Abstract
The hepatitis E virus (HEV) capsid protein has been demonstrated to be able to assemble into particles in vitro. However, this process and the mechanism of protein–protein interactions during particle assembly remain unclear. In this study, we investigated the assembly mechanism of HEV structural protein subunits, the capsid protein p239 (aa368–606), using analytical ultracentrifugation. It was the first to observe that the p239 can form particles in 4M urea as a result of supplementation with salt, including ammonium sulfate [(NH4)2SO4], sodium sulfate (Na2SO4), sodium chloride (NaCl), and ammonium chloride (NH4Cl). Interestingly, it is the ionic strength that determines the efficiency of promoting particle assembly. The assembly rate was affected by temperature and salt concentration. When (NH4)2SO4 was used, assembling intermediates of p239 with sedimentation coefficient values of approximately 5 S, which were mostly dodecamers, were identified for the first time. A highly conserved 28-aa region (aa368–395) of p239 was found to be critical for particle assembly, and the hydrophobic residues Leu372, Leu375, and Leu395of p239 was found to be critical for particle assembly, which was revealed by site-directed mutagenesis. This study provides new insights into the assembly mechanism of native HEV, and contributes a valuable basis for further investigations of protein assembly by hydrophobic interactions under denaturing conditions.
Keywords: particle assembly, urea, salt, hydrophobic interaction, hepatitis E virus, capsid protein
Introduction
Hepatitis E, which is a major form of infectious viral hepatitis with high rates of mortality in pregnant women, is caused by the hepatitis E virus (HEV).1 The HEV, which is the sole member of the genus Hepevirus within the family Hepeviridae, was first recognized in 1983.2 The spherical hepatitis E virion particle is 27–30 nm in diameter, as visualized by immune electron microscopy.2 Its 7.2 kb positive-sense RNA genome is encapsidated within a protein shell arranged with T = 3 icosahedral symmetry. The genome contains three open reading frames (ORF), and ORF2 encodes a capsid protein of 660aa residues that is responsible for most capsid-related functions, including particle assembly, host interaction, and immunogenicity.3
To date, there has been a lack of a robust cell culture system for HEV propagation. However, the expression of the ORF2 protein in heterologous systems has offered an excellent means to study HEV particle assembly in vitro.4–14 HEV virus-like particles (VLPs) with protruding spikes on the surface can be created using recombinant capsid proteins.4–6, 12–14 A capsid protein with an N-terminal truncation forms recombinant HEV (rHEV) particles that are smaller than the isolated native virions and exhibit T = 1 icosahedral symmetry.4–6 The crystal structures of HEV VLPs demonstrate that T = 1 particles share a common structural organization with virion-sized T = 3 particles and that each capsid protein contains four linear domains: the N domain (aa1–111), the S domain (aa118–313), the P1 domain (aa314–453) and the P2 domain (aa454–end).12–14 In addition, the N domain has been suggested to be involved in native particle assembly by binding to RNA.14 Recently, we analyzed the structure of the protruding domain (the E2s domain, aa459–602) of the capsid protein and found that the dimerization of this domain is essential for HEV–host interactions, including immunogenicity.15
The assembly mechanism of native HEV particles remains unclear, which is primarily due to the lack of an efficient cell system. The active principle of an efficient particulate recombinant hepatitis E vaccine (HEV 239) is a bacterially expressed recombinant peptide, p239, which corresponds to the aa368–606 region of HEV ORF2 of a genotype 1 strain of HEV.16 Previous studies have shown that p239 (aa368–606) is able to assemble into particles in vivo and could protect rhesus monkeys from HEV infection.5 The p239 particles could specifically bind and penetrate susceptible host cells in a manner similar to that of live viruses.17 These results suggest that the p239 particles share many common features with the native virus. Thus, the particles formed by p239 can serve as an excellent model for studies of viral assembly. In contrast, the N-terminal truncation of 26 residues from p239 disables the resultant E2 protein (aa394–606) from assembling into particles,10, 18 which suggests that these 26 residues are involved in the particle formation.
Urea is usually used to denature or dissociate proteins and particles.6, 19, 20 Interestingly, this study showed that p239 could form particles in urea when a salt [e.g., (NH4)2SO4] was added. To the best of our knowledge, this is the first report to demonstrate that intact VLPs can be assembled in the presence of urea. We also varied the incubation conditions, including the identity of the salt, the salt concentration, the temperature, and the incubation time, to further evaluate the influences of these conditions on p239 particle formation. The salt-mediated p239 assembly in 4M urea was characterized using various approaches, including analytical ultracentrifugation (AUC), electron microscopy (EM), dynamic light scattering (DLS), circular dichroism (CD), and gel filtration chromatography (GFC). The results indicated that the p239 assembly rate was largely affected by the ionic strength and the temperature. We also presented evidence to suggest that the hydrophobic core of p239 plays an important role in particle assembly.
Results
Particulate features of the p239 particles
Studies have previously reported that p239 particles share similar immunogenicity with native virions.5 Various structural analysis were performed to characterize the biochemical features of the p239 particles, which were obtained via previously described methods5 (Fig. 1). An EM study clearly revealed that the p239 particles with a diameter of approximately 25 nm possess a morphology similar to that of the T = 1 particles expressed in insect cells [Fig. 1(A)].4, 6 An AUC analysis, which was used to characterize the size distribution of the p239 particles, demonstrated that the p239 particles existed as a single major species with an s value of approximately 22.6 S [Fig. 1(B)]. GFC demonstrated that the particles were eluted at 13.7 min (kav = 0.1980) [Fig. 1(C)]. Moreover, DLS indicated that the diameter of the p239 particles was approximately 26 nm, which was consistent with the EM analysis [Fig. 1(D)]. Taken together, these results demonstrated the complete assembly and good homogeneity of the p239 particles.
Figure 1.
