SUMMARY
Selecting a suitable membrane-mimicking environment is of fundamental importance for the investigation of membrane proteins. Non-conventional surfactants, such as amphipathic polymers (amphipols) and lipid bilayer nanodiscs, have been introduced as promising environments that may overcome intrinsic disadvantages of detergent micelle systems. However, structural insights into the effects of different environments on the embedded protein are limited. Here we present a comparative study of the hepta-helical membrane protein bacteriorhodopsin in detergent micelles, amphipols and nanodiscs. Our results confirm that non-conventional environments can increase stability of functional bacteriorhodopsin, and demonstrate that well-folded heptahelical membrane proteins are in principle accessible by solution-NMR methods in amphipols and phospholipid nanodiscs. Our data distinguishes regions of bacteri-orhodopsin that mediate membrane/solvent contacts in the tested environments whereas the protein’s functional inner core remains almost unperturbed. The presented data allow comparing the investigated membrane mimetics in terms of NMR spectral quality and thermal stability required for structural studies.
INDRODUCTION
The in vitro investigation of membrane protein structure and function is inherently related to the choice of a suitable membrane-mimicking environment. While great progress has been made in the elucidation of membrane protein structure in detergent micelles using solution-state NMR (Kang and Li, 2011; Kim et al., 2009; Nietlispach and Gautier, 2011), detergents are often detrimental to protein structure and may not (fully) support its functional form, in particular if soluble domains are present that may be unfolded by detergents. Non-conventional surfactants such as amphipathic polymers (amphipols) (Popot et al., 2011) or lipid bilayer nanodiscs (Denisov et al., 2004) have lately received increased attention as promising tools for the investigation of membrane proteins (Gorzelle et al., 2002; Raschle et al., 2010; Zoonens et al., 2005). Advantages of using non-conventional membrane mimetics include the exceptionally good refolding properties of amphipols for heptahelical membrane proteins such as G protein-coupled receptors (GPCRs) (Dahmane et al., 2009) or the absence of detergent as well as the more native-like environment provided by nanodiscs. Additionally it has been demonstrated that the use of non-conventional surfactants can increase protein stability and improve the accessibility of the functional form (Popot, 2010). Initial NMR studies on β-barrel proteins indicate that amphipols (Zoonens et al., 2005) as well as nanodiscs (Gluck et al., 2009; Raschle et al., 2009; Shenkarev et al., 2009) may also be useful for NMR structural studies of polytopic membrane proteins. However, recent results obtained for a class A GPCR suggest that high-resolution solution-state NMR structural studies of heptahelical membrane proteins in nanodiscs are restricted to the extramembranous part of the protein (Park et al., 2011). Moreover our fundamental understanding of the effects that different membrane mimetics have on protein structure and dynamics is still very limited.
The heptahelical transmembrane protein bacteriorhodopsin (bR) offers ideal biophysical properties such as molecular size, topology, stability as well as its characteristic color (indicative of intact tertiary structure) to study the effects of different membrane mimetics on its structure and stability. As a light-activated proton pump bR consists of two moieties, the 27 kDa protein bacterioOpsin (bO) and the retinal, a vitamin A metabolite, covalently bound to a lysine side chain of bO. Due to its exceptionally high abundance as part of the purple membrane in Halobacterium salinarum, structural studies of bR so far have been solely carried out on the functional protein extracted from its native environment. The bR structure itself has been extensively studied, in particular by x-ray and electron crystallography (Edman et al., 1999; Hirai and Subramaniam, 2009; Luecke et al., 1998; Luecke et al., 1999a, b; Oka et al., 2000), as well as solid-state NMR (Bajaj et al., 2009; Engelhard et al., 1989; Harbison et al., 1983; Harbison et al., 1984; Saito et al., 2000). Additionally, a partial resonance assignment of bR in n-dodecyl-β-D-maltopyranoside (DDM) micelles using solution-state NMR techniques has been published (Schubert et al., 2002). While extraction of/from the native purple membrane is a very powerful strategy, it has the downside that cofactors such as the covalently bound ligand or coordinated lipids are often co-purified (Patzelt et al., 1997). The latter will interfere with the measurements of effects of different membrane-mimicking environments on protein structure and function by introducing a bias most likely towards the native conformation. Heterologous cell-free protein expression offers an interesting option for the production of “co-factor free” bO (Cappuccio et al., 2008).
In the following we present a comparative study of bR in DDM micelles, A8-35 amphipols (Popot et al., 2011) as well as in lipid bilayer nanodiscs formed using 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipids and the membrane scaffold protein MSP1D1 (Denisov et al., 2004). To avoid artifacts originating from imprecise starting conditions (i.e. coordinated lipids) we used cell-free methods (Cappuccio et al., 2008; Schwarz et al., 2007) to express bO in the absence of detergents, lipids or lig-and. Refolding of the protein in a well-defined setting hence enabled a direct evaluation of specific features of the different membrane mimetics. Moreover the in vitro expression simplified the production of (selectively) isotope-labeled samples. Our results confirm that non-conventional surfactants can increase membrane protein stability, and provide a first experimental reference of NMR accessibility of the hep-tahelical transmembrane protein bR in amphipols and nanodiscs. While the presented approach may serve as a (favorable-case) reference for future studies of membrane proteins including GPCRs, the obtained NMR insights which report on changes in chemical environment and/or on protein structure additionally may help to understand the effects of different membrane mimetics on the embedded bR proteins.
