Abstract
Objectives
High-salt intake has been demonstrated in link to hypertension, and cardiovascular diseases could be programmed in fetal origins. We determined the influence of high-salt diet during pregnancy on the development of the heart.
Methods
Fetal cardiac structures, cell cycle, renin–angiotensin system (RAS), and epigenetic alternations in the heart following maternal high salt intake during pregnancy were examined.
Results
Following exposure to high salt, disorganized myofibrillae and mitochondria cristae loss were found in the fetus, S-phase for cardiac cells was enhanced, plasma angiotensin II decreased, and cardiac angiotensin II increased in the fetus. Angiotensin II-increased S-phase in the fetal cardiac cells was primarily via AT1 receptor mechanisms. AT2 receptor mRNA and protein in the fetal heart were not affected, whereas AT1 receptor protein, AT1a, and AT1b mRNA were increased. DNA methylation was found at the CpG sites that were related to AT1b receptors in the fetal heart. Cardiac AT1 receptor protein in the adult offspring was also higher following exposure to prenatal high salt.
Conclusion
The results suggest a relationship between high-salt diet in pregnancy and developmental changes of the cardiac cells and renin–angiotensin system.
Keywords: AT1/AT2 receptor, cell cycle, DNA methylation, high-salt diet, renin–angiotensin-system
Introduction
High-salt intake is linked to hypertension [1], and recent progress has been made to demonstrate that hypertension could be programmed in fetal origins [2]. Although women tend to take more salt during pregnancy [3], whether and to what extent that maternal intake of high salt affects the fetal heart is unknown. In the present study, we determined the primary cardiac cells from the fetuses exposed to high salt during pregnancy. In response to high salt, the renin–angiotensin system (RAS) plays an important role in cardiovascular homeostasis, body fluid balance, and renal regulation [4]. Two major subtypes of angiotensin II (Ang II) receptors, subtype 1 of Ang II receptor (AT1R) and subtype 2 of Ang II receptor (AT2R), have been detected in neonatal and adult hearts [5]. AT1R could be subdivided into AT1a and AT1b [6]. Although AT1R is critical in physiological functions, including vascular regulation and fluid homeostasis [7], both AT1R and AT2R have been shown to mediate cell proliferation and differentiation [8]. Fetal RAS have been suggested to mediate development and apoptosis in the heart via those Ang II receptors [9,10]. However, it is still unclear whether prenatal exposure to high-salt diet could affect the local RAS and cardiac cells in the fetal heart in utero. To address this question is useful in exploring mechanisms related to an altered RAS during disease development in fetal origins.
Epigenetic regulation of gene promoters is mainly established during development and retained throughout the lifespan. DNA methylation plays an important role in cell differentiation by silencing expression of specific genes during development. Methylation can also induce transcriptional silencing by blocking the binding of transcription factors or through promoting the binding of methyl CpG-binding protein [11]. Alterations in methylation associated with cell differentiation and functional changes could be initiated during in-utero development [11]. The present study used the bisulfite pyrosequencing method in analyses of DNA methylation in the fetal heart under the condition of exposure to high salt during pregnancy.
Methods
Animals
Pregnant Sprague–Dawley rats were housed in a controlled environment of 22°C and a 12-h light–dark cycle. They were randomized to either normal-salt diet (NSD, 1% NaCl) or high-salt diet (HSD, 8% NaCl) [12,21] from gestational day 3–21 (n = 8, each group), and were allowed access to tap water. Food and water intake during pregnancy were measured daily. Maternal urine was collected daily for measuring urine volume and sodium. Protocols were approved by the Institutional Animal Care Committee.
Experimental design
Blood values and hormones
At gestational day 21, pregnant rats were anesthetized with sodium pentobarbital (20 mg/kg, intraperitoneally). Maternal blood was collected from the abdominal aorta. Fetuses (n = 6) were decapitated, and trunk blood was pooled in the same litter (n = 6–8). Blood samples were prepared for measuring sodium, osmolality, and the hormones. Fetal heart weight (n = 8) was measured, and then heart tissue was kept at −80°C before molecular analysis. The fetal left ventricle (LV) tissue was homogenized with 0.9% NaCl (10 ml/g), enzyme inhibitors contained 0.3 mmol/l Na2–edetic acid, 0.3 mmol/l dimercarptanol, and 0.3 mmol/l oxine sulfate, and the homogenates were boiled and centrifuged. Plasma and myocardial Ang II concentrations were measured by radioimmunoassay using a commercial kit (Beijing Sinouk Institute of Biological Technology, Beijing, China). The sensitivity of each assay was 10 pg/ml and 100 pg/g, respectively. The intra and inter-assay variations were 4 and 5%.
Cardiac structures with transmission electron microscope
Fetal LV tissue (n = 5, each group) was fixed in 10% neutral formalin. After washing with distilled water, specimens were immersed in a 10% NaOH, then in 1% tannic acid, postfixed in 1% osmium tetroxide, dehydrated in graded concentrations of ethanol, and sectioned transversely or longitudinally. Sections were freeze dried, coated with gold, and observed under a transmission electron microscope.