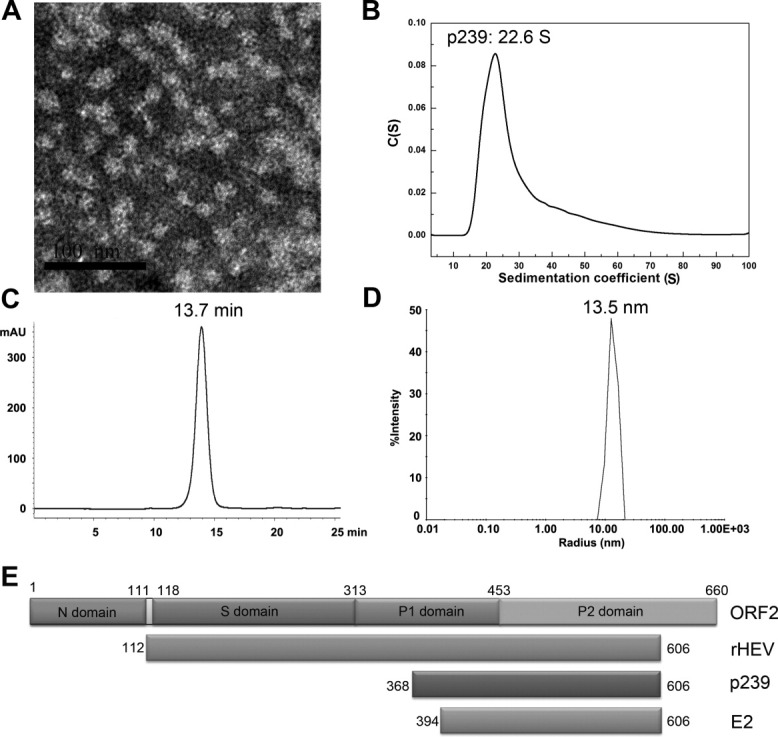
The characterization of the p239 particles. The p239 particles in PBS were analyzed using EM and determined to have a diameter of approximately 25 nm (A, bar = 100 nm). AUC demonstrated a sedimentation coefficient of approximately 23 S (B), and the GFC results revealed an elution time of 13.7 min (kav = 0.1980) (C). In addition, the DLS analysis showed a hydrated radius of approximately 13 nm (D), which is consistent with the EM data. The structural organization of HEV ORF2 and its different structural subunits containing cell-expressed rHEV and E. coli-expressed p239 and E2 were described (E).
The effect of (NH4)2SO4 on the overall refolding of p239
Aminonium sulfate, (NH4)2SO4, contributed to the self-assembly of p239 particles from 4M urea in our previous study.21 To investigate the exact role of (NH4)2SO4 in the p239 assembly procedure, refolded p239 particles with and without the (NH4)2SO4 pretreatment before renaturation dialysis were analyzed using AUC. The original unfolded p239 and E2 were initially characterized. Unfolded p239 was found to exist exclusively as a 1.7 S homodimer in 4M urea [Fig. 2(A)] with an apparent molecular mass of 45,722 ± 730 Da [Fig. 2(B)]. Similar to p239, E2 also existed as a homodimer in 4M urea with an s value of 2.0 S [Fig. 2(C)] and a molecular mass of 40,981 ± 1,523 Da [Fig. 2(D)]. These results are the first to demonstrate that the capsid particle of p239 assembles from its homodimer precursor. Thus, the dimerization of p239 is intrinsically strong in 4M urea as interpreted in the dimer crystal structure of its E2s moiety,15 which may not be involved in the particle assembly process.
Figure 2.
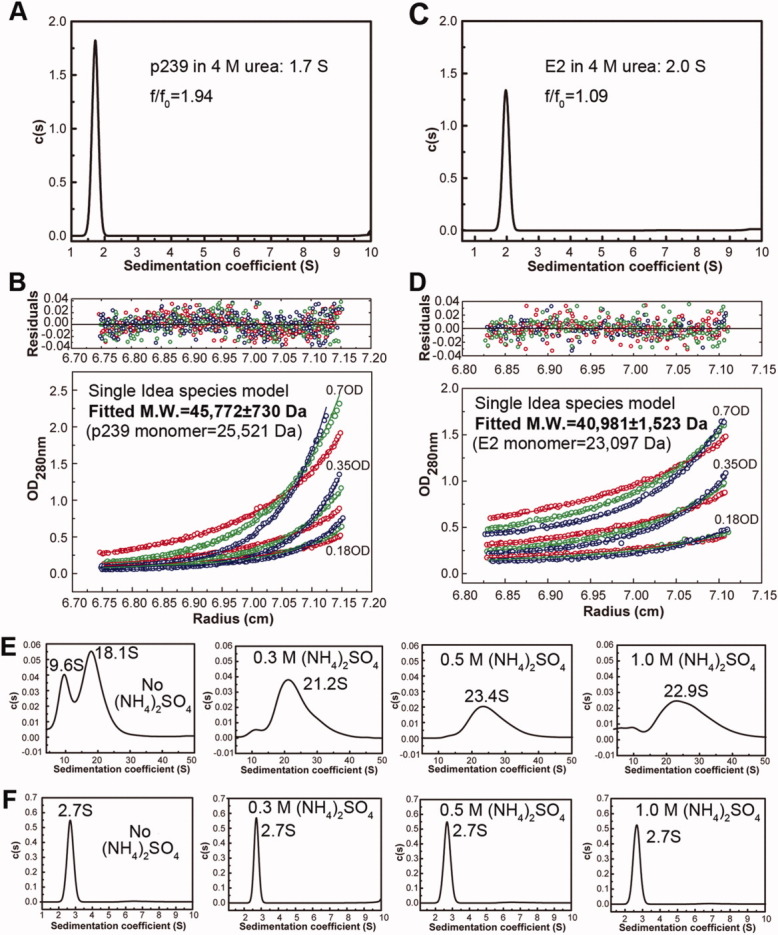
The size distribution of refolded p239 and E2 were investigated using AUC. SV (A) and SE (B) experiments show that the unfolded p239 existed as a dimer in urea. SV (C) and SE (D) experiments show that the unfolded E2 existed as a dimer in urea. The refolding of p239 (E) resulted in differences between the nontreated p239 [no (NH4)2SO4] and the (NH4)2SO4-treated p239 (the concentrations are labeled). The refolding of E2 (F) was similar between the nontreated E2 [no (NH4)2SO4] and the (NH4)2SO4-treated E2 (the concentrations are labeled). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
When refolded by overnight dialysis against PBS without (NH4)2SO4 pretreatment, p239 appeared to have two major species (9.6 S and 18.1 S) [Fig. 2(E)]. With respect to the original unfolded p239 in 4M urea (1.7 S), the larger components indicated that the assembly occurred, and the existence of two components indicated that the assembly was incomplete. When p239 was renatured with (NH4)2SO4 pretreatment step in the presence of 0.3, 0.5, or 1.0M (NH4)2SO4, single major component (s > 20) appeared, indicating the complete assembly [Fig. 2(E)]. In contrast, the assembly of E2 did not show any differences with or without the (NH4)2SO4 pretreatment step (a single major species with the same s value at 2.7 S) [Fig. 2(F)]. The RMSD of the best c(s) fitting was below 0.01, and the grayscale patterns of the residual bitmaps indicated a high quality fitting (data not shown). These results suggested that (NH4)2SO4 facilitated the hierarchical assembly of p239.