RESULTS AND DISCUSSION
Effects of (cofactor free) in vitro expression
Initially we compared the resonance assignments of bR in DDM micelles extracted from the native membrane (Schubert et al., 2002) to the cell-free expressed and refolded bR (also in DDM micelles) (see Figure S1). The close similarity of the observed peak positions strongly indicates that our refolded protein closely resembles the tertiary structure of the membrane-extracted protein and allows a direct transfer of most peak assignments (Schubert et al., 2002). Interestingly a cluster of noticeable chemical shift alternations is found for residues around Lys 172 (Figure 1 and Figure S1). In the crystal structure (Luecke et al., 1999b) direct interactions of the Lys 172 and Val 173 side chains to the head groups of (co-crystallized) lipids are found. Since these lipids are absent in the in vitro expression system our data suggest that specific lipids are co-purified when bR is extracted with detergents from the purple membrane (e.g. lipid 1 and/or lipid 2 facing helix F-G but most likely not lipid 3 facing helix A–G, Figure 1). This demonstrates the importance of using a well-defined system, such as offered by in vitro expression, to enable a bias-free investigation of the effects of different membrane mimetics on bR structure and stability.
Figure 1.
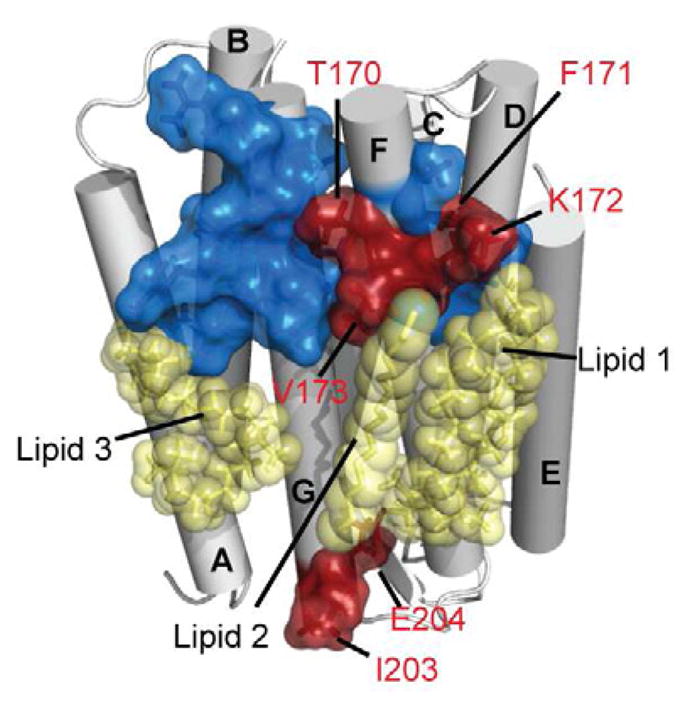
Differences between bR in DDM micelles when extracted from the native purple membrane (Schubert et al., 2002) or when refolded after cell-free expression. Residues in the transmembrane region which experience chemical shift alterations due to the different sample preparation are highlighted in red (on the crystal structure (Luecke et al., 1999b)). Blue residues do not display any significant difference. Lipids (fragments) as present in the crystal are shown in yellow. See Figure S1 for experimental data and more details.
Biophysical properties of the different membrane mimetics
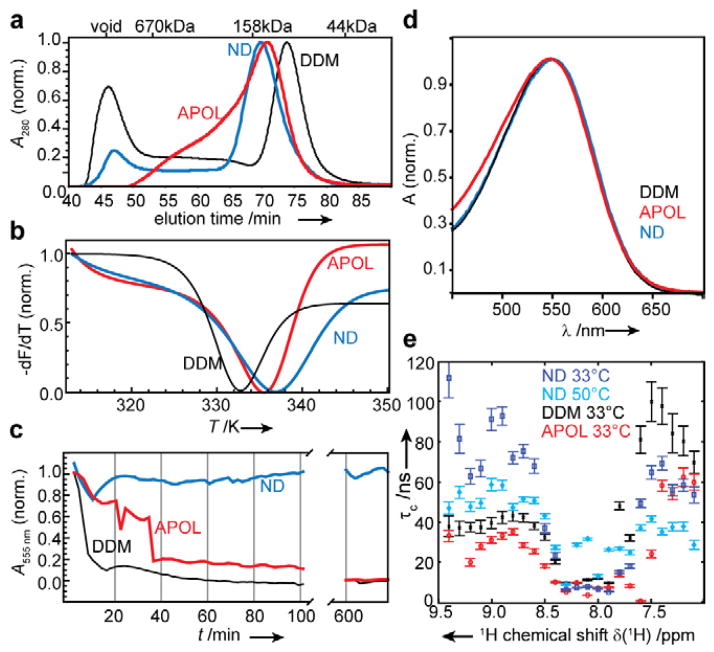
Figure 2 summarizes biophysical properties of cell-free expressed bR in the tested environments. Size exclusion chromatography (SEC) clearly indicates that the particle size of bR in detergent micelles (bR-DDM) is significantly smaller (about 60%) than in the amphipol (bR-APOL) and nanodiscs (bR-ND) environments, which themselves are comparable in size (Figure 2a). However, bR-ND has a more homogenous size distribution than bR-APOL. Interestingly thermal denaturation, measured by thermofluor assays (Matulis et al., 2005), shows that the micelle offers the least stable environment for bR (Figure 2b). We also measured the lifetimes of functional bR by time-resolved absorption spectroscopy at 58°C (Figure 2c). It is clearly evident that the functional state of bR is best preserved in the lipid bilayer nanodisc environment (t1/2@58°C > 14 h), followed by the amphipols ((t1/2@58°C = 32 min) and the detergent micelles (t1/2@58°C = 8 min). This finding is supported by the observation that long-time storage (several months) at 5°C led to significant reduction of absorption at 550 nm for bR-DDM samples while bR-APOL and bR-ND samples did not show any decay during this time (data not shown). The absorption profile of bR in the different environments is highly similar (Figure 2d), indicative of a largely conserved protein core. In addition a shift in absorption maximum between the dark and light adapted state (an essential prerequisite of the protein’s function) could be detected for all environments (see Figure S2). Table 1 summarizes the derived biophysical properties.