Cardiac cells
Fetal heart tissue (n = 6–7) was minced and incubated in phosphate-buffered saline (PBS) containing 0.8% trypsin and 0.1% collagenase II (Sigma, St. Louis, Missouri, USA) for 30 min. After digestion, cells in the supernatant were transferred into a tube containing Dulbecco’s medium cell culture with 15% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO, Fort Worth, Texas, USA). Then samples were centrifuged at 800g. The cell pellets were resuspended in the cell culture medium. The cells were cultured for 2 weeks. The cells used for testing must show typical beating patterns of cardiac cells under a microscopy. Cardiomyocytes were isolated from fibroblasts with fluorescence-activated cell sorting (FACS) method as cells were collected and fixed in 80% cold alcohol for 30 min, and permaeabilized by incubation for 10 min in 0.25% Triton X-100/PBS at room temperature (RT). Then cells were incubated with rabbit polyclonal anti-Troponin I (1 μg/l ×106 cells)(Santa Cruz Biotech, Santa Cruz, California, USA) in a covered ice bucket, away from light, for 30 min. Troponin I-stained cardiomyocyte cells were then analyzed using FACS (Becton Dickinson Bioscience, San Jose, California, USA) and collected into a tube for G1 and S-phase analysis.
The secondary cultured cardiac cells were maintained for 24 h in serum-free medium. Then, 10−6 mol/l Ang II (Sigma) was added. To determine the effects of angiotensin receptors, cultures were pretreated with 10−6 mol/l losartan (AT1R antagonist) or 10−6 mol/l PD123319 (AT2R antagonist) (Sigma) 15 min prior to addition of Ang II. In another two groups, 10−6 mol/l losartan or 10−6 mol/l PD123319 was added in cultures. Following incubation with the drugs for 24 h, 2 ×106 cells were collected and fixed at −20°C in 70% ethanol for analysis of cell cycle.
Cell cycle
Cells were centrifuged at 800g for 4 min, 100 μl of 200 μg/ml DNase-free and RNaseA were added, and incubated at 37°C for 30 min. Then, 100 μl of 1 mg/ml propidium iodide was added and incubated for 10 min at RT. Samples were then placed in 12 × 75 mm Falcon tubes and read on Becton Dickinson FAC Star PLUS.
Western blotting
Lysates were matched for protein, separated by SDS-PAGE, and transferred to nitrocellulose membranes, which were probed with AT1R or AT2R antibodies (Santa Cruz Biotech). Western blots were processed using enhanced chemiluminescence (Amersham plc, Amer-sham, UK), and data were quantified with blue fluorescence imaging on PhosphorImager. Relative AT1R and AT2R signal intensity in the fetal heart was normalized with β-actin. Another two groups of pregnant rats (fed with either 8% HSD or NSD from gestational day 3 to 21) were allowed to give birth, the heart of the male adult offspring (4 months after birth, n = 5–6) were used for western blotting analysis.
Real-time PCR
Total RNA was isolated, quantified, and reverse transcribed. All Q-PCR assays were performed using IQ iCycler (Bio-Rad Laboratories, Inc., Hercules, California, USA). Samples were incubated in a thermocycler at 95°C for 2 min. There were 45 cycles of PCR amplification performed consisting processes of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. Assays must show nonspecific product or should be excluded from data analysis. For confirmation of amplification presence and purity, the PCR product was substantiated on a 1.5% agarose gel. Quantitative values were obtained from the cycle number (CT value) at which the increase in fluorescent signal associated with an exponential accumulation of PCR products was detected. The amount of target gene was normalized to the reference 18S to obtain the relative threshold cycle (ΔCT), and then related to the CT of the control to obtain the relative expression level (2−ΔΔCT) of target gene. Primer sequences are shown in Table 1.
Table 1.
Relative gene primers sequence
| Gene | Accession | Primer | Product length (bp) |
|---|---|---|---|
| AT1a | NM030985 | F: 5′-cctgtcactccacctcaaa (position 3113) R: 5′-aaccctctgttctacggc (position 3305) |
110 |
| AT1b | NM031009 | F: 5′-aagggcggtaggaaagag (position 976) R: 5′-ctgggcattatccgtgac (position 1189) |
117 |
| AT2 | NM012494 | F: 5′-tccgctttggcttgtccattgct (position 317) R: 5′-tcccagcgcagcttgtagtcgtg (position 206) |
110 |
| 18s rRNA | NM11188 | F: 5′-cttagttggtggagcgatttgtctg (position 1353) R: 5′-gttattgctcaatctcgggtggc (position 1500) |
148 |
|
| |||
| AT1b Pyrosequencing | F: 5′-attttttgttgtttgggatttagg R: 5′-cattccaaacccaaataaacctat-biotin SeqF: 5′-ttaatttatttagtaaaggg |
||
Genomic DNA and bisulphite modification
Genomic DNA was isolated from the fetal heart using DNAsol reagent (Tiangen Biotech, Beijing, China). DNA samples were then digested with restriction enzymes EcoRV and Bgl, deproteinized with phenol/chloroform, and precipitated with ethanol. DNA was treated with sodium bisulfate, precipitated with ethanol, and dissolved in 50 μl water. PCR conditions included: 94°C for 3 min, then 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, and finally one cycle of 7 min for 72°C. The reaction mixture contained 1× PCR buffer, 0.2 mmol/l deoxyribonucleotide triphosphates, 2 mmol/l MgCl2, 1 μmol/l primers, 0.5 U of Taq Gold DNA polymerase (Takara, Tokyo, Japan), and 2 μl DNA template.
