Abstract
Activation of NF-κB (nuclear factor of kappa light chain gene enhancer in B cells) in response to DNA damage is considered to contribute to repair of genetic lesions, increased cell survival and cytokine release. The molecular mechanisms orchestrating this cytoplasmic event involve core components of the nuclear DNA damage response machinery, including ATM-kinase (ataxia telangiectasia mutated kinase) and PARP-1 (poly (ADP-ribose) polymerase 1). The physiological consequences of defective NF-κB activation in this context, however, remain poorly investigated. Here we report on the role of the ‘p53-induced protein with a death domain', PIDD, which appears rate limiting in this process, as is PARP-1. Despite impaired NF-κB activation, DNA damage did not increase cell death or reduce clonal survival of various cell types lacking PIDD, such as mouse embryonic fibroblasts or stem and progenitor cells of the hematopoietic system. Furthermore, lymphomagenesis induced by γ-irradiation (IR) was unaffected by deficiency for PIDD or PARP-1, indicating that loss of DNA damage-triggered NF-κB signalling does not affect IR-driven tumorigenesis. However, loss of either gene compromised cytokine release after acute IR injury. Hence, we propose that NF-κB's most notable function after DNA damage in primary cells is related to the release of cytokines, thereby contributing to sterile inflammation.
Keywords: PIDD, NF-κB, DNA damage
By controlling the expression of specific target genes, transcription factors can regulate the adaptive response of a cell to pathophysiological stress, such as the one triggered by DNA damage. Next to the tumour suppressor p53, members of the NF-κB family of transcription factors are activated by genotoxic stress, co-regulating the transcriptional response to DNA damage (reviewed in Hayden and Ghosh1). Although activation of NF-κB in response to ligands of the TNF family or bacterial products is well characterised, its activation upon DNA damage is only partly understood. A series of post-translational events, including sumoylation, ubiquitination, phosphorylation and nuclear-cytoplasmic shuttling of NF-κB essential modifier (IKKγ/NEMO), appear critical,2 but the order and the nature of molecules fine-tuning these events still need to be unravelled in detail. Cell line studies define a nuclear poly (ADP-ribose) polymerase 1 (PARP-1), ataxia telangiectasia-mutated (ATM) kinase, protein inhibitor of activated STAT4 (PIASy) and NEMO signalosome, as well as different cytoplasmic multi-protein complexes containing partially overlapping components, as critical.3, 4, 5 In addition, linear ubiquitin chains assembled on NEMO by LUBAC were most recently shown to be essential for DNA damage-induced NF-κB activation.6
On the basis of overexpression studies and RNA interference in model cell lines, PIDD was described as an additional possible intermediate in DNA damage-mediated activation of NF-κB. Initially, PIDD has been identified as a protein involved in the induction of apoptosis upon DNA damage in a p53-dependent manner.7, 8, 9 It contains seven leucine-rich repeats (LRR) at the N-terminus followed by two ZU5 domains, structural motives of unknown function present in ZO-1 and Unc5-like netrin receptors, and a death domain (DD) at the C-terminus. Upon low-level DNA damage, PIDD is autoproteolytically cleaved at serine 446,10 which leads to the generation of an N-terminal fragment of yet unknown function and the C-terminal fragment PIDD-C. This fragment reportedly forms a complex with NEMO and the serine/threonine kinase RIP1, considered essential in some but not all settings leading to NF-κB activation.11 The newly formed protein complex shuttles from the cytoplasm to the nucleus, where NEMO is sumoylated, presumably in a PIASy-dependent manner.12 After further phosphorylation by ATM, NEMO is exported back to the cytoplasm where it is monoubiquitinated and thereby enables activation of the IKK complex.13, 14, 15, 16, 17 Once active, this complex phosphorylates IκBα, which is subsequently K48-ubiquitinated and thereby marked for degradation by the proteasome, leading to NF-κB release, nuclear translocation and target gene transcription. Some of these genes are involved in the repair of DNA damage, such as Growth Arrest and DNA damage 45 (GADD45) or Breast Cancer 2 susceptibility protein (BRCA2), cell survival, including B-cell lymphoma-extra large (Bcl-xL) and inhibitor of apoptosis proteins (IAPs), or inflammation, with TNFα, IL-6 or IL-8 as the most prominent targets.18
After excessive DNA damage, the PIDD-C fragment can be further autoproteolytically cleaved at serine 588 to give rise to the PIDD-CC fragment.10 This fragment can associate with RIP1-associated ICH-1/CED-3-homologous protein with death domain (RAIDD) and the protease caspase-2 to form the ‘PIDDosome', suggested to serve as an activating platform for caspase-2 triggering cell death.19 However, even in PIDD-deficient cells the activation and processing of caspase-2 in response to various apoptotic triggers was observed, suggesting alternative modes of caspase-2 activation20, 21
Despite these various suggested functions of PIDD, the lack of any obvious phenotype of PIDD-knockout mice and the partly conflicting data regarding its involvement in caspase-2 activation and cell death leave its precise physiological role undefined.21, 22, 23 In this study, we present evidence that PIDD is critical for proper activation of NF-κB and cytokine production in response to genotoxic stress.