Figure 2.
Biophysical properties of cell-free expressed bR refolded into different soluble membrane-mimicking environments. a) Size exclusion chromatograph of bR in DDM micelles (bR-DDM, black), in A8-35 amphipols (bR-APOL, red) and in DMPC lipid bilayer nanodiscs (bR-ND, blue). b) Thermal denaturation curves of bR in the same environments, measured by fluorescence intensity of Sypro Orange (Sigma-Aldrich). First derivatives of fitting curves are shown (see Figure S2 for full experimental data). c) Lifetimes of bR in different environments as measured by time-resolved absorption spectroscopy at a wavelength of 550 nm (indicative of intact tertiary structure) at 58°C. d) Absorption profile of bR in the different environments. To enable a better comparison, data in a) - d) were normalized (also see Figure S2 for experimental data on light/dark adaptation). e) NMR analysis of bR rotational correlation times τc as a function of proton resonance frequency in indicated environments and temperatures. Values were determined using the TRACT experiment (Lee et al., 2006).
Table 1.
Overview of biophysical and NMR spectroscopic properties of bR in different membrane mimetics.
| TM[a]/°C | t1/2@58°C[b]/h | τC[c]/ns | MW NMR | MW SEC | |
|---|---|---|---|---|---|
| DDM | 59 | 0.13 | 41.4 | 128 | 108 |
| Amphipol | 62 | 0.53 | 31.7[d] | 93[d] | 151 |
| Nanodisc | 64 | > 14 | 85.6 55.8[e] |
287 180[e] |
166 |
Melting temperature as determined by thermofluor assay.
Half-life of functional bR as determined by time-resolved absorption spectroscopy at 58°C.
As determined by TRACT analysis of proton region 8.7–9.1ppm at 33°C.
Note that the TRACT data of bR-APOL may not be representative due to effects of sample heterogeneities (see text for more details).
Data recorded at 50°C.
To probe the NMR accessibility of bR in the different environments we measured rotational correlation times using the TRACT (Lee et al., 2006) experiment. We found a very variable set of peak intensities within each environment suggesting the presence of large variations in local protein dynamics. To investigate this dynamic range in more detail we calculated the correlation times of bR as a function of proton frequency (Figure 2e). This dissection easily exposes a considerable dynamic range in all tested environments and suggests that great care should be taken when reporting on the overall correlation time of the particle. In particular for α-helical proteins, which in general show only small 1H chemical shift differences between signals from loop and α-helical secondary structures, an analysis as shown in Figure 2e may therefore be preferable. Notably, the correlation times determined for bR-APOL are significantly shorter than expected from the SEC, which we attribute to the heterogeneous environment of the amphipols and the intrinsic tendency of the TRACT analysis to generate lower τc values if fast and slowly relaxing species overlap in the spectrum. Indeed additional data recorded on selectively labeled bR-APOL, which allows determination of a small subset of residue-specific rotational correlation times, show larger τc values for residues within the helical core in bR-APOL (see Figure S3). Making use of the excellent thermal stability provided by the nanodiscs, TRACT measurements at 50°C (Figure 1e, light blue) show a clear reduction of τC values as well as the dynamic range, thus improving the NMR accessibility considerably.
Interestingly, previous NMR studies, in which bR was extracted from the native membrane and transferred to DDM micelles, reported a higher stability in this environment than found here (Brouillette et al., 1989; Patzelt et al., 1997; Schubert et al., 2002). This may be explained by differences in buffer conditions as well as the absence of coordinated lipids in our preparation. The latter may suggest that detergent-lipid mixtures may be more effective in stabilizing membrane proteins. As an alternative to screening a large set of different detergents and detergent-lipid mixtures one may consider the use of am-phipols, which may simplify the screening due to their intrinsic heterogeneity that potentially mimics a broad range of conditions while only using one compound.