Pyrosequencing
PCR and sequencing primers for pyrosequencing were designed using PSQ Assay design software (Biotage AB, Uppsala, Sweden). One of the PCR primers was biotinylated with the treated strands being purified and sequenced using the PSQTM 96MA 2.1 instrument (Bio-tage AB). The primer sequences are listed in Table 1. Calibration curves were recorded using five mixtures of PCR products (0, 25, 50, 75, and 100% methylation) from several methylation and unmethylation at the gene promoter regions of the angiotensin receptor. Samples for DNA methylation studies were tested at Southern Gene Company (Shanghai, China) in a blind manner.
Statistical analysis
One-way analysis of variance method and t-test were used in the determination of differences between the control and treated groups. All data were expressed as mean ± SEM.
Results
Body weight and food intake
HSD did not affect body weight and food intake in maternal rats. However, maternal water intake, urine volume, and urine sodium concentrations were increased (Table 2).
Table 2.
The effect of high-salt diet on body weight and food intake in maternal rats
| NSD | HSD | |
|---|---|---|
| Body weight [Δ (GD21 – GD3), g] | 102 ± 15 | 118 ± 18 |
| Food intake (g/100 g BW) | 9.4 ± 2.1 | 10.2 ± 1.2 |
| Water intake (ml/100 g BW) | 13.4 ± 1.8 | 29.9 ± 4.5* |
| Urine sodium (mmol/100 g BW per min) | 0.9 ± 0.1 | 3.7 ± 0.5* |
| Urine volume (ml/100 g BW) | 6.4 ± 1.0 | 27.8 ± 2.9* |
P <0.05, HSD vs. NSD. BW, body weight; GD, gestational day; HSD, high-salt diet; NSD, normal-salt diet.
Fetal heart
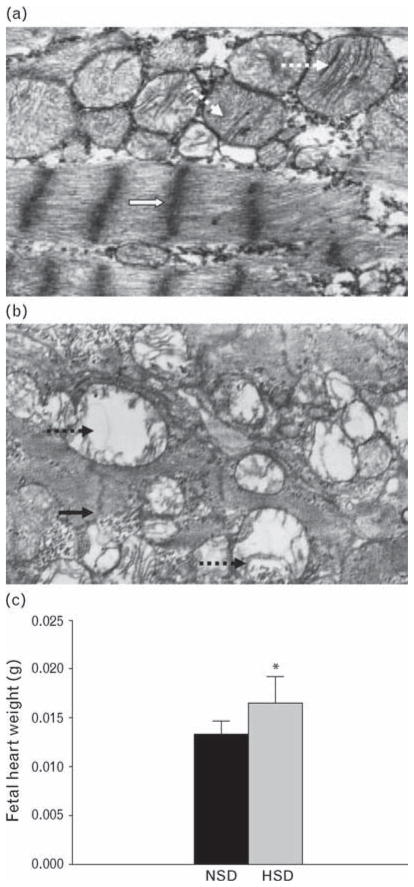
Prenatal treatment with HSD induced ultrastructural changes in the fetal heart, including disorganized myofibrillae, small mitochondria, cristae loss, and vacuolization (Fig. 1a and b). Fetal heart weight was increased compared with that of the control (t = 1.76, P <0.05) (Fig. 1c).
Fig. 1.
Fetal left ventricle tissue under electron microscope (×11 000) and fetal heart weight. (a) NSD, open arrows show normal arrangement of myofibrillae, dotted arrows show large bulk of mitochondria with abundant cristae. (b) HSD, solid arrows show disorganized myofibrillae, solid dotted arrow shows small bulk mitochondria with cristae loss, and vacuolization. (c) Fetal heart weight at gestational day 21. *P <0.05. HSD, high-salt diet; NSD, normal-salt diet.
