. [44]. Moreover, the GIRs for WT vs. B2Tg are not significantly different (Fig. 2D), but they are different for fasting glucose (Fig. 2C). Because there were differences for fasting glucose but not for GIR, these data suggest that expression of Bmal2 may have some effects on glucose/insulin relationships that are not restricted to insulin action. Nevertheless, the notable result is that transgenic expression of Bmal2 overcame the elevated post-clamp insulin deficit and insulin resistance of the B1ko strain. The results shown in Fig. 2 are not due to a non-specific stress to the mutant strains. Our clamp procedure differs from other clamp procedures that involve restraint and handling, and it has been demonstrated to be stress free [33]. In this regard, it is notable that serum corticosterone levels (an indicator of stress) are low in the three mutant strains (Fig. 2F).
Fig. 2.
Hyperinsulinemic-euglycemic clamps on mice at 25 h in dim red light (= CT13 for WT, B2Tg, and B1ko/B2Tg mice). Arterial glucose levels (A) and glucose infusion rates (B) during insulin clamps for wild-type (WT), Bmal2 transgenic (B2Tg), Bmal1 knockout (B1ko), and Bmal1ko/Bmal2Tg (B1koB2Tg) mice. (C) arterial glucose levels for mice subjected to a 5-h fast (average of times -10 min and 0 min prior to the initiation of insulin infusion). (D) glucose infusion rates during the last 50 minutes of the clamps. Arterial insulin (E) and corticosterone (F) levels during the clamps (Open bars: Basal, Solid bars: Clamp period). Data are shown as mean ± SEM (4–7 mice per group), *p < 0.05, **p < 0.01, ***p < 0.001 compared with WT mice (panels C&D, one-way ANOVA with LSD), compared with basal levels (panels E&F, two-tail unpaired T test). # p< 0.05 compared among four genotypes (one-way ANOVA). Insulin (p < 0.001) but not corticosterone (p=0.557) levels between the basal and clamp conditions were significantly different among the four genotypes as analyzed by two-way ANOVA. Statistically significant differences among the four genotypes were revealed by one-way ANOVA for both clamp insulin levels (p = 0.011, high in B1ko) and clamp corticosterone levels (p = 0.016, lower in B1ko). Two-way ANOVA analyses of strain X phase interaction indicated a significant difference for insulin levels (p = 0.013), but not for corticosterone levels (p = 0.674).
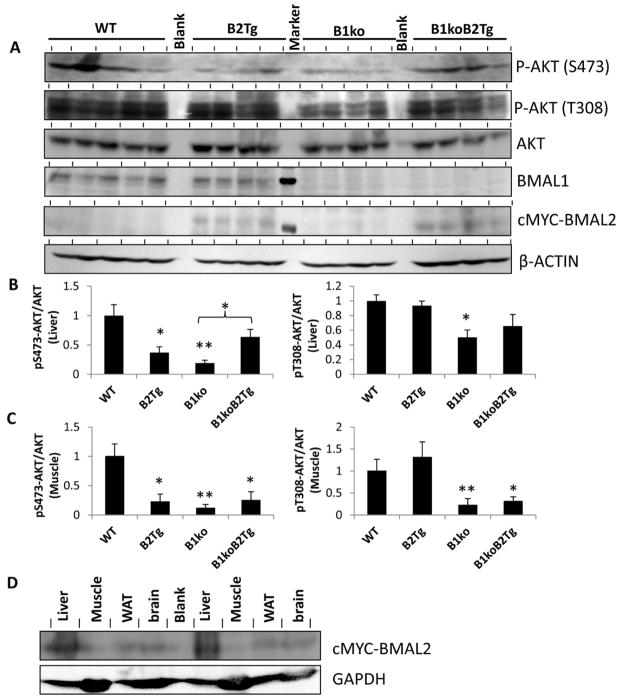
Insulin resistance is associated with the activity of the serine/threonine protein kinase Akt pathway through the PI3K/PIP3/Akt signaling cascades, and Akt and TOR signaling are also important for circadian timing [45]. The reduced GIR of the B1ko strain is correlated with attenuated activity of the Akt pathway in response to insulin action, as shown by the significantly decreased Akt phosphorylation at S473 and T308 residues in liver and muscle extracts of B1ko mice (Fig. 3). Constitutive expression of Bmal2 can reverse the attenuation of Akt phosphorylation in the liver, particularly on the S473 residue (Fig. 3B), which would be predicted to reactivate the Akt pathway. There does not appear to be a significant rescue of Akt phosphorylation in muscle extracts of the B1ko/B2Tg mice (Fig. 3C, S2). This observation is consistent with the fact that expression of BMAL2 in the B1ko/B2Tg mouse is significant in liver, brain, and white adipose tissue (WAT), but not in muscle (Fig. 3D). Moreover the low dose of insulin used during the clamp allowed us to detect any modulation of the impact of insulin on adipose tissue and liver, which are more sensitive to insulin than is muscle. It is possible that a higher dose of insulin would have allowed us to detect reversal of insulin signaling defects in muscle of B1ko/B2Tg mice. Therefore, there is a correlation between the rescue of circadian rhythmicity by Bmal2 expression in the B1ko mouse and the restoration of wild-type metabolic function.