Specific salt effect on p239 assembly
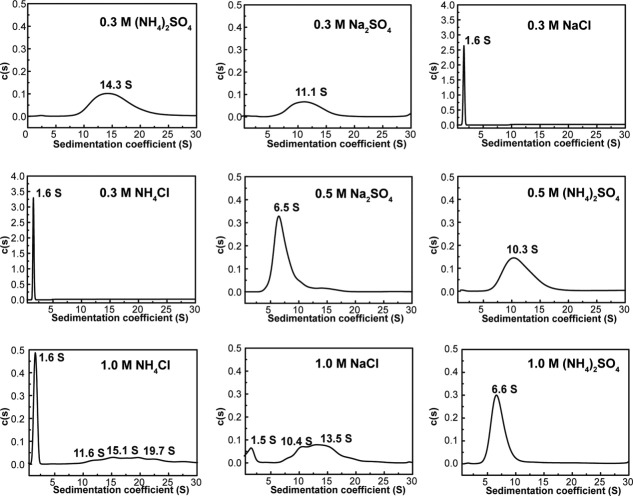
AUC sedimentation velocity (SV) experiment allows discerning wide scale of soluble species in solutions in terms of the sedimentation coefficient of the molecular or its ensemble form, which span from dimeric, oligomeric to particulate forms. It is unprecedented that a protein, p239 dimer, initiates particle assembly with the presence of (NH4)2SO4 in 4M urea as shown in Figure 3 (NH4)2SO4 panel. To further investigate whether ionic strength instead of specific salts facilitating particle assembly, p239 was treated with three other kinds of salts (Na2SO4, NaCl, and NH4Cl) in successively increased concentration 0.3, 0.5, and 1.0M, and incubated at 37°C overnight and analyzed using AUC in the presence of 4M urea. The addition of 1.0M Na2SO4 resulted in the precipitation of p239, and no difference was observed between the 0.3M and 0.5M treatments with NaCl and NH4Cl. Thus, the AUC profiles of the 1.0M Na2SO4, 0.5M NaCl, and 0.5M NH4Cl treatments are not shown.
Figure 3.
Sedimentation coefficient distribution analysis of p239 with different salt treatments in 4M urea. The salts and their concentrations are labeled in the panels. The assembly of p239 was activated by incubation with 0.3M (NH4)2SO4, 0.3M Na2SO4, 1.0M NaCl, or 1.0M NH4Cl, indicating the minimum ionic strength required for particle assembly.
As shown in Figure 3, a variation in the salt concentrations has the most striking effect on the activation of the p239 assembly. For Na2SO4 and (NH4)2SO4, 0.3M was sufficient to initiate the p239 assembly, and larger components (>2 S) were observed. In contrast, the treatments of p239 with 0.3M NaCl or NH4Cl did not result in aggregation, and their AUC profiles remained the same as that of unfolded p239 (1.6 S). Some irregular larger components were observed after 1.0M NaCl or NH4Cl was added, which indicated that a critical ionic strength was required to initiate particle assembly because the ionic strength is similar among 0.3M Na2SO4, 0.3M (NH4)2SO4, 1.0M NaCl, and 1.0M NH4Cl. In the presence of 1.0M NaCl, the unfolded component (with a sedimentation coefficient of 1.6 S) made up approximately 50% of the sample, whereas the 1.6 S component appeared to be the dominant species after the 1.0M NH4Cl treatment (over 90%), which indicated an incomplete assembly of p239 and different ion effects on the assembly. The relative effects of the salts on particle assembly activation are ranked in the sequence: Na2SO4 > (NH4)2SO4 > NaCl > NH4Cl.
The assembly process of (NH4)2SO4-treated p239 in urea
Although HEV particles can assemble in vitro, the actual steps required for the protein to assemble into the final particle have not been analyzed. Therefore, a series of experiments was performed to observe the (NH4)2SO4-mediated p239 assembly process in 4M urea.
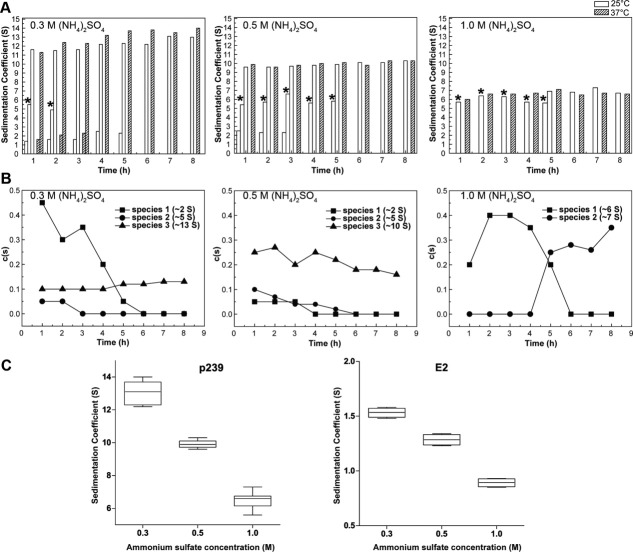
The states that exist during the (NH4)2SO4-mediated assembly procedure (1–8 h) were analyzed using AUC (Supporting Information Fig. S1). The time-scale sedimentation coefficient analysis of p239 is shown in a histogram, which is indicated in the figure legend of Figure 4(A). During the treatment with 0.3M (NH4)2SO4, p239 primarily existed as three sedimenting fractions (1.6 S, ∼5 S, and 12–13 S) during the first 2 h. The approximately 5 S species disappeared at 3 h, and p239 became exclusively a single species (12–13 S) after 6 h of incubation. During the 37°C incubation with 0.3M (NH4)2SO4, the state of p239 changed from two sedimenting components (∼2 S and ∼12 S) to a single component (13–14 S) after 4 h of incubation. Interestingly, we observed a difference during the treatment with 0.5M (NH4)2SO4 that was dependent on the incubation temperature. After incubation at 25°C, three species (∼2 S, ∼5 S, and ∼10 S) existed in the first 3 h and then became one species after 6 h. The extended existence of the approximately 5 S species for 5 h may be due to the stronger interaction formed in the intermediate. Incubation at 37°C led to an entirely different assembly pattern: only a predominant rapidly sedimenting fraction (∼10 S) existed during the entire procedure, which indicated that the complete assembly occurred rapidly. Regarding the treatment with 1.0M (NH4)2SO4, the same assembly pattern [i.e., only one species (6–7 S) during the entire procedure] was observed during the 37°C incubation. During the incubation at 25°C, p239 reached its homogeneity after 6 h, with a transition of two species (∼6 S and ∼7 S) during the 5 h incubation [Fig. 4(A)].