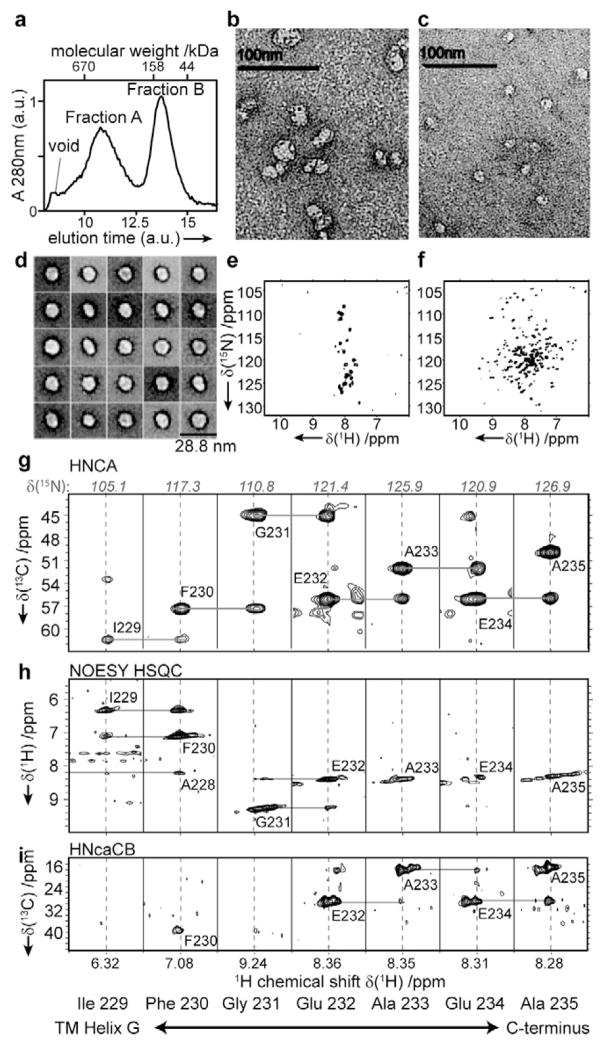
In our hands amphipol refolding was straightforward, and unlike the detergent and nanodiscs refolding protocols did not require thorough optimization of refolding conditions. This corroborates previous findings that amphipols provide excellent refolding properties for heptahelical membrane proteins (Dahmane et al., 2009) and suggests that amphipols may be particularly interesting for systems where refolding by other methods fails. Still, the heterogeneity of the amphipols may be disadvantageous for further analysis including NMR studies. It could hence be beneficial to transfer the amphipol-refolded protein into a different environment before NMR data acquisition. Here the preferred target systems may include detergent micelles for better spectroscopic properties or lipid bilayer nanodiscs for a more native and stable environment. Transfer (see e.g.(Popot et al., 2011) ) of bR from amphipols into detergent micelles as well as into nanodiscs was well achievable. However, under the conditions tested, amphipol-refolded bR transferred into nanodiscs showed the occurrence of a second species characterized by a significantly higher particle radius. Notably these species were also present after (not-optimized) refold-ing and when bR was directly incorporated into nanodiscs during cell-free expression (Figure 3a). Their occurrence largely depends on the lipid-to-MSP1D1 ratio and has been observed before (Bayburt et al., 2006; Cappuccio et al., 2008). EM images of negatively stained preparations show that the additional species consists of larger discs as well as of clusters of regular-sized discs (Figure 3b). NMR spectra of this species (Figure 3e) only show a subset of resonances (i.e. predominantly from C-terminal residues). Still both species represent correctly folded bR as evident by the characteristic color of the samples. Hence the limited dispersion found in the spectrum of fraction A is not indicative of unfolded protein but indicates that the size-limit of conventional solution-state NMR methods has been reached. Therefore, if high-resolution NMR studies of proteins in nanodiscs should be carried out, sample preparation has to be carefully optimized, and only the smallest species should be selected for data acquisition. Here, this species exhibits a well-dispersed 1H-15N-correlated TROSY-HSQC spectrum (Figure 3f). In line with the well-resolved NMR signal, negative-stain EM images of the smaller species (Figure 3c and d) show homogenous nanodiscs with a diameter of about 10 nm. Noteworthy our data also illustrate that functionality of a polytopic membrane protein in nanodiscs cannot be excluded solely based on a badly dispersed NMR spectrum.
Figure 3.
Characterization of bR-ND. a) Size exclusion chromatography of bR directly incorporated into nanodiscs during cell-free expression. Representative negative-stained EM images of fraction A (b) and of fraction B (c). d) Representative class averages obtained with fraction B (see Figure S3 for full set). TROSY-HSQC NMR spectra of fraction A (e) and fraction B (f). g–i) Set of 3D TROSY NMR data recorded on bR in DMPC nanodiscs after optimization of refolding conditions. g) HNCA, h) NOESY-HSQC, i) HNcaCB). Strip plots for residues from the end of helix G towards the C terminus are shown. 3D spectra were recorded at 48°C. (See supporting information Figure S3 for more details.)
NMR spectroscopic properties
Approximately 90% of the expected peaks of bR in nanodiscs could be resolved in a TROSY based HNCO spectrum (see Figure S3 for a more detailed analysis). This suggests that even the transmembrane region of a well-folded and well-behaving heptahelical membrane protein in nanodiscs is accessible by solution-state NMR. Figures 3g–i display strip plots of a set of conventional TROSY based 3D experiments recorded on bR-ND. The selected region shows the transition from the structured region of bR (end of helix G) to the unstructured C terminus. This transition can be clearly followed in the spectra by the decreased chemical shift dispersion (Figures 3g,h) and the absence of sequential (as well as i - i+2) NOE peaks (Figure 3h) for residues subsequent to Glu 232. While the spectra confirm that most resonances of the protein backbone can be resolved in the nanodisc environment, Figure 3i also clearly demonstrates that signal intensities in 3D experiments involving an INEPT carbon-carbon magnetization transfer step, such as found in Cα-Cβ (Figure 3i) or CO-Cα experiments, are strongly reduced with increased structural order. Indeed no Cβ signal could be detected for any of the residues expected in the transmembrane region. For a protein the size of bR, however, these chemical shift measurements are critical for a residue-specific assignment. Our data suggest that residue assignments in the non-conventional systems may hence also benefit from alternative approaches such as combinatorial labeling (Hefke et al., 2011) or carbon detection (Bermel et al., 2010; Richter et al., 2010).