Results
Attenuated NF-κB activation and target gene transcription upon DNA damage in the absence of PIDD
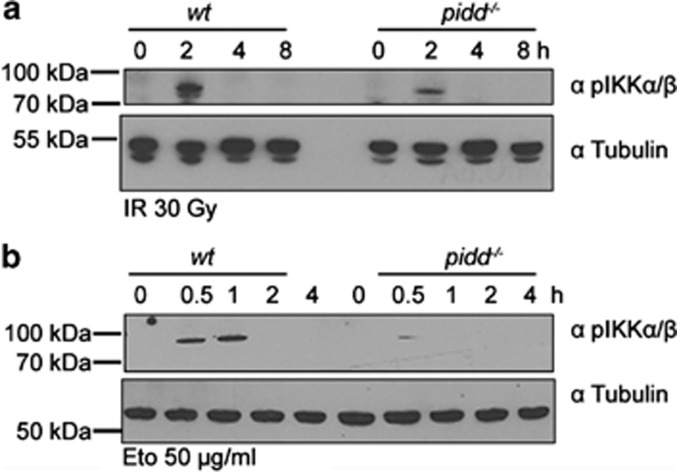
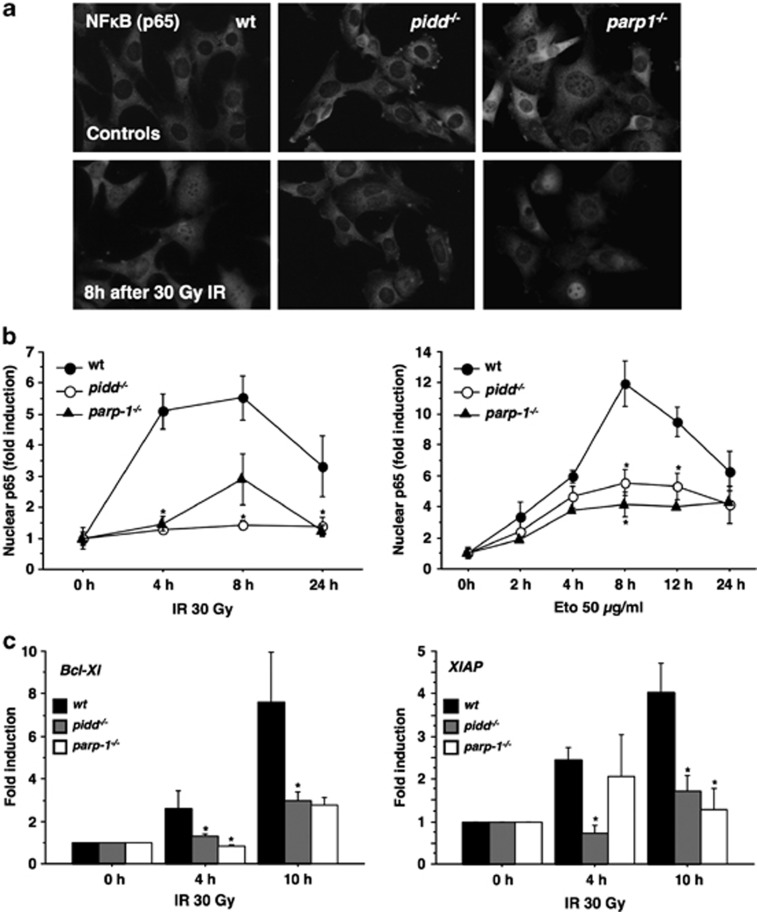
Activation of the NF-κB pathway upon DNA damage requires the assembly of the canonical IKK complex that is phosphorylated and activated by TGF β-activated kinase 1 (TAK1), inducing IκBα phosphorylation and its proteasomal degradation. Phosphorylation of IKKα/β and IκBα was indeed observed in SV40 MEF after exposure to 30 Gray (Gy) of γ-irradiation (IR) or treatment with the topoisomerase II inhibitor etoposide. In contrast, these phosphorylation events were less pronounced in MEF lacking PIDD (Figures 1a and b and Supplementary Figures 1a and b). In line with published data, MEF lacking PARP-14 or RIP124 also showed signs of impaired NF-κB activation in response to DNA damage (Supplementary Figures 1c and d). Consistent with our western blot data, immunofluorescence analysis revealed translocation and accumulation of p65/RelA in the nucleus of wt cells after either IR or etoposide treatment. In contrast, a strongly diminished accumulation of nuclear p65 was seen in PIDD-deficient MEF (Figures 2a and b). Similar observations were made in MEF lacking PARP-1 (Figures 2a and b) or RIP1 (Supplementary Figure 1e), again in support of published results.4, 24
Figure 1.
PIDD is required for DNA damage triggered NF-κB activation. Western blot analysis was performed using SV40 MEF extracts derived from wt and PIDD-knockout mice after exposure to 30 Gy IR (a) or 50 μg/ml etoposide (b), and subsequent culture for the indicated times. The membranes were probed for phosphorylated (Ser176/180) IKKα/β as well as for tubulin, which served as a loading control
Figure 2.