Blood values and hormones
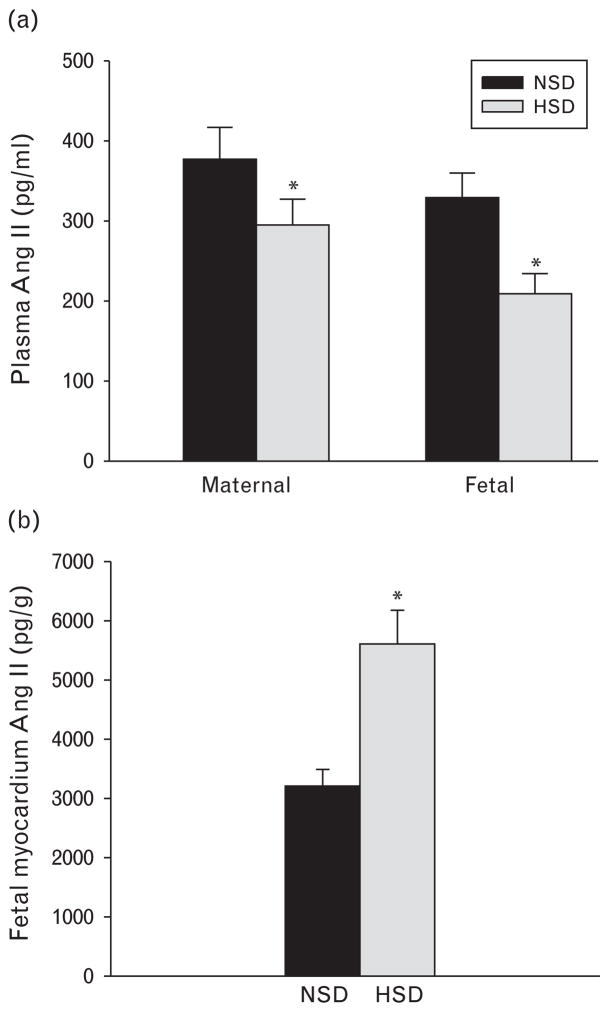
There were no significantly differences of blood Na+ concentrations and osmolality in both maternal and fetal rats between the control and HSD group (Table 3). HSD caused plasma Ang II decrease in both the mothers and fetuses (Fig. 2a), and Ang II in the fetal myocardium was significantly increased (Fig. 2b).
Table 3.
The effects of high-salt diet on maternal and fetal plasma Na+ and osmolality
| NSD
|
HSD
|
|||
|---|---|---|---|---|
| Maternal | Fetal | Maternal | Fetal | |
| Blood sodium (mmol/l) | 133 ± 14 | 118 ± 8 | 135 ± 18 | 122 ± 15 |
| Plasma osmolality (mosm/l) | 298 ± 14 | 284 ± 15 | 305 ± 21 | 302 ± 10 |
HSD, high-salt diet; NSD, normal-salt diet.
Fig. 2.
The effect of prenatal exposure to high-salt diet on the hormones. (a) Maternal and fetal plasma Ang II was decreased following high salt during pregnancy. (b) Fetal heart Ang II was increased following exposure to high salt during pregnancy. *P <0.05. Ang II, angiotensin II; HSD, high-salt diet, NSD, normal-salt diet.
Cell cycle
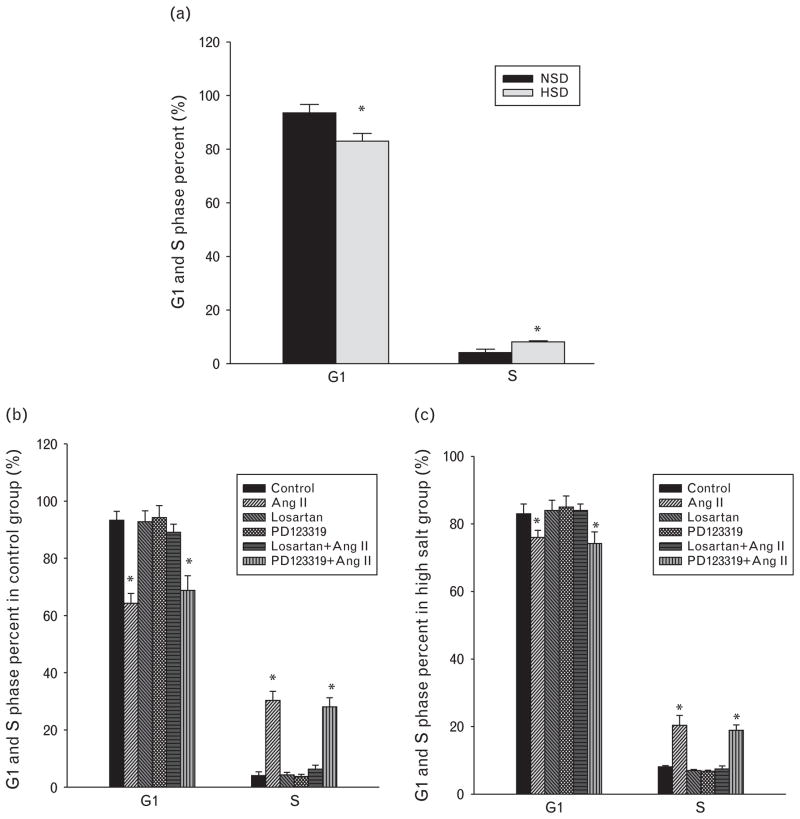
In cultured primary cardiac cells (n = 5–7), G1 phase percentage was significantly decreased, whereas S phase percentage was increased in the primary heart cells from the HSD group compared with those from the control (Fig. 3a). Ang II decreased G1 phase percentage and increased S phase percentage in primary cardiac cells (Fig. 3b and c). Losartan, AT1R antagonist, reversed the changes of G1 and S phases induced by Ang II. PD123319, AT2R antagonist, did not block the changes induced by Ang II in the cardiac cells (Fig. 3b and c). Losartan or PD123319 alone did not affect percentages of G1 or S phase (Fig. 3b and c).