Fig. 3.
Regulation of AKT pathway signaling by Bmal1/2. (A) Immunoblots from liver extracts for phospho-AKT (p-AKT S473 and T308), AKT (total), BMAL1, cMYC-BMAL2 (cMYC-tagged BMAL2 is the version of BMAL2 expressed in the B2Tg and B1ko/B2Tg mice, ref. 16), and β-ACTIN of mice after hyperinsulinemic-euglycemic clamps. Each lane comes from a separate mouse (n = 5 for WT, n = 4 for the other groups). The lane between the B2Tg and the B1ko samples in the BMAL1 and cMYC-BMAL2 blots shows a molecular weight standard indicating 75 kD. (B) Densitometric analyses of the data shown in panel A for liver extracts. Expression of AKT-pS473 and AKT-pT308 were normalized to total AKT. (C) Densitometric analyses of the data for muscle extracts analyzed and plotted as in panel B (see Fig. S2 for the raw immunoblot data). For panels B and C, the value of WT was set as 1.0, and values are expressed as mean ± SEM of integrated intensity. (D) Expression of cMYC-BMAL2 in various tissues of B1ko/B2Tg mice. Results from two representative mice are shown for liver, muscle, white adipose tissue (WAT) and brain tissues. The blot for cMYC-BMAL2 (upper blot) is compared with a blot for a control protein (GAPDH, lower blot). In all panels, *p < 0.05, ** p < 0.01 compared with WT or as indicated.
Clock-disrupted mice are obesity prone
In addition to insulin resistance and elevated fasting arterial glucose (Figs. 1 and 2), B1ko mice exhibit abnormal metabolic phenotypes related to fat accumulation. Young mice of the four genotypes placed on HFD do not show significant differences in total body weight after two months, but there are significant differences of body composition in the B1ko strain (Fig. 4A–C). Specifically, the B1ko mice have a significantly elevated percentage of body fat (Fig. 4A) with no significant difference in lean mass (Fig. 4B, 4C). Constitutive expression of the Bmal2 gene rescued the B1ko phenotype nearly to WT levels (Fig. 4A–C). Food intake was essentially the same among these four genotypes (Fig. 4E), but B1ko mice had significantly lower and B2Tg mice had significantly higher levels of activity as compared with WT and B1ko/B2Tg mice (Fig. 4D). The restoration of B1ko phenotypes to WT levels in the B1ko/B2Tg strain appears to be correlated with the rescue of rhythmicity and not to some other effect of Bmal2 expression, as can be observed in the B1ko/B2Tg mice. We have previously reported that the rescue of wheel-running rhythmicity in B1ko/B2Tg mice fed with regular chow is complete for some mice, but not for others [30]. We continue to observe this phenomenon of rhythmicity differences with high-fat diet (HFD) fed mice whose activity is monitored by the infrared sensor method that measures total activity more accurately than wheel running [30]. Fig. S3 depicts this phenomenon as a comparison of rhythmic B1ko/B2Tg-1 mice with arhythmic B1ko/B2Tg-2 mice. When these two groups of mice of the same genotype are separated on the basis of their rhythmicity profiles as assessed by periodogram analyses (Fig. S3), we found that the arhythmic B1ko/B2Tg-2 mice exhibited a significantly higher fat mass (p = 0.03) and lower total activity levels (p = 0.04, Table S1).
Fig. 4.