Defective nuclear translocation of p65 and target gene induction in the absence of PIDD or PARP-1. (a, b) MEF were irradiated with 30 Gy or treated with 50 μg/ml etoposide (Eto), cultured for the indicated times and fixed with paraformaldehyde. After immunostaining for p65 and image acquisition, individual cells were scored for nuclear localisation of p65 in a blinded manner. Fold induction of nuclear p65 compared to untreated cells was calculated for the indicated genotypes. Data represent mean±S.E.M.; n=3–4; *P<0.05 versus wt. (c) MEF were harvested after IR with 30 Gy at different time points and cDNA was prepared. Quantitative RT-PCR was performed and fold induction calculated compared to untreated cells. Data represent mean±S.E.M.; n=4–9; *P<0.05 versus wt
Once in the nucleus, NF-κB activates transcription of numerous genes, some of which relate to cell survival that should facilitate DNA repair. To investigate this transcriptional response, we monitored mRNA levels of the anti-apoptotic Bcl-2-family protein Bcl-xL and the IAP family member XIAP, two well-established NF-κB targets, by qRT-PCR on cDNA derived from cells exposed to IR. In wt MEF, we observed a clear induction of Bcl-xL and Xiap, as early as 4 h after DNA damage, with even higher levels noted 10 h after treatment (Figure 2c). In comparison, there was a clear delay in the transcription of these target genes in PIDD-deficient MEF. A similar overall trend was noted in PARP-1-deficient MEF (Figure 2c).
In contrast, treatment of MEF with TNFα led to a rapid degradation of IκBα in wt and PIDD-deficient MEF (Supplementary Figure 2a), and the relative increase of nuclear p65 was only slightly less pronounced while cells lacking RIP1 showed a much stronger defect (Supplementary Figure 2b). Together, this suggests that NF-κB activation in response to TNFα is largely intact in the absence of PIDD, whereas its activation after DNA damage appears to be compromised at a rate comparable to that observed in the absence of PARP-1 or RIP1.