Fig. 3.
The comparison of G1 and S phase percentages between the normal-salt and high-salt diet groups (a), and the effect of angiotensin II and its receptor antagonists (losartan and PD123319) on G1 and S phase percentages in the control (b) and high-salt (c) diet groups. *P <0.05. Ang II, angiotensin II; HSD, high-salt diet; NSD, normal-salt diet.
AT1a, AT1b, and AT2 gene
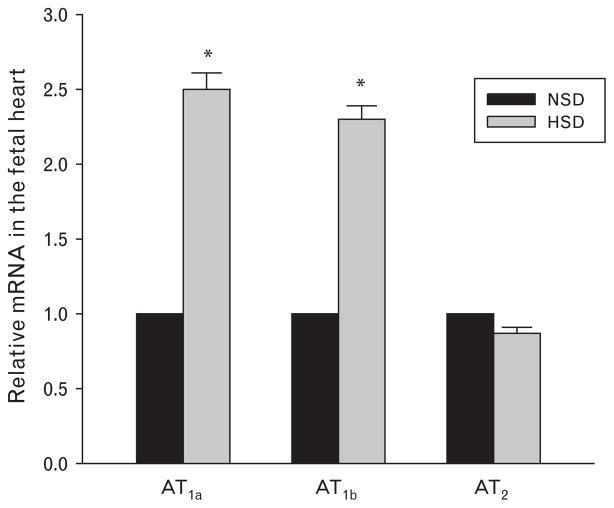
Real-time PCR showed that AT1a, AT1b, and AT2 mRNA were present in the fetal heart at gestational day 21 in rats. Prenatal treatment of HSD caused an increase of both AT1a and AT1b mRNA expression in the fetal heart, whereas AT2 mRNA was not changed (Fig. 4).
Fig. 4.
AT1a and AT1b mRNA, but not AT2 mRNA, expression were increased following prenatal exposure to high-salt diet. *P <0.05, HSD, high-salt diet; NSD, normal-salt diet.
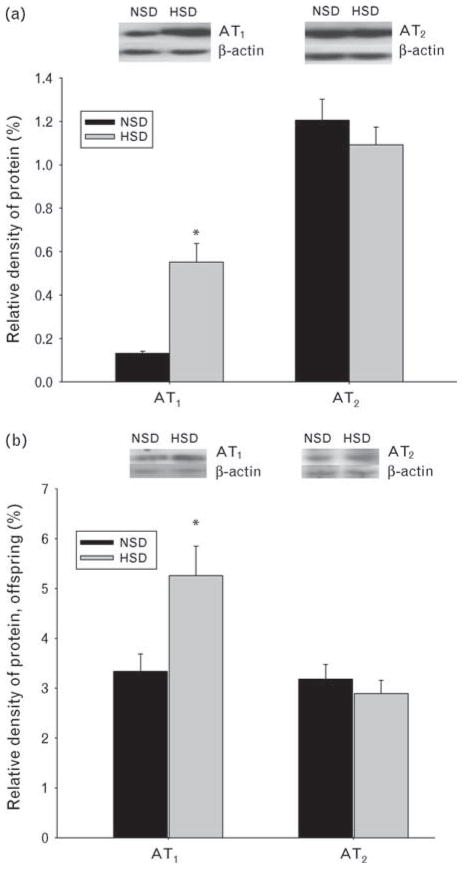
Ang II receptor protein
Maternal HSDs induced an increase of AT1R protein expression in the fetal heart. However, AT2R protein remained unchanged (Fig. 5a). In addition, AT1R protein in the offspring heart (prenatally exposed to high salt) was also significantly increased (Fig. 5b).
Fig. 5.
(a) Subtypes 1 and 2 of angiotensin II receptor protein expressions in the fetal heart following prenatal exposure to high-salt diet. (b) AT1R and AT2R AT2 receptor protein expressions in the offspring heart at gestational day 21 following prenatal exposure to HSD. *P <0.05. AT1R, subtype 1 of angiotensin II; AT2R, subtype 2 of angiotensin II; HSD, maternal high-salt diet in pregnancy; NSD. maternal normal-salt diet in pregnancy.
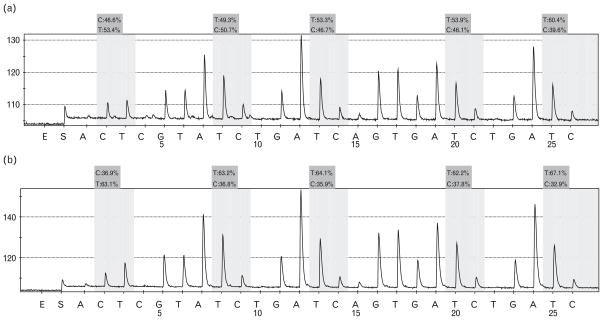
DNA methylation
Methylation percentage was calculated by average degree of methylation at five CpG sites formulated in pyrosequencing. In the maternal HSD group, average methylation was lowered (average 36.9, 36.8, 35.9, 37.8, and 32.9%), and the five affected sites were linked to the AT1b promoter. In comparison, in the sample from the NSD group, five CpG sites showed normal levels of methylation (average 46.6, 50.7, 46.7, 46.1, and 39.6%) (Fig. 6).