Effects of different environments on the bR structure
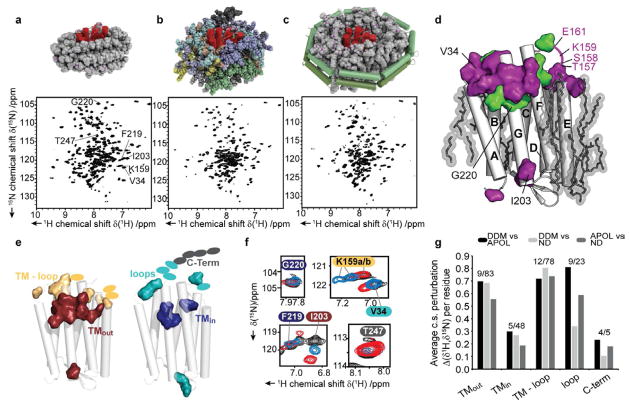
Figure 4 compares 2D TROSY-HSQC spectra of bR in the three different environments (see Experimental Procedures for more information on isotope labeling and NMR parameters). As anticipated from the smaller particle size of the detergent micelles, the spectrum of bR-DDM (Figure 4a) shows best resolution and sensitivity. However, bR-APOL (Figure 4b) and bR-ND (Figure 4c) also give decent spectra with a sizable number of resolved peaks at 40°C (bR-DDM = 216; bR-APOL = 178; bR-ND = 150). Due to the applied labelling in total 216 residues should be visible in the spectrum; however peak splitting and water exchange will increase or decrease the number of expected peaks. Note that the spectrum of bR-ND at 50°C (Figure S3) contains 209 resolved peaks. The relatively good spectral quality in all tested environments enables a direct chemical shift comparison for several residues, indicative for structural modifications or direct interactions with the environment. Using the previously published partial resonance assignment of bR in DDM micelles (Schubert et al., 2002) in combination with our 3D spectra as well as several selectively labeled samples, a set of chemical shift changes can be readily identified. Although the subset of residues for which such a sequential assignment can be obtained in all environments is limited (39 in total, see supporting information SI6 for complete list), our data indicate that the residues in the protein core region are less affected by the changes in the environments (Figure 4d, green). In contrast many residues in the loop regions experience strong chemical shift alterations in different environments (Figure 4d, purple).
Figure 4.
Effects induced by different membrane-mimetic environments on local bR structure. a-c) Illustration of bR in the different environments and corresponding TROSY-HSQC NMR spectra. All spectra were recorded at 40°C on cell-free expressed and refolded bR. The illustration in a) shows bR (red) in micelles with 126 detergent molecules. In b) 8 amphipol molecules are shown in different shading. The nanodisc shown in c) is comprised of 100 DMPC molecules and two copies of MSP1D1 (green). (Note that the shown particles are drawn to scale to illustrate the different environments and are not energy minimized.) d) Overall chemical shift perturbations for assigned residues mapped on the structure of bR (green: no perturbation, purple: changes in at least one environment). Lipids as identified in the crystal structure (Luecke et al., 1999b) are shown in transparent grey. (See supporting information Figure S4 for complete residue-specific, pairwise analysis). e) Classification of regions interacting differently with the surrounding membrane mimetic. Only residues for which chemical shift changes could be assigned in all environments (39 in total) are highlighted. Residues not present in the x-ray structure are indicated as ellipses in matching colors. f) Example of selected peaks for all classes (colors of residue labels indicate the respective classes shown in e); the color code for the NMR spectra is: black=bR-DDM; blue=bR-ND; red=bR-APOL). g) Average chemical shift perturbation per residue for the different classes. Numbers above bars indicate the amount of assigned/total residues per class. The pairwise differences between the three environments are shown separately.
To investigate the effects of the different environments in more detail, we classified the assigned residues according to their expected interaction with the surfactant, based on their location in the bR structure (Luecke et al., 1999b) (Figure 4e, see Figure S4 for more details on class selection). For example, residues within the transmembrane region and with surfactant-facing side chains (TMout) are likely to directly interact with the different environments. Indeed while larger chemical shift changes are observed for the assigned residues in this class, the residues assigned within the transmembrane region with protein-facing side chains (TMin) are much less affected (Figure 4g). These results indicate that no considerable structural rearrangements occur on the backbone level within the transmembrane helices and that the different environments affect outward-facing residues the most which then effectively shield the protein core. Note that our chemical shift data would be consistent with the view that bR has a very stable core that is not perturbed by the different tested environment and which hence may force the environment to adapt and provide an adequate hydrophobic coverage. Larger chemical shift perturbations are also found for residues on the direct TM–loop interface. These residues are mostly sensitive to changes in the detergent/lipid headgroup region as well as to differences in length of hydrophobic coverage. The observed chemical shift changes indicate a pronounced exposure of these residues to the different membrane environments. The observed chemical shift differences report either on direct interactions with the environment or on structural changes of the protein. Interestingly, the chemical shift deviations found in the TM–loop interface are not (completely) transferred to the neighboring loop region when comparing bR-DDM and bR-ND. This suggests that predominantly this TM-loop interface region is in contact with (the headgroups of) the membrane mimetic. However, bR-APOL shows larger perturbations that may indicate (unspecific) interactions of the amphipol chains with the loop region. Notably, peak splitting as observed for residues in the EF loop in DDM micelles (Schubert et al., 2002) is also observed for bR in nanodiscs but not (or largely reduced) for bR in amphipols. Solid-state NMR data obtained on bR in its native membrane also did not show peak splitting for this region indicating that the amphipols may be able to support the native structure of this specific loop better than DDM or DMPC. Finally residues in the unstructured C-terminal part of bR are largely unaffected by the different environments, suggesting that no sizable interaction between the terminus and any of the surfactant is present.
CONCLUSION
Our data demonstrate that well-behaving heptahelical membrane proteins can in principle be studied in an amphipol as well as a nanodiscs environment using 2D and 3D TROSY based solution-state NMR. However, careful sample optimization should be carried out and additional non-conventional approaches such as selective labeling may be necessary for more detailed insights. Here we used cell-free protein expression to minimize bias introduced by native cofactors (e.g. coordinated lipids) and to facilitate specific labeling. Our results may assist future studies on (well-folded) heptahelical membrane proteins and serve as a reference for NMR accessibility of this important class of proteins in the tested non-conventional membrane mimetics. We could show that the functional state of bR is most stable in the non-micellar environments suggesting that NMR structural studies of less stable membrane proteins (such as GPCRs) may strongly benefit from the use of these non-conventional surfactants.