Normal DNA damage response in the absence of PIDD
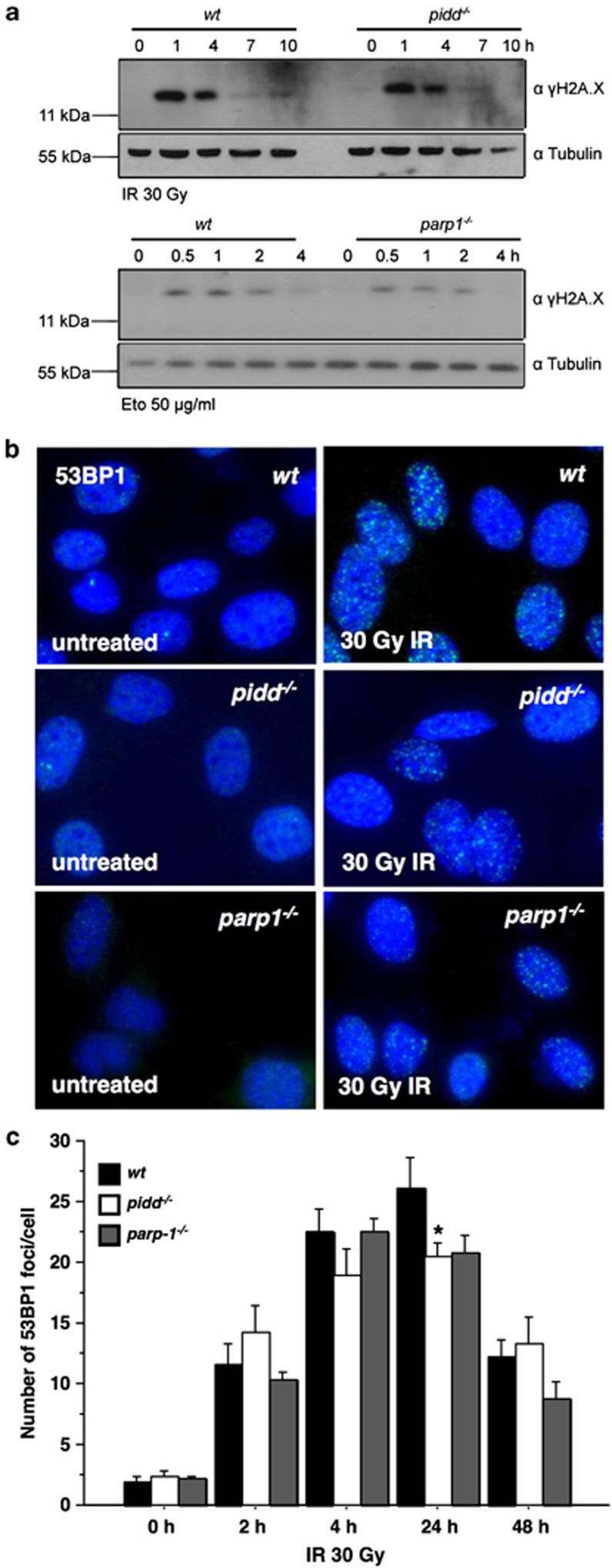
To rule out the possibility that our observations were due to a general DNA damage response defect in PIDD-deficient cells, we analysed accumulation of the γ-phosphorylated form of the histone 2A variant, γH2A.X, after IR, a well-known DNA damage marker. Western blot analysis demonstrated the accumulation of comparable amounts of γH2A.X, induced and cleared with similar kinetics in wt and PIDD-deficient MEF. A similar response was noted in PARP-1-deficient MEF (Figure 3a). As an additional readout for the ability of these cells to mount a successful DNA damage response, we quantified the number of p53-binding protein 1 (53BP1) foci after exposure of MEF to IR. In accordance with the results on H2A.X phosphorylation, both wt and PIDD-deficient MEF displayed a comparable increase and clearance rate (Figures 3b and c). PARP-1-deficient MEF showed a response similar to that of wt and PIDD-deficient MEF (Figure 3b and c).