Figure 4.
Time-scale sedimentation coefficient distribution analysis of p239 with treatments of (NH4)2SO4. The components of the p239 treatments are shown in histogram form (A), and the two column types representing the components at 25°C and 37°C are labeled. The concentration of (NH4)2SO4 is also labeled. The c(s) distribution of the different species that were present following the 25°C incubation is shown in a line and symbol form (B). The sedimentation coefficient distributions of p239 and E2 following different (NH4)2SO4 treatments are shown in a box chart (C). *, assembly intermediates with sedimentation coefficient around 5 S.
The time-scale c(s) distribution analysis of the 25°C incubation of p239 is shown in Figure 4(B) to identify the quantity of different species in solution. Regarding the treatments with 0.3M (NH4)2SO4 and 0.5M (NH4)2SO4, three species existed in the beginning, including unfolded p239 (∼2 S), the assembly intermediate (∼5 S) and the end assembly product of 0.3-M-mediated p239 (∼13 S) and 0.5-M-mediated p239 (∼10 S). The unfolding species existed predominantly in the first 4 h, after which point it rapidly disappeared. The end product was the predominant species during the entire procedure with 0.5M (NH4)2SO4 treatment. A small amount of the assembly intermediate existed in the beginning, but this species disappeared. During the treatment with 1.0M (NH4)2SO4, no unfolded p239 or the assembly intermediates were observed, which indicated an acceleration in assembly. However, a transition from the approximately 6 S species to the approximately 7 S species was observed after 5 h.
These results revealed that the (NH4)2SO4-mediated assembly of p239 was concentration- and temperature-dependent. At a given temperature, p239 assembled slower at a lower (NH4)2SO4 concentration. Conversely, in the presence of a fixed salt concentration, p239 assembled more rapidly at a higher temperature (37°C). An incubation period was a prerequisite for the complete assembly of p239 with ideal stability and homogeneity. As shown in Figure 4(C), the sedimentation coefficient of E2 decreased when treated with increasing concentrations of (NH4)2SO4. E2 could not assemble in any conditions. Thus, the decreased sedimentation coefficient was merely a result of the ion effect on the AUC analysis. Interestingly, p239 exhibited a similar concentration-dependent tendency as E2, which indicated that p239 reached the same assembly state with different (NH4)2SO4 treatments, although the size distributions were different.
Particle formation of p239 in urea
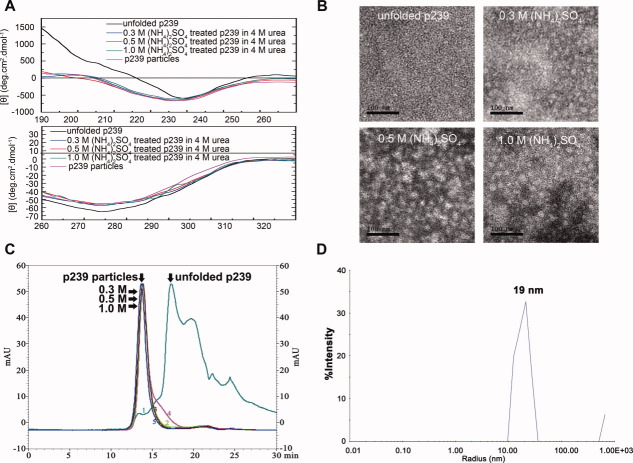
To test the hypothesis that p239 reached the same final assembly with different ammonium treatments, other analysis methods were applied to investigate the final states of p239 in urea, including EM, GFC, CD, and DLS (Fig. 5). Both the far-UV CD spectrum and the near-UV CD spectrum [Fig. 5(A)] of p239 following (NH4)2SO4 treatment were similar to those of the p239 particles in PBS; however, there was a considerable change compared with the unfolded p239, which suggested similar secondary and tertiary structures between the (NH4)2SO4-treated p239 and the p239 particles. Particles were observed in all (NH4)2SO4-treated p239 samples (0.3, 0.5, and 1.0M) in urea by EM analysis [Fig. 5(B)]. Based on the GFC data, the major peaks of 0.3M, 0.5M, and 1.0M (NH4)2SO4-treated p239 in 4M urea all eluted at a similar time as the p239 particles (kav = 0.1980). Unfolded p239 exhibited a delayed elution of the major peak compared with the p239 particles. A comparison between the (NH4)2SO4-treated p239 and the p239 particles confirmed the presence of the p239 particles in 4M urea [Fig. 5(A–C)]. The DLS results also confirmed the particle formation of p239 [Fig. 5(D)] following the 0.3M (NH4)2SO4 treatment (p239 existed as particles with a radius of ∼19 nm). Taken together, these results demonstrated that p239 could form particles in urea.
Figure 5.