In addition, our data reveal that while the overall fold and in particular the inner core of bR is not significantly altered, the different environments have a clear effect on the surfactant-exposed region as well as the TM-loop interface. This is likely a consequence of the different headgroups at the aqueous/lipophilic border, interactions with free detergent/amphipol molecules as well as changes in hydrophobic coverage and direct interactions at the protein-surfactant interface. Our results identify regions in the membrane protein that are affected by the different membrane mimetics. This localized environment-protein adaptation does not impact the functional core of the protein in accordance with very similar absorption profiles of the chromophore in the different environments. In contrast to bR being part of a well-defined membrane setting (i.e. the purple membrane), many less stable membrane proteins experience large variations in the lipid environment during their lifetimes (e.g. by being transferred to various cell organelles or during different stages of the cell). Notably our experimental insights are in agreement with the hypothesizes that the lipid-exposed surfaces of membrane proteins have evolved to maintain correct structure and function in changing environments and that even bR, which normally does not experience significant alternations in lipid composition in its native environments, can tolerate changes in its membrane environment.
However, in contrast to bR where its cofactor location coincides with its hydrophobic inner core, the membrane environment might play a more pronounced role for membrane proteins whose functional features are mediated by surface exposed loop regions. Our results emphasize that the choice of a suitable membrane mimetic may be in particular important in these systems in order to support the active conformation of the embedded membrane protein.
EXPERIMETNAL PROCEDURES
bO cloning and expression
The bO gene was isolated from Halobacterium salinarum strain JW-3 (a gift from Judy Herzfeld, Brandeis University) and transferred into the pIVEX2.4d expression vector using restriction-free cloning. The resulting construct comprises a 6x or 10x N-terminal His tag followed by a Factor Xa cleavage site. The restriction-free cloning allowed to minimize the number of artificially introduced residues to the following (N-terminal): MSGSHHHHHHSSGIEGRGRLILHM and MSGSHHHHHHHHHHSSGIEGRM (followed by the WT bO1–248 sequence) for the 6x and 10x His tag construct, respectively. We did not observe any effect of His tag cleavage in the 2D TROSY-HSQC spectra. The bO cell-free protein expression was carried out using an E. coli based system following published procedures (Schwarz et al., 2007). E. coli cell extract as well as T7 polymerase were produced following the given protocols. Dialysis mode reactions were carried out in the absence of retinal and surfactants. The resulting pellet was washed with S30 buffer and directly refolded or stored at −20°C.
Protein refolding
Refolding of bO into DDM micelles was carried out by resuspending the protein pel-let with DDM-refolding buffer (50 mM sodium phosphate pH 7, 1 M NaCl, 5% w/v DDM, 100 μM retinal). Refolded bR-DDM was purified using a Ni-NTA agarose column followed by gel filtration. Re-folding of bO into A8-35 amphipols was carried out by resuspending the bO pellet in SDS-buffer (50 mM sodium phosphate pH 7.5, 20 mM SDS). Retinal (to a concentration of 100 μM) and amphipols (to 2.2 % w/v) were added, the mixture was kept at room temperature for 30 min and SDS was precipitated by the addition of KCl to a final concentration of 150 mM. Refolded bR-APOL was purified using a Ni-NTA agarose column (using a buffer with 0.08% amphipols) followed by gel filtration (using amphipol-free buffer). Amphipols were synthesized in collaboration with FB reagents following published protocols (Gohon et al., 2004). A8-35 was selected based on its well characterized biophysical properties, its applicability for a wide range of membrane proteins as well as its excellent refolding properties for hep-tahelical membrane proteins (Dahmane et al., 2009; Popot et al., 2011; Tribet et al., 1996). Several strategies were tested to produce bR-ND. Best yields were obtained when bO in SDS-buffer was directly refolded into DMPC nanodiscs, by adding soluble MSP1D1 (with a cleaved-off His tag), SDS-solubilized DMPC and retinal (to a concentration of 100 μM). The ratio of bR:MSP1D1:DMPC was set to 1:6:420, with the MSP1D1:DMPC concentration being particularly critical. SDS was removed using Bio-beads SM2 (Biorad). Purification (including removal of excess of empty nanodiscs) was carried out using a Ni-NTA agarose column followed by gel filtration. Notably direct refolding from the protein pellet also simplifies amid proton back exchange due to the absence of a well-established hydrogen bonding network during washing and initial refolding steps, which are carried out in non-deuterated buffer. Refolding itself was optimized for each environment and refolding yields of about 50% could be obtained in each case. However, bR yields after purification were about 2x less for bR-ND than for bR-DDM and bR-APOL.
Electron microscopy and additional biophysical measurements
EM sample preparation, data collection and analysis were carried out as described previously (Raschle et al., 2009). Class averages (50 classes) were generated by iterative alignment and classification of 3000 particles. Analytical size exclusion chromatography was carried out using a Superdex 200 column. Protein eluted from the Ni-NTA agarose column was concentrated (Millipore, 30 kDa cut-off filter) before injection. 50 mM sodium phosphate buffer (pH 7.5, 150 mM NaCl) was used. For bR-DDM buffer was supplemented with 0.1% DDM. Thermofluor assays were carried out using a qPCR system (Life Technologies 7900HTT). Sypro-Orange (Sigma-Aldrich, 5000x) was used as a dye (diluted to 5x). bR concentration was about 10 μM in the different environments. Absorption spectra were recorded using a Carry 50 photo-spectrometer (Ag-ilent) and ultra-micro cuvettes (BRAND). Samples were kept in the dark for at least 12 h prior to measurements of the dark-adapted state. Time-resolved absorption measurements were carried out simultaneously for the different environments using a transparent 96-well plate and a SpectraMax M5 Plate reader, preheated to 58°C. Wells were sealed with adhesive film to limit evaporation of the samples. Buffer-only samples were used to determine and subtract condensation effects.