Figure 3.
Normal DNA damage response in PIDD-deficient MEF. (a) wt, PIDD and PARP-1-deficient MEF were treated for the indicated times with 30 Gy or 50 μg/ml (Eto) and lysates subjected to immunoblotting for γH2A.X or tubulin as loading control. (b) MEF were irradiated with 30 Gy for the indicated times and fixed. After staining for 53BP1 and image acquisition, the number of foci per cell was determined. (c) Data represent mean±S.E.M.; n=3 with at least 50 cells examined per treatment in a blinded manner
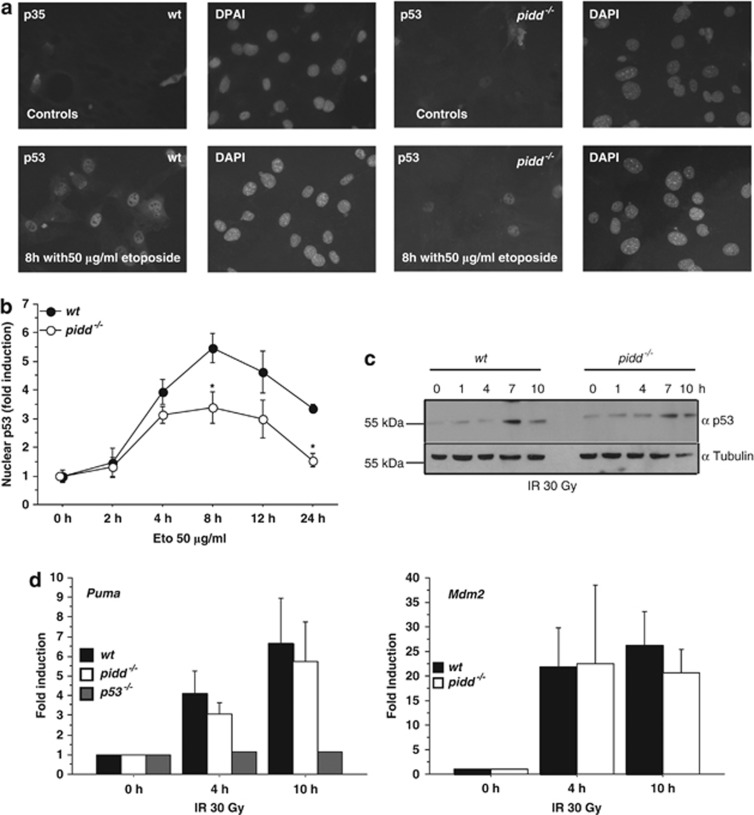
We further monitored the translocation and accumulation of p53 in response to genotoxic stress in the absence of PIDD. Despite the presence of the SV40 large T antigen that stabilizes p53 owing to direct binding in the cytoplasm, both wt and PIDD-deficient MEF showed increased nuclear localisation of p53 in response to DNA damage caused by etoposide. This effect was somewhat less pronounced in PIDD-deficient cells (Figures 4a and b). However, p53 stabilisation after IR by immunoblotting was found to be comparable (Figure 4c), as was the induction of p53 target genes such as Puma or Mdm2 (Figure 4d).
Figure 4.
Activation of p53 is not affected by loss of PIDD. (a, b) MEF were treated with 50 μg/ml etoposide and immunostained for p53. Fold increase of nuclear p53 compared to untreated cells was determined in wt and PIDD deficient MEF. Data represent mean±S.E.M.; n=3–4; *P<0.05 versus wt. (c) MEF were irradiated with 30 Gy and western blot analysis was performed with antibodies detecting p53 or tubulin serving as loading control. (d) Transcriptional upregulation of the p53 target genes Puma and Mdm2 was determined by qRT-PCR. p53-deficient MEF were included as control. Data represent mean±S.E.M.; n=5–10; means of n=2 for p53−/−
Together, these findings suggest that PIDD is not critically involved in the regulation of p53 accumulation, translocation, target gene transcription or the assembly of DNA damage response proteins at sites of genotoxic lesions. Along similar lines, phosphorylation of c-Jun N-terminal kinase (JNK), another signalling intermediate activated after DNA damage,25, 26 occurred unperturbed in response to etoposide treatment in cells lacking PIDD (Supplementary Figure 2c), indicating an overall intact DNA damage response machinery in MEF.