Structural analysis of the (NH4)2SO4-treated p239 in urea. The far-UV and near-UV CD spectra (A) of the p239 particles (purple line), the unfolded p239 (black line), the 0.3M (blue line), 0.5M (red line), and 1.0M (NH4)2SO4-treated p239 in urea (green line) were compared. The EM observation of the unfolded p239, the 0.3M, 0.5M, and 1.0M (NH4)2SO4-treated p239 in urea were studied as labeled (B). The gel filtration profiles of p239 (C) following different treatments indicated the particle formation: unfolded p239 (1, olive green line, equilibration buffer: 10 mM PB7.5 + 4M urea), 0.3M (NH4)2SO4-treated p239 in urea [2, light-green line, equilibration buffer: 10 mM PB7.5 + 4M urea + 0.3M (NH4)2SO4], 0.5M (NH4)2SO4-treated p239 in urea [3, brown line, equilibration buffer: 10 mM PB7.5 + 4M urea + 0.5M (NH4)2SO4], 0.5M (NH4)2SO4-treated p239 in urea [4, magenta line, equilibration buffer: 10 mM PB7.5 + 4M urea + 1.0M (NH4)2SO4], and p239 particles in PBS (5, blue line, equilibration buffer: PBS). mAU, milliabsorbance units. DLS also demonstrated that the 0.3M (NH4)2SO4-treated p239 in urea formed particles with a radius of approximately 19 nm (D). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The critical role of hydrophobic interactions in particle assembly
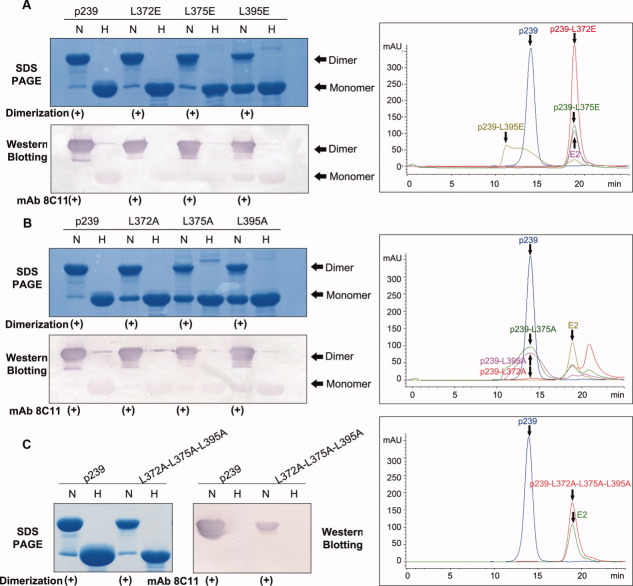
The domain (aa368–395) located at the N terminus of p239 is not present in E2; thus, this domain is likely involved in particle formation. The highly conserved sequence of hydrophobic amino acids among the 28 residues of this domain suggests that the hydrophobic interaction might be the dominant molecular interaction during particle assembly (Supporting Information Table S1). Because particle assembly could be triggered by the addition of salts (Fig. 3), and conserved hydrophobic residues have been verified to be required for the capsid particle assembly of the hepatitis virus,22 we wanted to determine the role of the hydrophobic residues in the N terminus during the particle assembly process. Three hydrophobic leucine residues (Leu372, Leu375, and Leu395) were replaced with Glu individually to perturb the hydrophobic interaction to investigate their strategic action contributed for particle assembly, and the mutants L372E, L375E, and L395E were generated. For the sake of folding maintenance monitoring, Ala replacement on the interest sites was performed as control and constructs L372A, L375A, and L395A were produced. SDS-PAGE was used to verify the dimerization of the mutants. The mutants were efficiently expressed, and they all mainly migrated as dimer and reacts with neutralizing mAb 8C11 in Western blotting, which demonstrates that the mutations on L372, L375, and L395 not lead unfavorable effect on protein folding [Fig. 6(A,B) left panel]. A GFC analysis was performed to further determine the nature of the assembly of the mutants, and we observed that the mutants L372E, L375E, and L395E failed to form p239-like particles [Fig. 6(A) right panel]. Although Ala replacement maintained the assembly ability of p239 [Fig. 6(B) right panel], the assembly ratio of particle component with respect to the dimer form decreased (Supporting Information Fig. S3), which suggested that the reduced hydrophobicity of side chain via Ala replacement leads to the lower particle assembly efficiency. To further confirm the important role of these three residues in the particle assembly, a mutant with triple-site mutation (L372A, L375A, and L395A) was constructed and its particle formation was checked. The results showed that the similar function of triple-site Ala mutation to that of single-site Glu mutation, which maintained dimerization architecture and mAb 8C11 reactivity [Fig. 6(C) left panel] but failed to form p239-like particles [Fig. 6(C) right panel]. These results suggested that the hydrophobic nature of the three leucine residues (Leu372, Leu375, and Leu395) is crucial for particle assembly, which can be triggered by salt addition. Furthermore, the weight of these three strategic leucine residues involved in particle assembly were quantitated and ranked as Leu372 > Leu375 > Leu395 (Supporting Information Fig. S3A).
Figure 6.
Mutational studies of the N-terminal region of p239. (A) SDS-PAGE, Western blotting, and GFC analysis of three single-site Glu mutants (p239-L372E, p239-L375E, and p239-L395E). (B) SDS-PAGE, Western blotting and GFC analysis of three single-site Ala mutants (p239-L372A, p239-L375A, and p239-L395A). (C) SDS-PAGE, Western blotting, and GFC analysis of the triple-site Ala mutant (p239-L372A-L375A-L395A). The lanes labeled with H indicate samples in the reduced condition that were heated to 100°C for 3 min, and these samples were primarily resolved as monomers. The lanes labeled with N indicate samples under non-reducing conditions, which only contained 0.1% SDS, and the samples were not heated. p239 and its mutants were reacted with the neutralizing mAb 8C11 of HEV. Western blotting analysis showed that all the mutants remained the strong reactivity with mAb 8C11 with respect to the prototype p239. Three single-site Ala mutants could partially assemble into particles with similar elution time to p239 particles, while three single-site Glu mutants and the triple-site Ala mutant failed to form p239-like particles. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
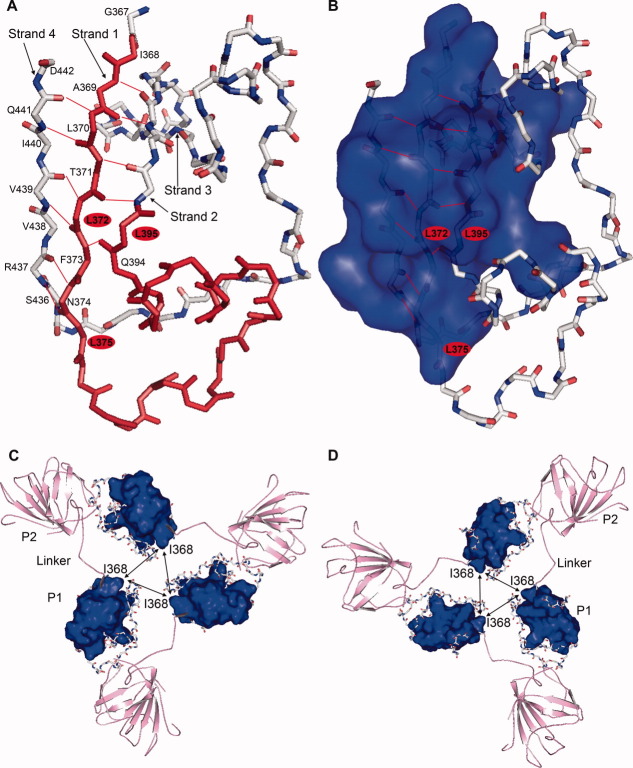
A structural analysis provided new insight into the importance of the N-terminal domain (aa368–395) [Fig. 7(A,B)]. This region (aa368–395) stabilized the other two strands of the β-sheet via hydrogen bonds, formed a tight anti-parallel β-sheet architecture and promoted the formation of a hydrophobic core. Leu372, Leu375, and Leu395 were directly engaged in the formation of the hydrophobic core, which consists of three strands of β-sheets. Leu372 is involved in the intramolecular hydrogen bonding contacts between two β-sheets, whereas Leu375 and Leu395 did not exhibit any hydrogen bonding contacts. A similar hydrophobic core has been observed in the case of other anti-parallel β-sheet folding proteins.23, 24 Because a hydrophobic force often plays an important role in protein–protein interactions, we hypothesized that this hydrophobic core may be crucial for particle assembly. The hydrophobic core may be involved in the trimer formation of the P1 domain near the 3-fold axis [Fig. 7(C,D)], and the trimer formation is responsible for the particle formation. Leu372, Leu375, and Leu395 may be important for maintaining the structure of the hydrophobic core; therefore, mutating the leucine residues to hydrophilic residues destroyed the hydrophobic core and perturbed the particle assembly.
