Abstract
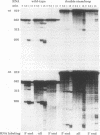
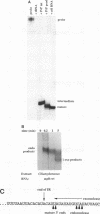
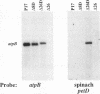
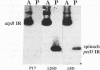
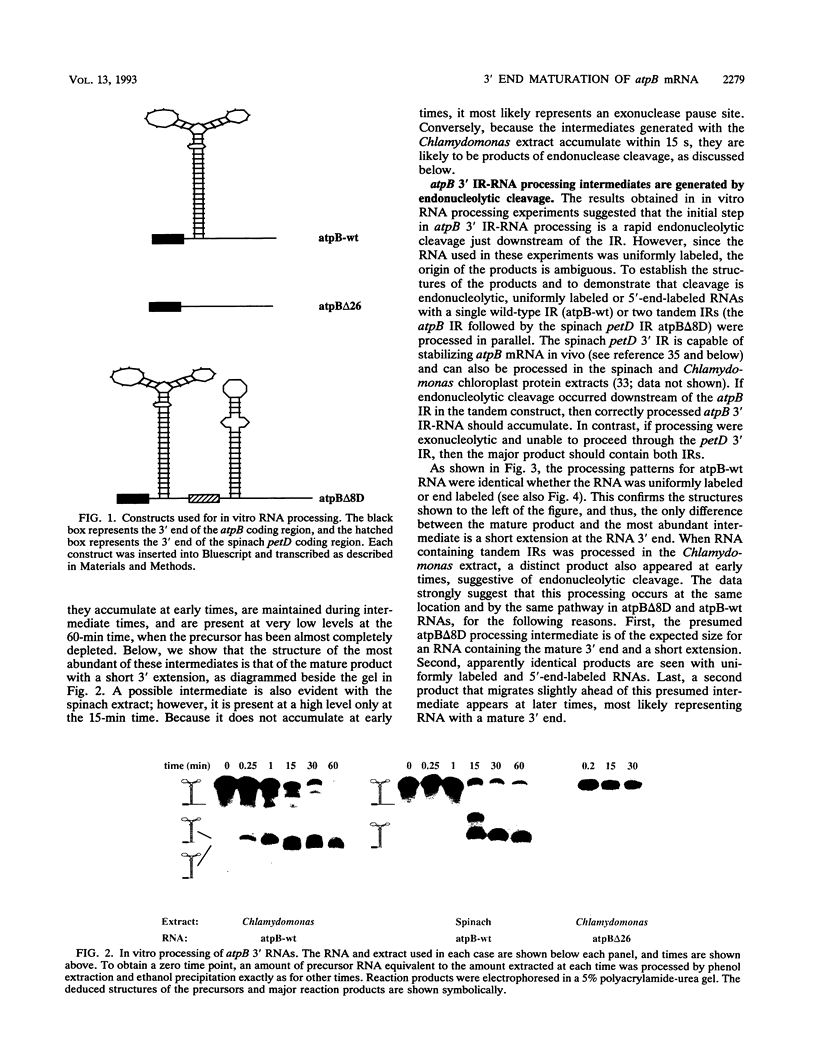
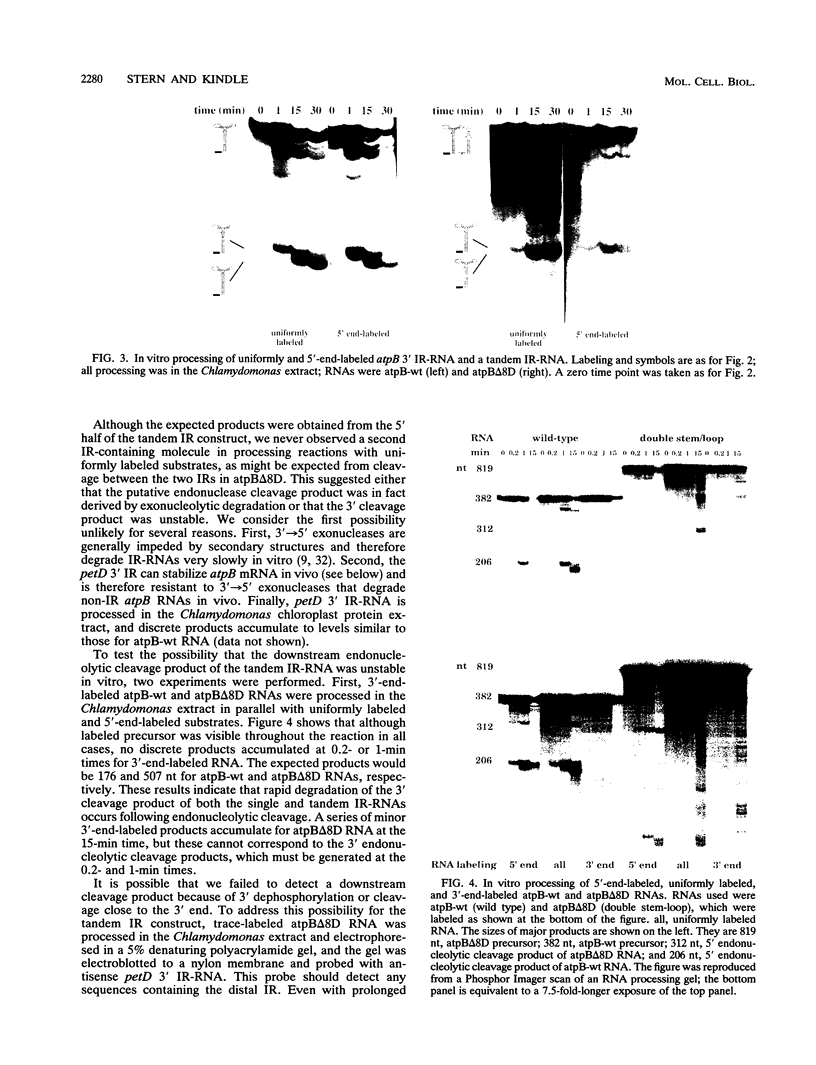
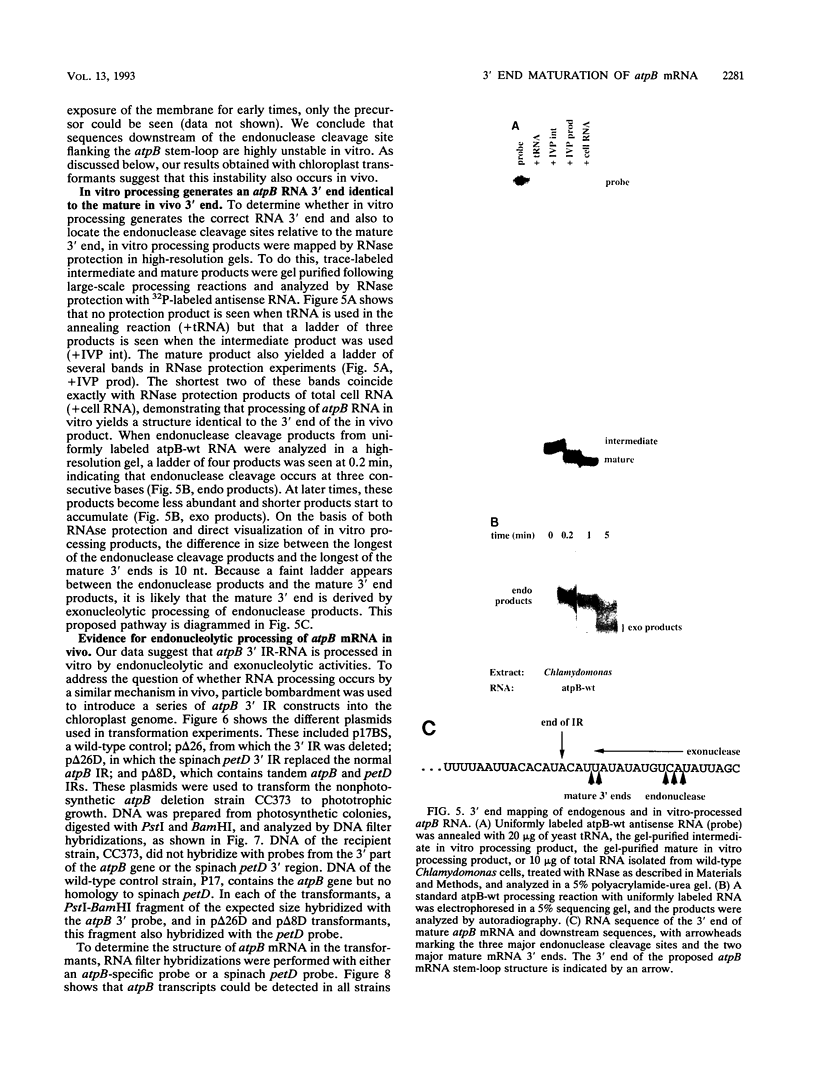
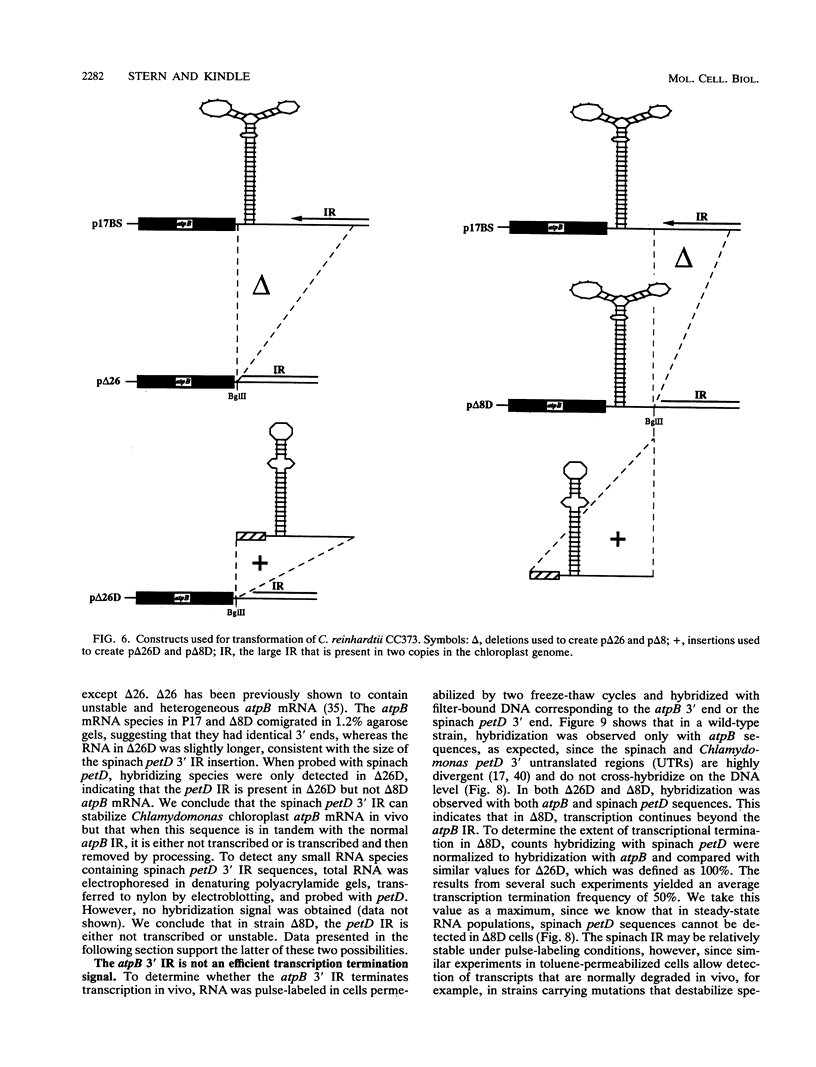
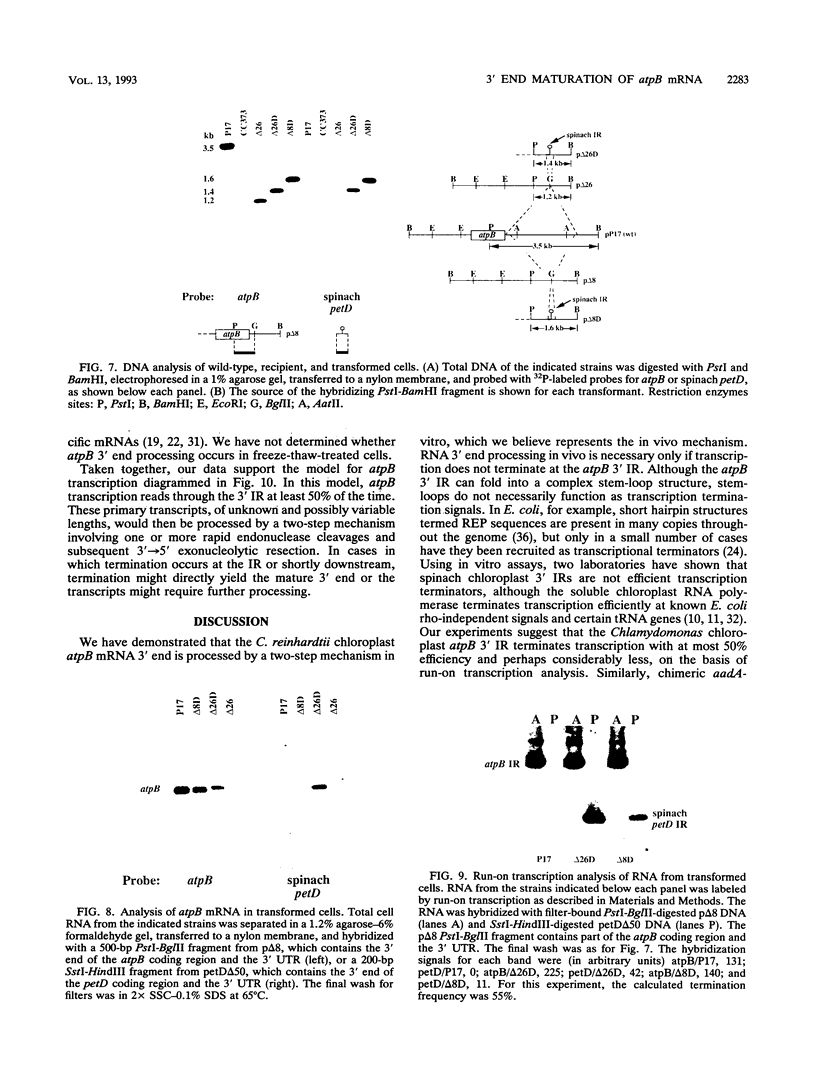
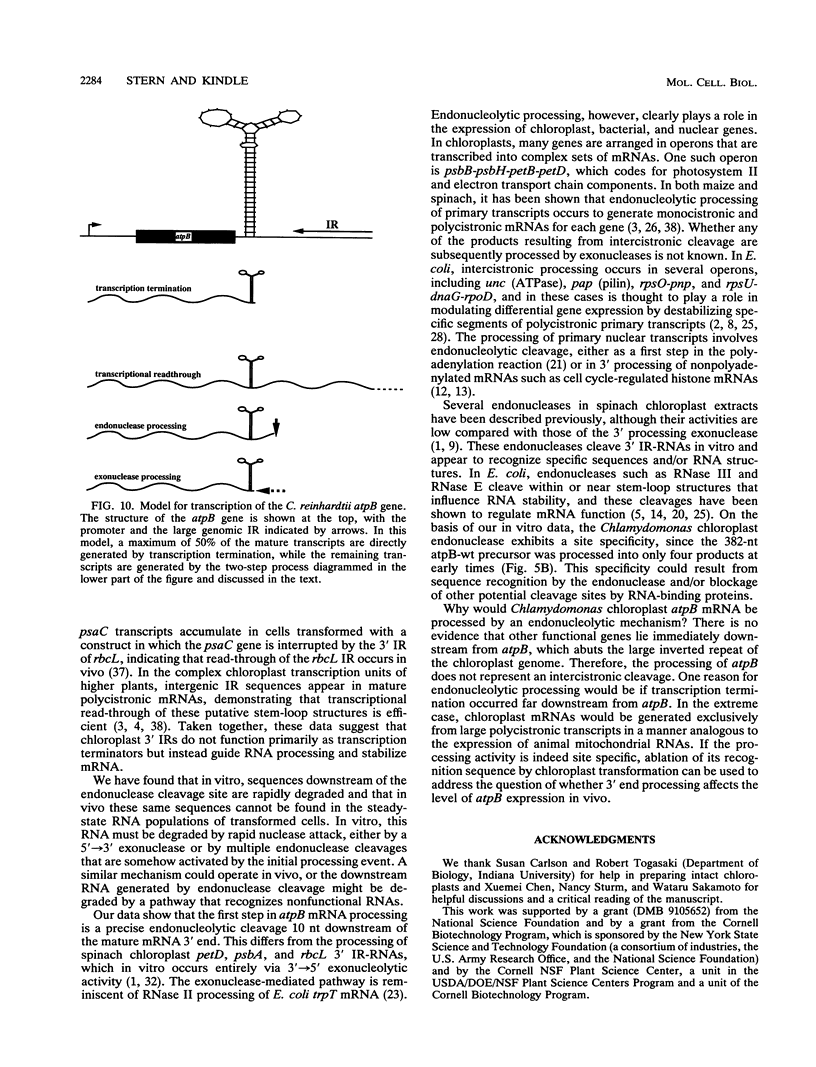
Inverted repeat (IR) sequences are found at the 3' ends of most chloroplast protein coding regions, and we have previously shown that the 3'IR is important for accumulation of atpB mRNA in Chlamydomonas reinhardtii (D. B. Stern, E.R. Radwanski, and K. L. Kindle, Plant Cell 3:285-297, 1991). In vitro studies indicate that 3' IRs are inefficient transcription termination signals in higher plants and have furthermore defined processing activities that act on the 3' ends of chloroplast transcripts, suggesting that most chloroplast mRNAs are processed at their 3' ends in vivo. To investigate the mechanism of 3' end processing in Chlamydomonas reinhardtii chloroplasts, the maturation of atpB mRNA was examined in vitro and in vivo. In vitro, a synthetic atpB mRNA precursor is rapidly cleaved at a position 10 nucleotides downstream from the mature 3' terminus. This cleavage