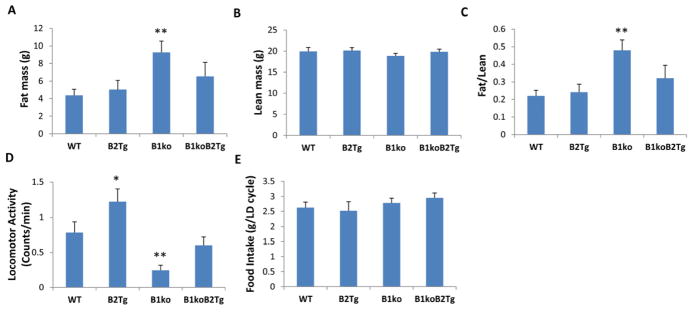
Body composition, food consumption, and locomotor activity in four different strains of mice fed a high-fat-diet (HFD). Mice were fed HFD starting at an age of one month and maintained under LD 12:12 (12 hr light: 12 hr dark, lights on 6:00 am–6:00 pm).
(A) Fat mass of these mice at age 3 months (2 months on HFD), and (B) Lean mass of these mice at age 3 months (2 months on HFD). Each bar represents mean ± SEM (n = 12–16/genotype).
(C) Ratio of fat mass to lean mass.
(D) Locomotor activity recorded by infrared sensors in these mice at 3 months of age in LD. (mean ± SEM, n = 7–10/genotype).
(E) Daily food intake during one 24 h LD cycle (mean ± SEM, n = 7–10/genotype).
*p<0.05 and **p<0.01 compared with WT mice (one-way ANOVA with LSD).
To confirm the conclusion that clock-disrupted mice are obesity prone [22,24], we tested another way to disrupt the circadian system in WT animals. In particular, WT mice can be rendered arhythmic (or their rhythms are severely disrupted) by transferring them to constant light (LL), whereas they remain strongly rhythmic in LD or constant darkness (DD) (Fig. 5D). Arhythmia induced by light exposure also strongly suppresses the expression of Bmal1 expression [46]. Therefore, we compared LL as another way to effect rhythmicity and metabolism through Bmal1 expression. As further support for the association of high body fat and low lean mass with the absence of rhythmicity, WT mice fed HFD were compared in LD 12:12, DD, or LL conditions. The body fat is significantly higher in arhythmic WT mice in LL after feeding with HFD for three months as compared with mice in LD (Fig. 5A), whereas the lean mass is not significantly altered in mice maintained in LL for three months (Fig. 5B), resulting in a significant increase of fat mass to lean mass ratio in WT mice exposed to LL (Fig. 5C). While the amount of total locomotor activity appears to be lower for mice in LL as compared with LD, this difference is not significant (Fig. 5E), and these WT mice in LL accumulated more body fat (Fig. 5A) in spite of a trend towards decreased food intake (Fig. 5F). Consequently, LL leads to disrupted rhythmicity and possibly to decreased locomotor activity, which results in fat accumulation. This phenomenon is not restricted to mice; our results confirm observations of a diabetes-prone rat strain that developed metabolic dysregulation after exposure to constant light for 10 weeks [47].
Fig. 5.
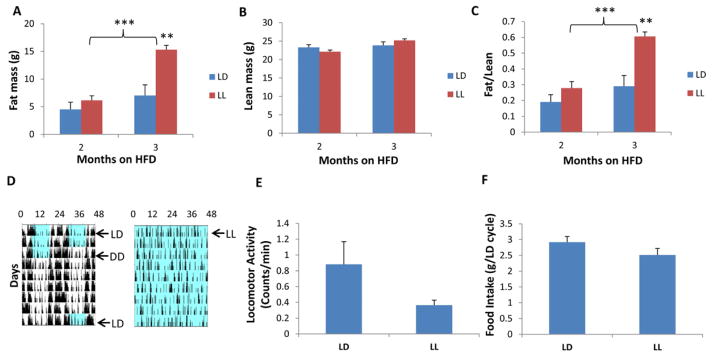
Body composition, food consumption, and locomotor activity in high-fat diet fed wild type male mice under light-dark (LD 12:12) or constant light (LL) conditions. Mice were fed RC until they were one month old, then transferred to HFD at one month under either regular LD 12:12 (lights on 6am–6pm) or LL.
(A) Fat mass, and (B) lean mass of 3 and 4 month-old WT male mice. Mice were fed with high-fat diet under either LD 12:12 (blue) or LL (red). (C) Ratio of fat mass to lean mass. Each bar represents mean ± SEM.