Figure 7.
Structural model of the N-terminal domain (aa367–442) and the trimer of p239. (A) Backbone structure of the N-terminal domain of p239 with the residues, strands, and hydrogen bonds labeled. (B) Backbone structure the N-terminal domain of p239 with the surface of the hydrophobic core shown in blue. In A and B, the leucine residues in the hydrophobic core chosen for the mutagenesis are identified using red ovals. The buried hydrogen bonds are shown in red, whereas the other hydrogen bonds are not shown. (C) and (D) Structural model of the p239 trimer from an inside particle view (C) and an outside particle view (D). The P1 domain is shown using a backbone representation, and the surface of the hydrophobic core is shown in blue. The P2 domain and the hinge linker connecting the P1 and P2 domains are shown in pink. The hydrophobic interactions between the p239 monomers are indicated by arrows. The N-terminal region of p239 played an important role in stabilizing the folding of Strand 2 and Strand 4 and determined the formation of a small hydrophobic core that was critical for the trimer formation. The structural model was derived from the crystal structure of genotype 3 HEV ORF2 (PDB ID: 2ZTN). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
Most recombinant ORF2 proteins form VLPs when expressed in eukaryotic cells, which makes it difficult to elucidate the individual steps involved in particle assembly.4, 6, 12, 13 Our p239 protein was expressed by E. coli and assembled into particles via a refolding step in vitro, which allowed for assembly studies of HEV.
The effect of salt on p239 particle assembly
In this study, we successfully demonstrated that p239 underwent an association step to result in particle formation after treatment with (NH4)2SO4 in urea (Fig. 5). The finding that the appearance of particles was unaffected by urea was consistent with the ability of native virions to survive in the intestinal tract.25, 26
These results suggest that the assembly rate primarily depends on the temperature, and to a lesser extent, on the concentration of the salt. To ascertain the importance of salts, we applied three different salts in the assembly experiments. These salts were all able to activate the particle assembly of p239, and multivalent salts [(NH4)2SO4 and Na2SO4] had stronger effects, which were likely due to the larger ionic strength at the same concentration compared with the monovalent salts (NaCl and NH4Cl). A previous structural study on HEV VLPs demonstrated that salts and acidic pH conditions induce ORF2 peptides to form particles during crystallization, and the hypothesis that salt may be crucial for particle assembly has previously been raised.12 This study verified that salt is involved in the assembly of HEV particles.
The identification of intermediates in the (NH4)2SO4-mediated assembly
The assembly intermediate of the HEV has not been captured. An analysis of the assembly process of p239 using AUC makes it possible to demonstrate the existence of an intermediate. The appearance and disappearance of a component with a sedimentation coefficient of approximately 5 S were identified using AUC during the assembly procedures of the 0.3 and 0.5M (NH4)2SO4-treated p239 at 25°C. Indeed, the extended existence of an intermediate in the presence of a higher (NH4)2SO4 concentration indicated that the stability of the intermediate may be dependent on the concentration of the salt [Fig. 4(B)]. Urea may also be essential for the identification of the assembly intermediate because it reduced the rate of the assembly and made it possible to record the assembly process.
Subunit interactions involved in particle formation
Three contacts that are essential for placing the capsid proteins into a T = 1 shell domain were characterized using a structural study, namely, the tight clusters at the 5-fold, 3-fold, and 2-fold axes.12, 13 The S domain was not included in p239, and we verified that dimerization was not involved in particle assembly [Fig. 2 and Supporting Information Fig. S2). Therefore, trimer formation may be the crucial contact for placing the capsid proteins into the VLPs. Based on the structural analysis, the clustering at the 3-fold axis results from weak hydrophobic interaction, thus, trimers could not be observed on the gel (Supporting Information Fig. S2).
The characteristics of p239 and E2 revealed that the N-terminal region is crucial for particle assembly. We performed further structural analysis of the region and found that the aa368–395 region, which contains hydrogen bonding contacts, stabilized the other two β strands and promoted the formation of a hydrophobic core (Fig. 7). To further determine the mechanism of p239 particle assembly, site-directed mutagenesis was utilized to disrupt the hydrophobic core of the P1 domain. Three mutations to amino acids involved in the hydrophobic core (i.e., L372E, L375E, and L395E) led to a failure in the particle formation process. These results confirm that the hydrophobic core is crucial for particle assembly. We speculate that the hydrophobic core is important for subunit interaction near the 3-fold axis via hydrophobic interactions.
In summary, we obtained properly assembled HEV particles in a solution of 4M urea that was supplemented with specific salts. Importantly, we observed that the particle assembly from capsid p239 was initiated by homodimerization of the protein, and the protein was further packed into oligomeric intermediates via hydrophobic interactions triggered by salts. We also showed that hydrophobic interactions among the capsid protein molecules are crucial in HEV particle assembly. Overall, we provide valuable insights regarding how protein assembly mediated by hydrophobic interactions may occur under denaturing conditions when a salt is used, and propose a putative assembly process of p239 T = 1 VLP in urea (Supporting Information Fig. S4). Currently, the HEV 239 vaccine has been approved with high efficacy and safety in a large-scale, randomized, double-blind placebo-controlled phase 3 trial.16 This study provides well-defined information for the VLP assembly process control during the manufacture of the vaccine.
Materials and Methods
Cloning of the p239 mutant genes
The three mutants were generated using site-directed PCR reactions, which were amplified from the p239 gene.5 The mutated DNA fragments were subcloned into the pMD 18-T vector using a TA cloning kit (TaKaRa, Dalian, China). The resulting plasmid was digested with NdeI and EcoRI for 2 h at 37°C. The flanking fragment was purified using agarose gel electrophoresis and a gel extraction mini kit (Watson, Shanghai, China) and ligated to a nonfusion pTO-T7 expression vector that had been previously27 linearized with NdeI and EcoRI. All of the clones were further characterized by sequencing and a restriction enzyme analysis.