Normal cell death responses despite impaired NF-κB activation in the absence of PIDD
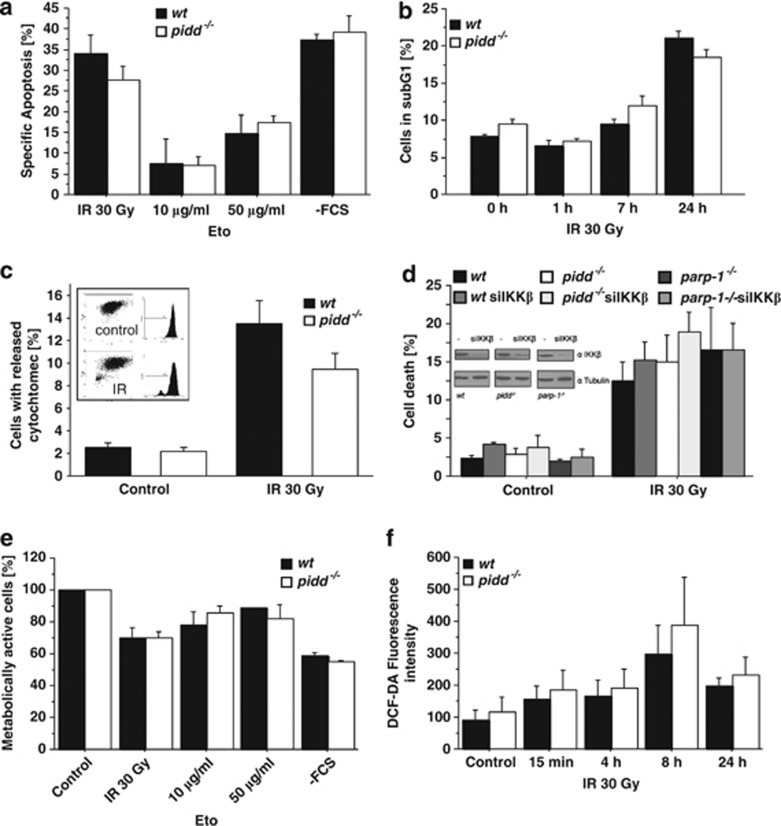
Next, we evaluated cell survival of wt and PIDD-deficient MEF in response to DNA damage in order to test whether cells lacking PIDD may actually become more sensitive to cell death initiation owing to failed or delayed NF-κB activation. However, we observed no difference in cell survival in response to graded doses of etoposide or IR, as monitored 24 h after treatment by flow cytometric analysis (Figure 5a and b). Both wt and PIDD-deficient cells also showed similar release rates of cytochrome c in response to IR, suggesting induction of apoptotic cell death (Figure 5c). Similar findings were made in MEF transformed with c-Myc/Ras (not shown). This was remarkable, given the fact that these treatments failed to induce proper NF-κB activation in PIDD-deficient cells. We therefore reasoned that the reduction in NF-κB activation caused by loss of PIDD might simply not suffice to sensitise SV40 MEF to IR-induced cell death. Hence, we also assessed whether RNAi-mediated ablation of IKKβ expression may overcome this problem, but in our hands knockdown of IKKβ also failed to significantly increase cell death in MEF lacking PIDD or PARP-1 (Figure 5d).
Figure 5.