is followed by exonucleolytic processing to generate the mature 3' end. In vivo run-on transcription experiments indicate that a maximum of 50% of atpB transcripts are transcriptionally terminated at or near the IR, while the remainder are subject to 3' end processing. Analysis of transcripts derived from chimeric atpB genes introduced into Chlamydomonas chloroplasts by biolistic transformation suggests that in vivo processing and in vitro processing occur by similar or identical mechanisms.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988 Sep;7(9):2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. Tissue-dependent plastid RNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell. 1989 Apr;1(4):437–445. doi: 10.1105/tpc.1.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belknap W. R. Partial Purification of Intact Chloroplasts from Chlamydomonas reinhardtii. Plant Physiol. 1983 Aug;72(4):1130–1132. doi: 10.1104/pp.72.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988 Jun 10;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Chen H. C., Stern D. B. Specific ribonuclease activities in spinach chloroplasts promote mRNA maturation and degradation. J Biol Chem. 1991 Dec 15;266(35):24205–24211. [PubMed] [Google Scholar]
- Chen L. J., Orozco E. M., Jr Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 1988 Sep 12;16(17):8411–8431. doi: 10.1093/nar/16.17.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. J., Rogers S. A., Bennett D. C., Hu M. C., Orozco E. M., Jr An in vitro transcription termination system to analyze chloroplast promoters: identification of multiple promoters for the spinach atpB gene. Curr Genet. 1990 Jan;17(1):55–64. doi: 10.1007/BF00313249. [DOI] [PubMed] [Google Scholar]
- Cotten M., Gick O., Vasserot A., Schaffner G., Birnstiel M. L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3' RNA processing reaction. EMBO J. 1988 Mar;7(3):801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R., Ellmeier W., Birnstiel M. L. Mature mRNA 3' end formation stimulates RNA export from the nucleus. EMBO J. 1991 Nov;10(11):3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehretsmann C. P., Carpousis A. J., Krisch H. M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992 Jan;6(1):149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- Gagné G., Guertin M. The early genetic response to light in the green unicellular alga Chlamydomonas eugametos grown under light/dark cycles involves genes that represent direct responses to light and photosynthesis. Plant Mol Biol. 1992 Feb;18(3):429–445. doi: 10.1007/BF00040659. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B. M., Zurawski G., Hallick R. B. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986;118:253–270. doi: 10.1016/0076-6879(86)18077-3. [DOI] [PubMed] [Google Scholar]
- Kindle K. L., Richards K. L., Stern D. B. Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1721–1725. doi: 10.1073/pnas.88.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka M. R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J. D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell. 1989 Sep 8;58(5):869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J Biol Chem. 1992 Jan 15;267(2):1054–1061. [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Monod C., Goldschmidt-Clermont M., Rochaix J. D. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet. 1992 Feb;231(3):449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Rock C. D., Barkan A., Taylor W. C. The maize plastid psbB-psbF-petB-petD gene cluster: spliced and unspliced petB and petD RNAs encode alternative products. Curr Genet. 1987;12(1):69–77. doi: 10.1007/BF00420729. [DOI] [PubMed] [Google Scholar]
- Régnier P., Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3' stabilizing stem and loop structure. J Mol Biol. 1991 Jan 20;217(2):283–292. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Kindle K. L., Stern D. B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of beta-glucuronidase translational fusions. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E. M., Hartz D., Gold L., Simoni R. D. Ribosome-binding sites and RNA-processing sites in the transcript of the Escherichia coli unc operon. J Bacteriol. 1989 Jul;171(7):3901–3908. doi: 10.1128/jb.171.7.3901-3908.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. Chloroplast mRNA 3' end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991 Jun;10(6):1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd H. S., Boynton J. E., Gillham N. W. Mutations in nine chloroplast loci of Chlamydomonas affecting different photosynthetic functions. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1353–1357. doi: 10.1073/pnas.76.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L. E., Berry-Lowe S., Schmidt G. W. Chloroplast RNA Stability in Chlamydomonas: Rapid Degradation of psbB and psbC Transcripts in Two Nuclear Mutants. Plant Cell. 1991 Feb;3(2):175–189. doi: 10.1105/tpc.3.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Chloroplast mRNA 3' end maturation is biochemically distinct from prokaryotic mRNA processing. Plant Mol Biol. 1989 Dec;13(6):615–625. doi: 10.1007/BF00016017. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Jones H., Gruissem W. Function of plastid mRNA 3' inverted repeats. RNA stabilization and gene-specific protein binding. J Biol Chem. 1989 Nov 5;264(31):18742–18750. [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Goldschmidt-Clermont M., Soen S. Y., Franzén L. G., Rochaix J. D. Directed chloroplast transformation in Chlamydomonas reinhardtii: insertional inactivation of the psaC gene encoding the iron sulfur protein destabilizes photosystem I. EMBO J. 1991 Aug;10(8):2033–2040. doi: 10.1002/j.1460-2075.1991.tb07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P., Herrmann R. G. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur J Biochem. 1988 Feb 1;171(3):551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- Woessner J. P., Gillham N. W., Boynton J. E. The sequence of the chloroplast atpB gene and its flanking regions in Chlamydomonas reinhardtii. Gene. 1986;44(1):17–28. doi: 10.1016/0378-1119(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Yu W., Spreitzer R. J. Sequences of trnR-ACG and petD that contain a tRNA-like element within the chloroplast genome of Chlamydomonas reinhardtii. Nucleic Acids Res. 1991 Feb 25;19(4):957–957. doi: 10.1093/nar/19.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]