Production and purification of p239, E2, and related mutant proteins
The plasmid-containing genes that encode the EV capsid protein E2, p239,5, 10 and the p239 mutants were transformed into E. coli ER2566 cells (Invitrogen). An overnight culture of the transformant was grown at 37°C in LB medium with 50 μg/mL kanamycin until the optical density value reached 0.8 OD at 660 nm. Isopropyl-β-D-thiogalactoside was added to the culture to a final concentration of 0.2 mM, and the culture was further incubated for 6 h. The bacterial cells harvested from the culture were lysed by sonication, and the proteins formed inclusion bodies. The inclusion bodies were separated from the cellular debris by treatment with 2% Triton X-100 at 37°C for 30 min, and the inclusion bodies were dissolved in 4M urea buffer [10 mM PB7.5 (phosphate buffer, pH7.5), 4M urea]. The proteins were further purified using an AKTA Explorer 100 FPLC chromatograph system (GE) with a Q Sepharose FF column, which was equilibrated in 10 mM PB7.5 + 4M urea, and the proteins were eluted in 10 mM PB7.5 + 4M urea + 50 mM NaCl. The purified proteins in 10 mM PB7.5 + 4M urea +50 mM NaCl were dialyzed against 10 mM PB7.5 + 4M urea overnight and were used for further biological analyses and salt-mediated assembly experiments.
Salt treatment
(NH4)2SO4, NH4Cl, NaCl, and Na2SO4 were purchased from Sigma and used without further treatment. All of the salts were dissolved in the 4M urea buffer (10 mM PB7.5, 4M urea) at a concentration of 3.0M. All of the aqueous solutions were sterilized by filtration through 0.22 μm Millipore filters prior to their use in the salt-mediated assembly experiments. All of the salt treatment experiments were carried out in 4M urea buffer unless otherwise stated.
The protein solution in 4M urea buffer (10 mM PB7.5, 4M urea) were mixed with the 3.0M stock buffer of the three salts to reach the final salt concentrations in the assembly experiments (0.3, 0.5, and 1.0M). Overnight incubation of p239 with (NH4)2SO4, Na2SO4, NH4Cl, or NaCl at concentrations of 0.3, 0.5, and 1.0M at 37°C were performed to analyze the specific effects of salt on particle assembly. In the assembly process study, p239 and E2 were incubated with 0.3, 0.5, and 1.0M (NH4)2SO4 at 25°C and 37°C, and the incubation times were 1–8 h in 1 h intervals. The final concentration of p239 protein subjected to assembly was 1 mg/mL determined by BCA assay.
Analytical ultracentrifugation
The SV was used to monitor the status of the salt-treated p239 as previously described.15 The samples were diluted to 1 OD at 280 nm in 1.2 cm lightpath, and the reference buffer corresponded with the different sample buffers. For example, we used 10 mM PB7.5 with 4M urea as the reference buffer for nontreated p239, 10 mM PB7.5 with 4M urea and 0.3M (NH4)2SO4 for the 0.3M (NH4)2SO4 treatments, and PBS for the refolding samples. The rotor speed was set to 20,000 rpm and 40,000 rpm for p239 and E2, respectively. The sedimentation coefficient was obtained using the c(s) method28 with the Sedfit software that was kindly provided by Dr. P. Schuck, National Institutes of Health (NIH) (http://www.analyticalultracentrifugation.com). The accurate molecular masses of p239 and E2 in urea buffer (i.e., the original state) were determined from the results of the SE experiments. p239 and E2 were both diluted to 0.7 OD, 0.35 OD, and 0.18 OD in 10 mM PB7.5 with 4M urea. The samples were centrifuged sequentially at 14,000 rpm, 17,000 rpm, 20,000 rpm, and 42,000 rpm for solute depletion. The data sets were processed as previously described using a nonlinear least squares fit.15, 29 In subsequent models, the monomer molecular mass was fixed at the value calculated from the p239 and E2 sequences (25,521 Da and 23,098 Da, respectively).
SDS-PAGE
The analysis of proteins using SDS-PAGE was performed according to the method of Laemmli with minor modifications. We used polyacrylamide gels with 12% acrylamide in the separating gel and 5% acrylamide in the stacking gel. The protein samples were mixed with equal volumes of 2× loading buffer (100 mM Tris–HCl, pH 6.8, 200 mM BME, 4% SDS, 0.2% bromophenol blue, and 20% glycerol). The sample mixtures were loaded onto the separating gel without being heated.
Gel filtration chromatography
The samples treated with different concentrations of (NH4)2SO4 were loaded onto a TSK Gel PW5000xl 7.8x300-mm column (TOSOH, Japan) equilibrated in corresponding sample buffers using an 1120 Compact LC HPLC system (Agilent Technologies, German). The samples were diluted to 1 mg/mL and the column flow rate was maintained at 0.5 mL/min, and we determined whether particles were formed by comparing the results to the p239 particles. Peak area of particle and dimer component could be integrated using peak area calculation, and the relative content of particle component for each protein was determined by area normalization to the sum of particle and dimer area.
Electron microscopy
The p239 particles were examined using negative-stain EM. A sample was diluted to 1 mg/mL, applied to a carbon-coated grid, and the sample was stained with 2% uranyl acetate after the removal of excess fluid. The samples were examined using a JEM2100HC transmission electron microscope (JEOL, Japan) operating at 200 kV to characterize the morphology of the p239 particles.
Circular dichroism spectrometry (CD)
The CD spectra were obtained using a Jasco J-810 spectropolarimeter, and the samples were placed in a 0.01-cm and 1.0-cm path-length cuvette for the far-UV and near-UV measurements, respectively. The spectra were the averages of three scans at a 50 nm/min scan speed, and the buffer baselines were subtracted. The data were collected from 190 nm to 260 nm for the far-UV measurement at a protein concentration of 4.5 mg/mL, whereas a protein concentration of 0.5 mg/mL was used for the 260–320 nm near-UV data collection. The CD ellipticity in degrees was converted to the mean residue molar ellipticity, [θ], using the formula
where θ is the ellipticity in mdeg, l is the optical path in cm, Cg is the concentration in mg/mL, M is the molecular mass, and n is the number of residues in the protein. The mean residue molar ellipticity is given in deg · cm2 · dmol−1.
Dynamic light scattering
The hydrodynamic size distributions of the p239 particles were measured using a DynaPro-MS/X (Protein Solutions,) DLS system.30 Samples (15 μL) were loaded into a 1.5-mm path length 12 μL quartz cuvette. Data were collected and analyzed with Dynamics software. The hydrodynamic radius (RH) of the particles was calculated through the Stokes-Einstein equation and 15 acquisitions were averaged.