NMR measurements
Isotope-labeled bR was produced by adding/replacing the respective amino acids in the in vitro cell-free expression system. The different isotope-labeled samples are shown in Table 1. All NMR samples were expressed under >90% D2O conditions. All samples were measured in the same NMR buffer (20 mM sodium phosphate, pH 7.3, 50 mM NaCl, 8% D2O, 0.03% NaN3). The pH value was selected due to limited solubility of the amphipols for pH < 7. Typical bR concentrations were in the range of 300 μM for ALGAL(D,N,C) labeled samples and 150 μM for the other specific labeled samples. NMR measurements were carried out at 33°C – 50°C, at proton resonance frequencies of 750 or 800 MHz. Duration of 2D experiments was in the order of 4 – 12 hours, 3D experiments were recorded in 3 – 4 days (each).
Supplementary Material
Table 2.
Overview of different isotope labeled samples used in this study.
| Sample name | Supplied labeled amino acids | Supplied unlabeled amino acids | Used for spectra shown in |
|---|---|---|---|
| ALGAL(D,N)-bR-DDM | (15N,2H)-ALGAL 16 mix[a] | Q,N,W,R,M,D,E | Figure 5a |
| ALGAL(D,N,C)-bR-DDM | (2H,15N,13C)-ALGAL 16 mix[b] | Q,N,W | Figure S1 |
| ALGAL(D,N)-bR-APOL | (15N,2H)-ALGAL 16 mix[a] | Q,N,W,R,M,D,E | Figure 5b |
| ALGAL(D,N,C)-bR-APOL | (2H,15N,13C)-ALGAL 16 mix[b] | Q,N,W | Figure S3c |
| ALGAL(D,N)-bR-ND | (15N,2H)-ALGAL 16 mix[a] | Q,N,W,R,M,D,E | Figure 5c, Figure S3g,h |
| ALGAL(D,N,C)-bR-ND | (2H,15N,13C)-ALGAL 16 mix[b] | Q,N,W | Figure 3g–h, Figure S3d–f |
| GIF-bR-DDM | 15N-Gly, U-(15N,13C)-Phe, U-(13C)-Ile | A,R,N,D,Q,E,H,L,K,M,P, S,T,W,Y,V | not shown |
| GIF-bR-APOL | 15N-Gly, U-(15N,13C)-Phe, U-(13C)-Ile | A,R,N,D,Q,E,H,L,K,M,P, S,T,W,Y,V | Figure S3a |
| KA-bR-DDM | U-(2H,15N,13C)-Lys, 1-13C-Ala | R,N,D,Q,E,G,H,I,L,M,F,P,S,T,W,Y,V | not shown |
Double-labeled (2H and 15N) ALGAL amino acid mix was purchased from Cambridge Isotopes and contains all amino acids (in different concentrations) except for the four amino-acids: Cys, Trp, Gln, Asn.
Triple-labeled (2H, 15N and 13C) ALGAL amino acid mix was purchased from Sigma Isotec and contains the same amino acids as the double-labeled mix (with slightly different concentrations). Note that bR does not contain any cys.
Highlights.
Bacteriorhodopsin (bR) is NMR accessible in nanodiscs and amphipols
Non-conventional surfactants offer increased stability for cell-free expressed bR
The bR-membrane interface is highly adaptable in the selected membrane mimetics
Applications and limitations of the environments for NMR studies are discussed
Acknowledgments
We thank Judy Herzfeld and Bob Griffin for providing the Halobacteria Salinarum used to isolate the bO gene as well as Bernd König and Julian Glück for providing the MSP-plasmid. Technical assistance by and helpful discussions with Dr. Tsyr-Yan Yu, Dr. Haribabu Arthanari, Dr. Rafael Luna, Dr. Sven Hyberts and Dr. Mayaan Gal are gratefully acknowledged. This project was supported by the NIH (GM094608, GM047467, EB002026); M.E. thanks the DAAD, F.H. the EMBO and the HFSP for postdoctoral fellowships.