(D) Representative locomotor activity patterns recorded by infrared sensors. Blue shading denotes illumination, while white denotes darkness. (left panel is LD 12:12 -> DD -> LD 12:12, right panel is LL).
(E) Activity levels are expressed as mean counts/min ± SEM.
(F) Daily food intake is expressed as mean grams intake per 24 h LD cycle ± SEM.
n = 5–7 per treatment, **p<0.01 and ***p<0.001 compared with mice in LD (two-tail unpaired T test).
High fat diet feeding affects subsequent clock properties of tissues in vitro
Therefore, lack of rhythmicity is correlated with metabolic dysfunction (Figs. 1–5; [2, 6]), and the expression of BMAL2 can rescue metabolic phenotypes associated with the knockout of Bmal1 in intact mice. Feeding HFD to mice leads to changes in the period of the (i) locomotor activity rhythm and (ii) cycling of central circadian clock genes in vivo [23]. Do isolated tissues that can express circadian rhythms in vitro also show correlations between expression patterns and “after-effects” of feeding HFD to the animals from which the tissues are collected? The clockwork in the suprachiasmatic nuclei (SCN) of the hypothalamus is the “master” pacemaker that entrains to environmental cycles and coordinates rhythms throughout the body [38, 48]. Outside of the SCN, circadian molecular machinery exists and oscillates in peripheral tissues–nearly every mammalian tissue expresses circadian rhythms of clock gene expression in vivo as measured by RNA microarrays and also when luminescence reporters of gene activity are recorded from explanted tissues in vitro [10, 14, 49]. Therefore, mammalian organisms are a “clockshop” of circadian oscillators distributed throughout the body [12].
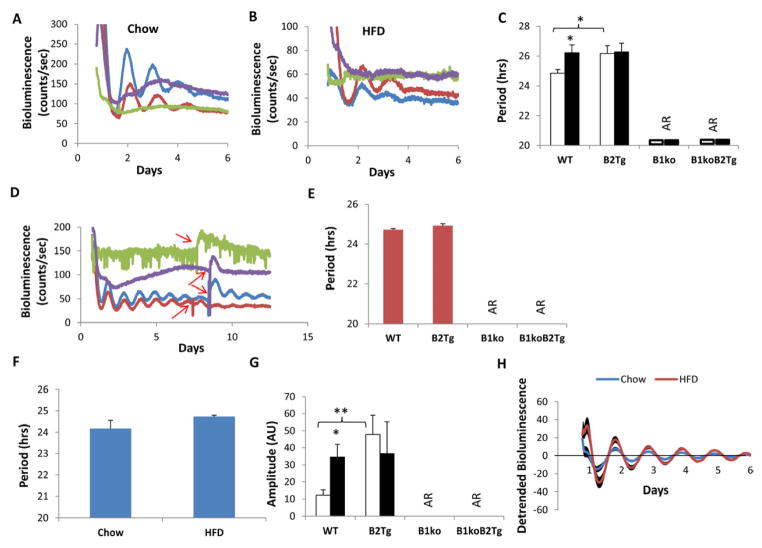
In our previous study, we reported that isolated SCN, lung, and liver slices from chow-fed animals exhibit circadian rhythms of luminescence in vitro from WT and B2Tg animals, but not from B1ko or B1ko/B2Tg mice [30]. However, damped oscillations in liver could be elicited by treatment with glucocorticoids ([30] and see Fig. S4). That study, however, did not test adipose tissue nor did it test tissues from HFD-fed mice. Fig. 6 shows that the PmPer2::mPER2-Luc reporter in white adipose tissue (WAT) from the B1ko mouse does not exhibit rhythmicity. Moreover, this was not rescued by Bmal2 expression in the B1ko/B2Tg mouse regardless of whether the mouse was fed chow (Fig. 6A) or a HFD (Fig. 6B) prior to sacrifice for the real-time luminescence assay. Therefore, even though expression of Bmal2 is able to rescue metabolic phenotypes of intact B1ko mice (Figs. 2–4), it appears to be less effective in rescuing B1ko phenotypes in isolated tissues of WAT (Fig. 6A–6B) or suprachiasmatic nuclei (SCN), lung, or liver [30]. An interesting observation, however, was an effect of HFD on the period of the Per2 rhythm in WAT for wild-type mice; the period in vitro was significantly longer in adipose tissue from WT mice that had been fed HFD as compared with chow (Fig. 6C). This effect was not observed in B2Tg mice, suggesting that constitutive expression of Bmal2 might compensate for the HFD effect on period in WAT (Fig. 6C). Nonetheless, damped in vitro oscillations can be stimulated by forskolin or glucocorticoid treatment of lung and liver of B1ko/B2Tg mice (Fig. S4 [30]).