Fig. 6.
DNA methylation detected in the fetal heart tissue. (a) Following prenatal NSD, (b) following prenatal HSD. Grey areas correspond to CpG sites. DNA methylation of AT1b promoter was decreased at the five sites following prenatal exposure to HSD. C, methylated cytosine; HSD, high-salt diet, NSD, normal-salt diet; T, unmethylated site.
Discussion
To the best of our knowledge, this was the first study on the influence of maternal HSDs on the development of the heart. We found that HSD during pregnancy influenced the development of the fetal RAS associated with fetal cardiac cellular changes. Expression of Ang II receptors in the fetal heart was selectively changed by high salt during pregnancy, and this change was linked to DNA methylation, and persisted in adulthood.
A body of studies reported that dietary high salt could induce ventricle hypertrophy [12] associated with abnormal mitochondria in adults [13]. Myocardial adaptive remodeling included an increase in mitochondria, addition of sarcomeres, and thickened Z lines [14]. In the present study, the ultrastructure changes in the fetal heart included disorganized myofibrillae, small mitochondria, mitochondria cristae loss, and vacuolization. Fetal heart weight was increased following exposure to HSD during pregnancy. The results indicated that HSD during pregnancy could cause heart changes with ultra-structural alterations during fetal life.
In examination of the cardiac development following exposure to HSD, analysis of cell cycle showed an increase of S phase accompanied by a decrease of G1 phase in the primary cardiac cells from the fetuses exposed to high salt, suggesting an increased DNA and possible cardiac cell growth following prenatal exposure to high salt. A question raised immediately was what mechanisms contributed to the increased cell cycle under the condition of salt diet during pregnancy. Previous studies [15,16] have shown that Ang II stimulated heart cell proliferation. Among several hormonal factors in causing cardiac hypertrophy, Ang II plays a role as a stimulating growth factor [17,18]. When Ang II was added to the cardiac cells, S phase was significantly increased, indicating that Ang II may affect fetal heart cell development. With pretreatment with losartan, Ang II-induced cellular changes were markedly reduced. In comparison, AT2R antagonist did not reverse Ang II-mediated cell cycle changes. This demonstrated that Ang II-stimulated cellular responses in the fetal cardiac cells were primarily via AT1R mechanisms. Although previous studies showed that Ang II induced adrenal cell proliferation via AT1R [19] and stimulated cellular growth in vitro in neonatal and adult myocytes [20], the present study was the first to demonstrate that Ang II can change cell cycle in the fetal myocytes via AT1R mechanisms. Importantly, the percentage of S phase was higher in the myocytes of the fetuses exposed to high salt, in association with an increase of local fetal Ang II in the heart, indicating that an increase of protein synthesis could be a possible cause for the observed changes in the fetal heart.
As Ang II receptors play an important role in the cardiovascular system [20], we examined both mRNA and protein of AT1R and AT2R in the fetal heart. The protein of AT1R, not AT2R, was significantly increased in the fetal heart following exposure to HSD. In addition, fetal AT1a and AT1b mRNA levels in the heart were also higher than that of the control, whereas AT2R mRNA was not changed. A previous study [8] showed that both AT1R and AT2R were mediated in cell proliferation and differentiation, indicating their roles in cell development. The higher level of AT1R could possibly contribute to the increased S phase in the primary heart cells. Importantly, the increased AT1R was also found in the heart of the offspring with history of prenatal exposure to high salt, as evidence that the changes observed could be reserved postpartum. Although further studies are required for detailed mechanisms due to limitations in our experiments, the present study provides novel information that high-salt intake during pregnancy could affect both protein and mRNA of AT1R in the fetal heart, and such changes could last after birth.
Changes of body fluids and electrolytes are major causes of secretion and release of hormones such as Ang II [24,25]. Although plasma Na+ and osmolality in both maternal and fetal rats were not changed, water intake and renal excretion (urine volume and Na+ concentrations) in pregnant rats were increased following high-salt intake as observed before [22,23]. HSD could affect cardiovascular systems and RAS, whereas plasma osmolality and Na+ concentration changes were adjusted by physiological regulations in excreting extra salt from the body. In the present study, plasma Ang II in both maternal and fetal rats was significantly decreased by high salt, indicating that high-salt intake suppressed the RAS in the peripheral circulation. High salt tends to inhibit circulating Ang II in adult animals [26,27]. The present study indicated that high-salt intake could also suppress fetal circulating Ang II that is important in body fluid homeostasis [28]. Notably, we found that HSD increased fetal Ang II in the heart associated with an increase of cardiac AT1R protein and AT1a/AT1b mRNA. This upregulation of the local RAS may contribute to cell changes in the fetal heart, and affect the development of the heart during fetal stages.