Glossary
Abbreviations
- AUC
analytical ultracentrifugation
- CD
circular dichroism
- DLS
dynamic light scattering
- EM
electron microscopy
- GFC
gel filtration chromatography
- HEV
hepatitis E virus
- NIH
National Institutes of Health
- ORF
open reading frames
- SV
sedimentation velocity
- VLP
virus-like particle
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Jaiswal SP, Jain AK, Naik G, Soni N, Chitnis DS. Viral hepatitis during pregnancy. Int J Gynaecol Obstet. 2001;72:103–108. doi: 10.1016/s0020-7292(00)00264-2. [DOI] [PubMed] [Google Scholar]
- 2.Balayan MS, Andjaparidze AG, Savinskaya SS, Ketiladze ES, Braginsky DM, Savinov AP, Poleschuk VF. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology. 1983;20:23–31. doi: 10.1159/000149370. [DOI] [PubMed] [Google Scholar]
- 3.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li TC, Yamakawa Y, Suzuki K, Tatsumi M, Razak MA, Uchida T, Takeda N, Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li SW, Zhang J, Li YM, Ou SH, Huang GY, He ZQ, Ge SX, Xian YL, Pang SQ, Ng MH, Xia NS. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine. 2005;23:2893–2901. doi: 10.1016/j.vaccine.2004.11.064. [DOI] [PubMed] [Google Scholar]
- 6.Xing L, Kato K, Li T, Takeda N, Miyamura T, Hammar L, Cheng RH. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology. 1999;265:35–45. doi: 10.1006/viro.1999.0005. [DOI] [PubMed] [Google Scholar]
- 7.Panda SK, Nanda SK, Zafrullah M, Ansari IH, Ozdener MH, Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purdy MA, McCaustland KA, Krawczynski K, Tam A, Beach MJ, Tassopoulos NC, Reyes GR, Bradley DW. Expression of a hepatitis E virus (HEV)-trpE fusion protein containing epitopes recognized by antibodies in sera from human cases and experimentally infected primates. Arch Virol. 1992;123:335–349. doi: 10.1007/BF01317268. [DOI] [PubMed] [Google Scholar]
- 9.He J, Tam AW, Yarbough PO, Reyes GR, Carl M. Expression and diagnostic utility of hepatitis E virus putative structural proteins expressed in insect cells. J Clin Microbiol. 1993;31:2167–2173. doi: 10.1128/jcm.31.8.2167-2173.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang JZ, Ng MH, Xia NS, Lau SH, Che XY, Chau TN, Lai ST, Im SW. Conformational antigenic determinants generated by interactions between a bacterially expressed recombinant peptide of the hepatitis E virus structural protein. J Med Virol. 2001;64:125–132. doi: 10.1002/jmv.1027. [DOI] [PubMed] [Google Scholar]
- 11.Tsarev SA, Tsareva TS, Emerson SU, Kapikian AZ, Ticehurst J, London W, Purcell RH. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 12.Guu TS, Liu Z, Ye Q, Mata DA, Li K, Yin C, Zhang J, Tao YJ. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci USA. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, Shima R, Moriishi K, Tsukihara T, Li TC, Takeda N, Miyamura T, Matsuura Y. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci USA. 2009;106:12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing L, Li TC, Mayazaki N, Simon MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, Cheng RH. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Tang X, Seetharaman J, Yang C, Gu Y, Zhang J, Du H, Shih JW, Hew CL, Sivaraman J, Xia N. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 2009;5:e1000537. doi: 10.1371/journal.ppat.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu FC, Zhang J, Zhang XF, Zhou C, Wang ZZ, Huang SJ, Wang H, Yang CL, Jiang HM, Cai JP, Wang YJ, Ai X, Hu YM, Tang Q, Yao X, Yan Q, Xian YL, Wu T, Li YM, Miao J, Ng MH, Shih JW, Xia NS. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet. 2010;376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]
- 17.He S, Miao J, Zheng Z, Wu T, Xie M, Tang M, Zhang J, Ng MH, Xia N. Putative receptor-binding sites of hepatitis E virus. J Gen Virol. 2008;89:245–249. doi: 10.1099/vir.0.83308-0. [DOI] [PubMed] [Google Scholar]
- 18.Ge S, Zhang J, Huang G, Pang S, Zhou K, Xia N. [The immuno-protect study of a hepatitis E virus ORF2 peptide expressed in E. coli] Wei Sheng Wu Xue Bao. 2003;43:35–42. [PubMed] [Google Scholar]
- 19.Mok YK, de Prat Gay G, Butler PJ, Bycroft M. Equilibrium dissociation and unfolding of the dimeric human papillomavirus strain-16 E2 DNA-binding domain. Protein Sci. 1996;5:310–319. doi: 10.1002/pro.5560050215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umashankar M, Murthy MR, Savithri HS. Mutation of interfacial residues disrupts subunit folding and particle assembly of Physalis mottle tymovirus. J Biol Chem. 2003;278:6145–6152. doi: 10.1074/jbc.M207992200. [DOI] [PubMed] [Google Scholar]
- 21.Li SW, Zhang J, He ZQ, Gu Y, Liu RS, Lin J, Chen YX, Ng MH, Xia NS. Mutational analysis of essential interactions involved in the assembly of hepatitis E virus capsid. J Biol Chem. 2005;280:3400–3406. doi: 10.1074/jbc.M410361200. [DOI] [PubMed] [Google Scholar]
- 22.Yu M, Miller RH, Emerson S, Purcell RH. A hydrophobic heptad repeat of the core protein of woodchuck hepatitis virus is required for capsid assembly. J Virol. 1996;70:7085–7091. doi: 10.1128/jvi.70.10.7085-7091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deechongkit S, Nguyen H, Jager M, Powers E, Gruebele M, Kelly J. β-Sheet folding mechanisms from perturbation energetics. Curr Opin Struct Biol. 2006;16:94–101. doi: 10.1016/j.sbi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Jager M, Deechongkit S, Koepf EK, Nguyen H, Gao J, Powers ET, Gruebele M, Kelly JW. Understanding the mechanism of β-sheet folding from a chemical and biological perspective. Biopolymers. 2008;90:751–758. doi: 10.1002/bip.21101. [DOI] [PubMed] [Google Scholar]
- 25.Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Emerson SU, Purcell RH. Hepatitis E virus. Rev Med Virol. 2003;13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 27.Luo WX, Zhang J, Yang HJ, Li SW, Xie XY, Pang SQ, Li SJ, Xia NS. Construction and application of an Escherichia coli high effective expression vector with an enhancer. Sheng Wu Gong Cheng Xue Bao. 2000;16:578–581. [PubMed] [Google Scholar]
- 28.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruce J, Berne RP. Dynamic Light Scattering. the United States: Courier Dover Publications; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.