Fig. 6.
Per2 expression measured as luminescence emanating from tissues of the PmPer2::mPER2-Luc reporter mouse [49]. WAT and SCN explants were dissected and recorded with a LumiCycle apparatus in vitro.
(A, B) Representative raw data of luminescence monitored in vitro from WAT of PmPer2::mPER2-Luc knock-in mice fed with either chow (panel A) or high-fat diet (HFD, panel B). Colors denote: WT (blue), B2Tg (red), B1ko (green) and B1ko/B2Tg (purple).
(C) Period of Per2::luc luminescence rhythms from WAT explant cultures as mean ± SEM (Chow: white, n = 6–8/genotype; High Fat Diet {HFD}: black, n = 5–7/genotype).
(D) Representative data of luminescence monitored in vitro from SCN slices of mice harboring PmPer2::mPER2-Luc and fed with high-fat diet (HFD). After 7–8 days in culture, the SCN slices were given a 0.5-hour pulse of 10 μM forskolin at the times indicated by red arrows (in vitro cultures were maintained at 36.5°C). Blue: WT, Red: B2Tg, Green: B1ko, Purple: B1koB2Tg.
(E) Period of SCN cultures from HFD-fed mice. Each bar represents mean ± SEM, n = 5–7/genotype.
(F) Period of SCN cultures from WT mice fed with chow or HFD. Each bar represents mean ± SEM, Chow: n=8, HFD: n=7.
(G) Amplitude of SCN cultures in arbitrary units (AU) from chow (open rectangle, n = 6–8/genotype) or HFD-fed mice (solid rectangle, n = 5–7).
(H) Detrended bioluminescence rhythms from cultured SCN slices of WT mice harboring PmPer2::mPER2-Luc and fed with chow or high-fat diet (HFD). Each trace represents mean ± SEM (Chow: n=8; HFD: n=7).
*p<0.05; **p<0.01 compared with chow fed mice or as indicated (2-tail unpaired T test).
The SCN of the hypothalamus are the central neural pacemaker of mammalian circadian clocks [50]. As we previously reported [30], luminescence activity from SCN slices of B1ko and B1ko/B2Tg mice was not rhythmic in vitro, and treatment with forskolin to synchronize the cells in those tissue slices did not reinitiate rhythmicity from tissues of those genotypes (Fig. 6D). In addition, although we found that feeding HFD to mice significantly lengthens the subsequent period of the Per2 rhythm from WAT cultures in vitro (Fig. 6C), we did not observe an effect of prior HFD feeding on the period of Per2 rhythms from SCN slices in vitro (Fig. 6D,E,F). However, HFD feeding does enhance the amplitude of the Per2 rhythm in WT SCN slices (Fig. 6G, H). This observation suggests that a diet-induced change in fatty acid metabolism of HFD-fed mice can alter clock properties, and this alteration persists in tissues isolated from those mice and subsequently assayed in vitro; the effect appears to be true for both SCN pacemaker neurons (amplitude but not period) and peripheral tissues such as WAT (period).
Discussion
Developed countries are facing an epidemic of interrelated metabolic diseases collectively referred to as the metabolic syndrome, the hallmarks of which include obesity, hyperlipidemia, hyperglycemia, insulin resistance, and hepatic steatosis [51]. These symptoms are all independent risk factors of Type 2 diabetes. By virtue of its pervasive control over metabolic pathways, the circadian system might be a tractable target for decreasing the prevalence of metabolic diseases, especially since the circadian system serves as the interface between internal rhythms and the cycles of the external world (light/dark, food availability, etc.). Indeed, studies in both mice and humans indicate that relatively non-invasive manipulations of the circadian patterns of activity and feeding can have significant effects on body weight and metabolism [3, 9, 22, 52]. The results reported here conclusively demonstrate the timing of insulin action is key; this conclusion is highly relevant for human vulnerability to hyperglycemia and hypoglycemia, and could alter strategies for insulin therapy.