Finally, we tested DNA methylation in the fetal heart tissue related to the subtype of angiotensin receptors. Most CpGs are methylated before birth [29], and retained throughout the lifespan. DNA methylation can induce transcriptional silencing by blocking the binding of transcription factors or through promoting the binding of methyl CpG-binding protein [12]. Promoter methylation is important for regulation of imprinted genes [29]. Thus, alterations in methylation in association with cell differentiation and functional changes could be established at different times during in-utero development [12]. As reported before [30], no methylation was found at CpG sites close to promoter regions at AT1a. We focused on CpG sites linked to AT1b promoter in the present study. We found a significant decrease of methylation in the fetal heart. The average degree of methylation at the five CpG sites linked to AT1b promoter was lower in the group exposed to HSD during pregnancy. Methylation affected promoters are critical for regulation of imprinted genes and functional changes [12,29]. This epigenetic change has been suggested as a molecular mechanism in disease programming during development. The data open new opportunities for further study on whether and to what extent this epigenetic change impacts on cardiovascular diseases in fetal origins.
In conclusion, the present study provides new insight on the relationship between high-salt intake during pregnancy and the development of the heart. An increase of S phase in the primary cardiac cells suggests a possible enhanced growth via AT1 pathway following prenatal exposure to high salt. The data demonstrated an altered local RAS or its receptors in the fetal and offspring heart affected by maternal salt diet. Interestingly, the epigenetic change at the CpG sites may contribute to the alteration of the Ang II receptors in the fetal heart. Together, the information is important for further understanding of programming diseases in fetal origins.
Acknowledgments
This work was partially supported by HL090920 grant, National Natural Science Foundation (no. 30871400 and 30973211), Jiangsu Natural Science grant (no. BK2009122 and 08KJB32001), and Suzhou grant (no. 90134602, SS08045, and SS08018). Thanks to Richard Wang (M.S., Cambridge, UK) for word editing.
Abbreviations
- Ang II
angiotensin II
- AT1R
subtype 1 of angiotensin II (Ang II) receptor
- AT2R
subtype 2 of angiotensin II (Ang II) receptor
- HSD
high-salt diet
- NSD
normal-salt diet
- PBS
phosphate-buffered saline
- RAS
renin–angiotensin system
References
- 1.Meneely GR, Battarbee HD. High sodium–low potassium environment and hypertension. Am J Cardiol. 1976;38:768–785. doi: 10.1016/0002-9149(76)90356-8. [DOI] [PubMed] [Google Scholar]
- 2.Langley-Evans SC, Sherman RC, Welham SJ, Nwagwu MO, Gardner DS, Jackson AA. Intrauterine programming of hypertension: the role of the renin–angiotensin system. Biochem Soc Trans. 1999;27:88–93. doi: 10.1042/bst0270088. [DOI] [PubMed] [Google Scholar]
- 3.Brown JE, Toma RB. Taste changes during pregnancy. Am J Clin Nutr. 1986;43:414–418. doi: 10.1093/ajcn/43.3.414. [DOI] [PubMed] [Google Scholar]
- 4.Crowley SD, Coffman TM. In hypertension, the kidney rules. Curr Hypertens Rep. 2007;9:148–153. doi: 10.1007/s11906-007-0026-2. [DOI] [PubMed] [Google Scholar]
- 5.Wollert KC, Drexler H. The renin–angiotensin system and experimental heart failure. Cardiovasc Res. 1986;43:838–849. doi: 10.1016/s0008-6363(99)00145-5. [DOI] [PubMed] [Google Scholar]
- 6.Rahman M, Kimura S, Nishiyama A, Hitomi H, Zhang G, Abe Y. Angiotensin II stimulates superoxide production via both angiotensin AT1A and AT1B receptors in mouse aorta and heart. Eur J Pharmacol. 2004;485:243–249. doi: 10.1016/j.ejphar.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 7.Rush JW, Aultman CD. Vascular biology of angiotensin and the impact of physical activity. Appl Physiol Nutr Metab. 2008;33:162–172. doi: 10.1139/H07-147. [DOI] [PubMed] [Google Scholar]
- 8.Regitz-Zagrosek V, Auch-Schwelk W, Neuss M, Fleck E. Regulation of the angiotensin receptor subtypes in cell cultures, animal models and human diseases. Eur Heart J. 1994;15:92–97. doi: 10.1093/eurheartj/15.suppl_d.92. [DOI] [PubMed] [Google Scholar]
- 9.Beinlich C, White G, Baker K, Morgan H. Angiotensin II and left ventricular growth in newborn pig heart. J Mol Cell Cardiol. 1991;23 :1031–1038. doi: 10.1016/0022-2828(91)91638-8. [DOI] [PubMed] [Google Scholar]
- 10.Brown GP, Venuto RC. Angiotensin II receptor alterations during pregnancy in rabbits. Am J Physiol. 1986;251:58–64. doi: 10.1152/ajpendo.1986.251.1.E58. [DOI] [PubMed] [Google Scholar]