ABBREVIATIONS
- APol
amphipathic polymer (amphipol)
- ND
nanodiscs
- DDM
n-dodecyl-β-D-maltopyranoside
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- MSP
membrane scaffold protein
- bR
bacte-riorhodopsin
- TROSY
transverse relaxation optimized spectroscopy
Footnotes
Additional NMR spectra, thermofluor raw data, light/dark adapted absorption profiles, full set of EM classes and detailed analysis of chemical shift perturbations.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proc Natl Acad Sci U S A. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Bermel W, Bertini I, Felli IC, Peruzzini R, Pierattelli R. Exclusively heteronuclear NMR experiments to obtain structural and dynamic information on proteins. Chemphyschem. 2010;11:689–695. doi: 10.1002/cphc.200900772. [DOI] [PubMed] [Google Scholar]
- Brouillette CG, McMichens RB, Stern LJ, Khorana HG. Structure and thermal stability of monomeric bacteriorhodopsin in mixed phospholipid/detergent micelles. Proteins. 1989;5:38–46. doi: 10.1002/prot.340050106. [DOI] [PubMed] [Google Scholar]
- Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, Kuhn EA, Chromy BA, Segelke BW, Rothschild KJ, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol Cell Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane T, Damian M, Mary S, Popot JL, Baneres JL. Amphipol-assisted in vitro folding of G protein-coupled receptors. Biochemistry. 2009;48:6516–6521. doi: 10.1021/bi801729z. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Edman K, Nollert P, Royant A, Belrhali H, Pebay-Peyroula E, Hajdu J, Neutze R, Landau EM. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature. 1999;401:822–826. doi: 10.1038/44623. [DOI] [PubMed] [Google Scholar]
- Engelhard M, Hess B, Emeis D, Metz G, Kreutz W, Siebert F. Magic angle sample spinning 13C nuclear magnetic resonance of isotopically labeled bacteriorhodopsin. Biochemistry. 1989;28:3967–3975. doi: 10.1021/bi00435a051. [DOI] [PubMed] [Google Scholar]
- Gluck JM, Wittlich M, Feuerstein S, Hoffmann S, Willbold D, Koenig BW. Integral membrane proteins in nanodiscs can be studied by solution NMR spectroscopy. J Am Chem Soc. 2009;131:12060–12061. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- Gohon Y, Pavlov G, Timmins P, Tribet C, Popot JL, Ebel C. Partial specific volume and solvent interactions of amphipol A8–35. Anal Biochem. 2004;334:318–334. doi: 10.1016/j.ab.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Gorzelle BM, Hoffman AK, Keyes MH, Gray DN, Ray DG, Sanders CR. Amphipols can support the activity of a membrane enzyme. J Am Chem Soc. 2002;124:11594–11595. doi: 10.1021/ja027051b. [DOI] [PubMed] [Google Scholar]
- Harbison GS, Herzfeld J, Griffin RG. Solid-state nitrogen-15 nuclear magnetic resonance study of the Schiff base in bacteriorhodopsin. Biochemistry. 1983;22:1–4. doi: 10.1021/bi00270a600. [DOI] [PubMed] [Google Scholar]
- Harbison GS, Smith SO, Pardoen JA, Winkel C, Lugtenburg J, Herzfeld J, Mathies R, Griffin RG. Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc Natl Acad Sci U S A. 1984;81:1706–1709. doi: 10.1073/pnas.81.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefke F, Bagaria A, Reckel S, Ullrich SJ, Dotsch V, Glaubitz C, Guntert P. Optimization of amino acid type-specific 13C and 15N labeling for the backbone assignment of membrane proteins by solution- and solid-state NMR with the UPLABEL algorithm. J Biomol NMR. 2011;49:75–84. doi: 10.1007/s10858-010-9462-4. [DOI] [PubMed] [Google Scholar]
- Hirai T, Subramaniam S. Protein conformational changes in the bacteriorhodopsin photocycle: comparison of findings from electron and X-ray crystallographic analyses. PLoS One. 2009;4:e5769. doi: 10.1371/journal.pone.0005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Li Q. Solution NMR study of integral membrane proteins. Curr Opin Chem Biol. 2011;15:560–569. doi: 10.1016/j.cbpa.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins. Prog Nucl Magn Reson Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Luecke H, Richter HT, Lanyi JK. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999a;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. J Mol Biol. 1999b;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- Nietlispach D, Gautier A. Solution NMR studies of polytopic alpha-helical membrane proteins. Curr Opin Struct Biol. 2011;21:497–508. doi: 10.1016/j.sbi.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Oka T, Yagi N, Fujisawa T, Kamikubo H, Tokunaga F, Kataoka M. Time-resolved x-ray diffraction reveals multiple conformations in the M-N transition of the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 2000;97:14278–14282. doi: 10.1073/pnas.260504897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ. Nanodiscs versus Macrodiscs for NMR of Membrane Proteins. Biochemistry. 2011 doi: 10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzelt H, Ulrich AS, Egbringhoff H, Dux P, Ashurst J, Simon B, Oschkinat H, Oesterhelt D. Towards structural investigations on isotope labelled native bacteriorhodopsin in detergent micelles by solution-state NMR spectroscopy. Journal of Biomolecular NMR. 1997;10:95–106. [Google Scholar]
- Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. Amphipols from a to z*. Annu Rev Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle T, Hiller S, Yu TY, Rice AJ, Walz T, Wagner G. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Kovacs H, Buck J, Wacker A, Furtig B, Bermel W, Schwalbe H. 13C-direct detected NMR experiments for the sequential J-based resonance assignment of RNA oligonucleotides. J Biomol NMR. 2010;47:259–269. doi: 10.1007/s10858-010-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Tuzi S, Yamaguchi S, Tanio M, Naito A. Conformation and backbone dynamics of bacteriorhodopsin revealed by (13)C-NMR. Biochim Biophys Acta. 2000;1460:39–48. doi: 10.1016/s0005-2728(00)00128-6. [DOI] [PubMed] [Google Scholar]
- Schubert M, Kolbe M, Kessler B, Oesterhelt D, Schmieder P. Heteronuclear multidimensional NMR spectroscopy of solubilized membrane proteins: resonance assignment of native bacteriorhodopsin. ChemBioChem. 2002;3:1019–1023. doi: 10.1002/1439-7633(20021004)3:10<1019::AID-CBIC1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Junge F, Durst F, Frolich N, Schneider B, Reckel S, Sobhanifar S, Dotsch V, Bernhard F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat Protoc. 2007;2:2945–2957. doi: 10.1038/nprot.2007.426. [DOI] [PubMed] [Google Scholar]
- Shenkarev ZO, Lyukmanova EN, Solozhenkin OI, Gagnidze IE, Nekrasova OV, Chupin VV, Tagaev AA, Yakimenko ZA, Ovchinnikova TV, Kirpichnikov MP, et al. Lipid-protein nanodiscs: possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Mosc) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- Tribet C, Audebert R, Popot JL. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci U S A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonens M, Catoire LJ, Giusti F, Popot JL. NMR study of a membrane protein in detergent-free aqueous solution. Proc Natl Acad Sci U S A. 2005;102:8893–8898. doi: 10.1073/pnas.0503750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.