While the fundamental concept of homeostasis would predict that insulin activity and glucose levels would be constant over the 24 h day, our data clearly show that mice are relatively insulin resistant in the middle of their subjective day (CT7), i.e., during the inactive portion of the daily cycle for mice (Figs. 1 and S1). Moreover, fasting glucose shows a circadian rhythm with the highest levels occurring during the insulin-resistant phase of inactivity. The “clockless” B1ko mice appear to be locked arhythmically into this insulin-resistant phase, as they demonstrate non-rhythmic insulin-resistance and enhanced fasting glucose levels. When circadian rhythmicity is rescued in the B1ko mouse by transgenic expression of the Bmal1 paralog, Bmal2 [30], the metabolic phenotypes of insulin resistance, fasting glucose levels, Akt signaling, and fat accumulation are concomitantly rescued (Figs. 2–4). Another way to induce arhythmicity and suppress Bmal1 expression–exposure to LL–also lowers total activity and stimulates fat accumulation (Fig. 5).
The influence of HFD on metabolism not only affects behavioral rhythms in [23], but it also alters molecular clock properties in WAT and SCN by a mechanism that persists for at least several days in these tissues when isolated and assayed in vitro (Fig. 6). Moreover, the rescue of metabolic phenotypes by expression of the Bmal2 gene may be mediated by the partial restoration of circadian rhythmicity in peripheral tissues such as liver and lung. In particular, the damped oscillation capability in the isolated tissues of the B1ko/B2Tg mouse (Figs. 6, S4 [30]) are likely to return to full-blown rhythmicity in vivo by virtue of systemic signals in the intact animal that synchronize and amplify the rhythms of peripheral tissues [53]. This is shown in Figure S4A, where cyclic cAMP signals elicited by forskolin restore robust rhythmic gene expression to fibroblasts (MEFs) derived from the B1ko/B2Tg mouse but not to fibroblasts from the B1ko mouse. Because B1ko/B2Tg-1 mice express rhythmic locomotor activity, they are likely to have rhythmic systemic signals so that peripheral tissues are also rhythmic and metabolism is rescued. On the other hand, B1ko mice do not express rhythmic behavior and are therefore B1ko peripheral tissues may not experience rhythmic systemic signals, resulting in metabolic defects.
The data reported here are consistent with a model derived from studies of epigenomic regulation of metabolic switching in mice [26]. During the night (active/feeding phase for mice), metabolic intermediates are largely channeled to lipid synthesis and storage, and we find this phase to be characterized by enhanced insulin activity and lower fasting glucose (Fig. 1). Conversely, during the day (inactive/fasting phase for mice), metabolism switches into a calorie-restricted, glucose-producing mode that is characterized by a higher fasting glucose and insulin resistance. Clockless B1ko mice appear to be locked in a metabolic state that resembles the day-phase of WT mice. This metabolic day/night switching may be accomplished in WT mice by circadian regulation of the histone deacetylase HDAC3 [26].
Our results have important implications for studies of insulin action in mice. In particular, we show that insulin action as measured by hyperinsulinemic-euglycemic clamps, performed in the absence of stress (i.e., mice are not handled, restrained, or have their tails cut) varies over the time span of the 24 h day, confirming and extending some previous studies of the interaction between circadian systems and metabolism where stress of handling and blood sampling might have been a factor [28, 29, 31,54]. Our current studies emphasize the need to apply precise control to the timing of metabolic experiments and to consider timing in the interpretation of results. Some of the variance of results amongst published studies that have been ascribed to diet, mutant background, etc., could be at least partially due to differing times of day when the hyperinsulinemic-euglycemic clamps were performed [54].
Because most previous hyperinsulinemic-euglycemic clamp studies of mice have been done in the day phase, it is possible that gene-dependent effects on insulin action and glucose metabolism that occur during the night phase have been missed. Therefore, the conclusions of this investigation indicate that results obtained from hyperinsulinemic-euglycemic clamps and other measurements of insulin action and glucose tolerance need to be considered in the context of the time of day/circadian phase to facilitate comparison to other studies. It would be most comprehensive to test insulin action and glucose tolerance at different times of day, e.g., mid-day and mid-night to improve the chance of capturing clock-dependent effects on metabolism.