- 11.Graham CB, Mark AH, Jo LSJ, Karen AL. Epigenetic regulation of transcription: a mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life? Br J Nutr. 2007;97 :1036–1046. doi: 10.1017/S0007114507682920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohlich ED, Chien Y, Sesoko S, Pegram BL. Relationship between dietary sodium intake, hemodynamics, and cardiac mass in SHR and WKY rats. Am J Physiol. 1993;264:30–34. doi: 10.1152/ajpregu.1993.264.1.R30. [DOI] [PubMed] [Google Scholar]
- 13.Marin-Garcia J, Goldenthal MJ. Mitochondrial cardiomyopathy. Pediatr Cardiol. 2006;16:28–30. doi: 10.1007/BF02310331. [DOI] [PubMed] [Google Scholar]
- 14.Hayden MR, Chowdhury N, Govindarajan G, Karuparthi PR, Habibi J, Sowers JR. Myocardial myocyte remodeling and fibrosis in the cardiometabolic syndrome. J Cardiometab Syndr. 1995;1:326–333. doi: 10.1111/j.1559-4564.2006.05626.x. [DOI] [PubMed] [Google Scholar]
- 15.Okubo SF, Niimura H, Nishimura F, Takemoto A, Fogo T, Ichikawa I. Angiotensin-independent mechanism for aldosterone synthesis during chronic extracellular fluid volume depletion. J Clin Invest. 1997;99:855–860. doi: 10.1172/JCI119249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michael D, Peter PS. ANG II-induced cell proliferation is dually mediated by c-Src/Yes/Fyn-regulated ERK1/2 activation in the cytoplasm and PKC-controlled ERK1/2 activity within the nucleus. Am J Physiol. 2006;291:1297–1307. doi: 10.1152/ajpcell.00617.2005. [DOI] [PubMed] [Google Scholar]
- 17.Gray MO, Long CS, Kallnyak JE. Angiotensin stimulates cardiac myocyte hypertrophy via paracrine release of TGF-(1 and edothelin-1 from fibroblasts. Cardiovasc Res. 1998;40:352–363. doi: 10.1016/s0008-6363(98)00121-7. [DOI] [PubMed] [Google Scholar]
- 18.Annarosa YL, Baosheng L, Wang X. Angiotensin II stimulation in vitro induces hypertrophy of normal and postinfarcted ventricular myocytes. Circ Res. 1998;82:1145–1159. doi: 10.1161/01.res.82.11.1145. [DOI] [PubMed] [Google Scholar]
- 19.McEwan PE, Vinson GP, Kenyon CJ. Control of adrenal cell proliferation by AT1 receptors in response to angiotensin II and low-sodium diet. Am J Physiol. 1999;276:303–309. doi: 10.1152/ajpendo.1999.276.2.E303. [DOI] [PubMed] [Google Scholar]
- 20.McEwan PE, Gray GA, Sherry L, Webb DJ, Kenyon CJ. Differential effects of angiotensin II on cardiac cell proliferation and intramyocardial perivascular fibrosis in vivo. Circulation. 1998;98:2765–2773. doi: 10.1161/01.cir.98.24.2765. [DOI] [PubMed] [Google Scholar]
- 21.Cowley AW, Jr, Skelton MM, Merrill DC. Osmoregulation during high salt intake: relative importance of drinking and vasopressin secretion. Am J Physiol. 1986;251:878–886. doi: 10.1152/ajpregu.1986.251.5.R878. [DOI] [PubMed] [Google Scholar]
- 22.Sanderford MG, Bishop VS. Angiotensin II acutely attenuates range of arterial baroreflex control of renal sympathetic nerve activity. Am J Physiol. 2000;279:1804–1812. doi: 10.1152/ajpheart.2000.279.4.H1804. [DOI] [PubMed] [Google Scholar]
- 23.Funder JW. Aldosterone action. Annu Rev Physiol. 1993;55:115–130. doi: 10.1146/annurev.ph.55.030193.000555. [DOI] [PubMed] [Google Scholar]
- 24.Ye P, Kenyon CJ, MacKenzie SM, Seckl JR, Fraser R, Connell JM, Davies E. Regulation of aldosterone synthase gene expression in the rat adrenal gland and central nervous system by sodium and angiotensin II. Endocrinology. 2003;144:3321–3328. doi: 10.1210/en.2003-0109. [DOI] [PubMed] [Google Scholar]
- 25.Franco M, Martínez F, Quiroz Y, Galicia O, Bautista R, Johnson RJ, Rodríguez-Iturbe B. Renal angiotensin II concentration and interstitial infiltration of immune cells are correlated with blood pressure levels in salt-sensitive hypertension. Am J Physiol. 2007;293:251–256. doi: 10.1152/ajpregu.00645.2006. [DOI] [PubMed] [Google Scholar]
- 26.Carillo BA, Beutel A, Mirandola DA, Vidonho AFJ, Furukawa LN, Casarini D, et al. Differential sympathetic and angiotensinergic responses in rats submitted to low- or high-salt diet. Regul Pept. 2007;140:5–11. doi: 10.1016/j.regpep.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Fyhrquist F, Metsärinne K, Tikkanen I. Role of angiotensin II in blood pressure regulation and in the pathophysiology of cardiovascular disorders. J Hum Hypertens. 1995;1:19–24. [PubMed] [Google Scholar]
- 28.Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 29.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 30.Bogdarina I, Welham S, King PJ, Burns S, Adrian JL. Clark epigenetic modification of the renin–angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]