From the work of Claude Bernard in the 19th century, the concept of homeostasis (Bernard’s milieu intérieur) as the maintenance of a constant internal environment is deeply ingrained in our thinking about how organisms work. The question then arises why is such a fundamental component of internal environment as glucoregulation rhythmic rather than constant? It is likely to be an inevitable and unavoidable consequence of evolving in an external environment that is profoundly rhythmic over the course of the daily cycle. Consequently, an organism whose responses are merely constant will be out-competed by organisms that anticipate the predictable changes in their external environment by rhythmically preparing and altering their internal environment. To optimally adapt to a rhythmic environment, organisms must rhythmically regulate their behavior, physiology, and gene expression [55]. These studies show that insulin action and glucose metabolism are interwined with an internal timekeeping system that evolved to accommodate the environmental rhythmicity.
Experimental Procedures
Complete experimental procedures are described in the Supplemental Experimental Procedures.
Animal care
Mice were housed on a 12:12-h light-dark cycle unless otherwise stated and fed chow (5001; Purina Mills, St. Louis, MO) containing 13.5% calories from fat. For high-fat diet (HFD, F3282; BioServ, Frenchtown, NJ) experiments, mice were fed HFD containing 60% calories as fat, beginning at one month of age. All animal experiments were approved by the Vanderbilt University Institutional Animal Care and Use Committee and were conducted according to that committee’s guidelines.
Hyperinsulinemic-euglycemic clamp
Detailed clamp procedures have been previously reported [33, 34] except for the entrainment and circadian procedures shown in Figure S1 that are unique to this study.
Determination of Akt phosphorylation by immunoblotting
Samples of liver and of vastus muscle harvested from mice of the CT13 group at the end of the hyperinsulinemic-euglycemic clamp procedure were processed for immunoblotting and probed for total Akt, phosphorylated Akt (S473 and T308), MYC-BMAL2, BMAL1, β-ACTIN, and GAPDH.
Locomotor behavior, body weight/fat composition, and food intake assays
Mice were fed HFD beginning at one month of age. After two months on HFD, some mice were singly housed in cages equipped with infrared sensors. Locomotor activity and food intake of the mice was measured at 2 months on HFD. Body fat composition was determined with an mq10 nuclear magnetic resonance analyzer (Bruker Optics).
Tissue culture and in vitro luminescence recording
The mice for tissue culture were fed HFD for 3 to 4.5 months or they were fed chow. The cultures of SCN and peripheral tissues were prepared as previously described [30, 49].
Statistics
Data are expressed as means ± SEM. Statistical analyses were performed by two-tail unpaired T test, one-way, and two-way ANOVA as indicated.
Supplementary Material
Highlights.
Insulin action shows a bona fide circadian rhythm
Mice are most resistant to insulin during their daily phase of relative inactivity
Disruption of circadian clocks predisposes animals to insulin resistance and obesity
Function of insulin sensitive tissue depends upon the rhythmic internal environment
Acknowledgments
We thank Dr. Julio Ayala for advice during the design of these experiments, Dr. Holly Resuehr for help with infrared recording of circadian locomotor activity. and Dr. David McCauley for statistical advice. We also thank Carlo Malabanan, MerryGay James, and Alicia Lorance of the Vanderbilt Mouse Metabolic Phenotyping Center for assistance and advice. The antibody to c-Myc was a gift of Dr. Heping Yan (Washington University Medical School) and the antibody to BMAL1 was a gift of Dr. Charles Weitz (Harvard University Medical School). Vanderbilt’s Mouse Metabolic Phenotyping Center was supported by the NIDDK (grant # DK059637) and this grant was additionally supported by grants to CHJ from the NIDDK (DHHS/NIH/NIDDK #5U24 DK076169-04, Subward # 20497-34), the NHLBI (R21HL102492-01A1), and Vanderbilt University’s Diabetes Research Training Center (2P60DK020593). Finally, SQS was partially supported by NARSAD Young Investigator Award # 17623.
Footnotes
Conflict of Interest Statement:
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer Associates, Inc; 2004. [Google Scholar]
- 2.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimble JM, Sutton GM, Bunnell BA, Ptitsyn AA, Floyd ZE. Prospective influences of circadian clocks in adipose tissue and metabolism. Nat Rev Endocrinol. 2011;7:98–107. doi: 10.1038/nrendo.2010.214. [DOI] [PubMed] [Google Scholar]
- 5.Antunes LC, Levandovski R, Dantas G, Caumo W, Hidalgo MP. Obesity and shift work: chronobiological aspects. Nutr Res Rev. 2010;23:155–68. doi: 10.1017/S0954422410000016. [DOI] [PubMed] [Google Scholar]
- 6.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Cauter E, Polonsky KS, Scheen AJ. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev. 1997;18:716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- 8.Knutson KL, Ryden AM, Mander BA, Van Cauter E. Role of sleep duration and quality in the risk and severity of type 2 diabetes mellitus. Arch Intern Med. 2006;166:1768–74. doi: 10.1001/archinte.166.16.1768. [DOI] [PubMed] [Google Scholar]
- 9.Buxton OM, Cain SW, O’Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA, Shea SA. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4:129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 13.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–20. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Woon PY, Kaisaki PJ, Bragança J, Bihoreau MT, Levy JC, Farrall M, Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104:14412–7. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32:658–62. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 18.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spégel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–8. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, Haim A, Nelson RJ. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A. 2010;107:18664–9. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci U S A. 2011;108:1657–62. doi: 10.1073/pnas.1018375108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity. 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and Metabolic Syndrome in Circadian Clock Mutant Mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int. 2010;27:1317–28. doi: 10.3109/07420528.2010.489166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010;20:316–21. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–4. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, Tezuka M, Kosuge Y, Ishige K, Ito Y, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1) induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6:e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayala JE, Bracy DP, McGuinness OP, Wasserman DH. Considerations in the design of hyperinsulinemic-euglycemic clamps clamps in the conscious mouse. Diabetes. 2006;55:390–7. doi: 10.2337/diabetes.55.02.06.db05-0686. [DOI] [PubMed] [Google Scholar]
- 34.Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, Wasserman DH, McGuinness OP. NIH Mouse Metabolic Phenotyping Center Consortium. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3:525–34. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3/Bmal1 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lall GS, Revell VL, Momiji H, Al Enezi J, Altimus CM, Güler AD, Aguilar C, Cameron MA, Allender S, Hankins MW, et al. Distinct contributions of rod, cone, and melanopsin photorecptors to encoding irradiance. Neuron. 2010;66:417–28. doi: 10.1016/j.neuron.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molusky MM, Li S, Ma D, Yu L, Lin JD. Ubiquitin-Specific Protease 2 Regulates Hepatic Gluconeogenesis and Diurnal Glucose Metabolism Through 11β-Hydroxysteroid Dehydrogenase 1. Diabetes. 2012;61:1025–35. doi: 10.2337/db11-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 39.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–82. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 40.Lowrey PL, Takahashi JS. Mammalian Circadian Biology: Elucidating Genome-Wide Levels of Temporal Organization. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripperger JA, Jud C, Albrecht U. The daily rhythm of mice. FEBS Lett. 2011;585:1384–92. doi: 10.1016/j.febslet.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 42.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 43.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 44.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–8. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grone BP, Chang D, Bourgin P, Cao V, Fernald RD, Heller HC, Ruby NF. Acute light exposure suppresses circadian rhythms in clock gene expression. J Biol Rhythms. 2011;26:78–81. doi: 10.1177/0748730410388404. [DOI] [PubMed] [Google Scholar]
- 47.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms. 2011;26:423–33. doi: 10.1177/0748730411416341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver R, Schwartz WJ. The Suprachiasmatic Nucleus is a Functionally Heterogeneous Timekeeping Organ. Methods Enzymol. 2005;393:451–65. doi: 10.1016/S0076-6879(05)93022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Beydoun MA, Liang L, Caballero B, Kumanyika SK. Will all Americans become overweight orobese? estimating the progression and cost of the US obesity epidemic. Obesity. 2008;16:2323–30. doi: 10.1038/oby.2008.351. [DOI] [PubMed] [Google Scholar]
- 52.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JAJ, et al. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab. 2012;15:848–60. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGuinness OP, Ayala JE, Laughlin MR, Wasserman DH. NIH experiment in centralized mouse phenotyping: the Vanderbilt experience and recommendations for evaluating glucose homeostasis in the mouse. Am J Physiol Endocrinol Metab. 2009;297:E849–55. doi: 10.1152/ajpendo.90996.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson CH, Kyriacou CP. Clock evolution and adaptation: whence and whither? In: Hall AJW, McWatters HG, editors. Endogenous Plant Rhythms. Oxford, UK: Blackwell Publishing Ltd; 2005. pp. 237–260. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.