PIDD deficiency does not sensitise to DNA damage. (a) MEF were treated as indicated and harvested after 24 h, followed by staining with AnnexinV-FITC and propidium iodide and analysed by flow cytometry. Specific apoptosis was calculated using the equation (induced apoptosis−spontaneous cell death)/(100−spontaneous cell death). Data represent mean±S.E.M.; n=4. (b) Cells treated with 30 Gy were fixed at the indicated time points and apoptosis was assessed by subG1-staining. Data represent mean±S.E.M.; n=3 (c) The percentage of wt and PIDD-deficient SV40 MEF with released cytochrome c was determined 24 h after IR with 30 Gy. Data represent mean±S.E.M.; n=3. (d) SV40 MEF of the indicated genotypes with or without knockdown of IKKβ were irradiated and cell death determined after 24 h by subG1 staining. Data represent mean±S.E.M.; n=3. Knockdown efficiency was determined by immunoblotting for IKKβ and tubulin as loading control. (e) Cells were seeded in 96 wells and treated as indicated. ATP content was assessed after 24 h with the CellTiter-Glo luminescent cell viability assay. Data represent mean±S.E.M.; n=4. (f) MEF were treated with 30 Gy IR for the indicated times and ROS production was determined using DCF-DA labelling and measuring the resulting fluorescence intensity. Data represent mean±S.E.M.; n=3
As a means to assess a possible contribution of non-apoptotic cell death, for example, by DNA damage-triggered reactive oxygen species (ROS) production,27 and subsequent necrosis that may trigger passive leakage of cytochrome c from mitochondria, we also monitored cellular ATP levels but again failed to observe any significant differences between genotypes (Figure 5e). In addition, we determined ROS production in response to IR, and observed a similar induction of ROS in both wt and PIDD-deficient MEF (Figure 5f).
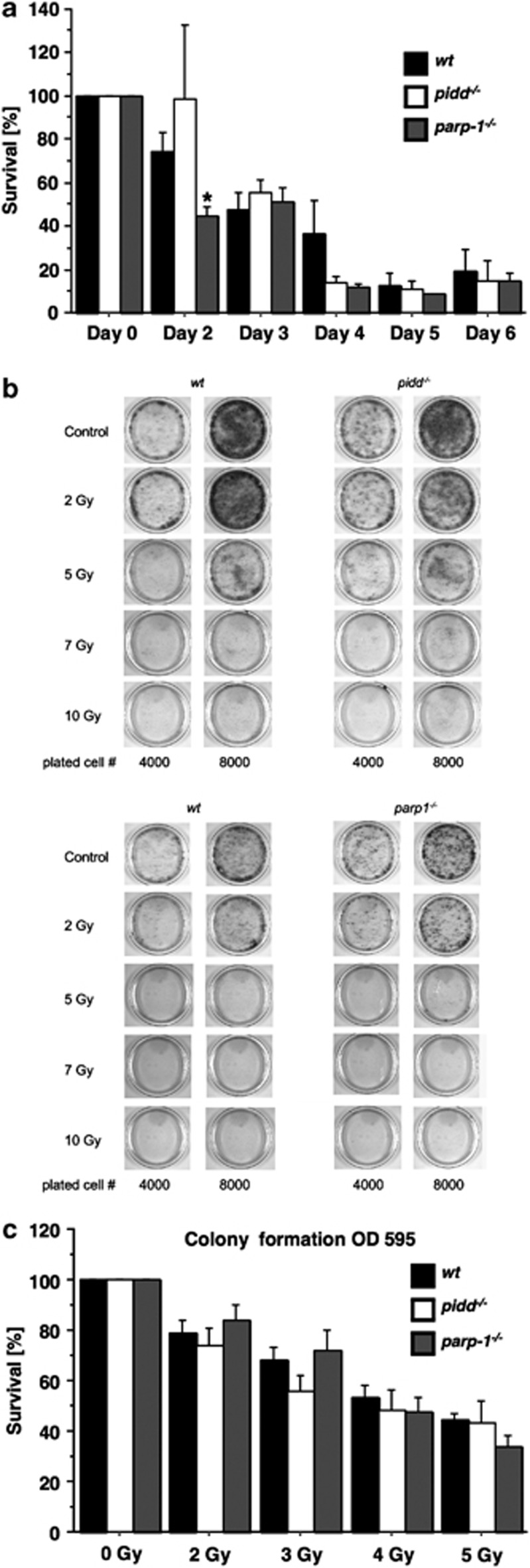
As delayed activation of NF-κB or IKKβ knockdown in MEF had no effect on cell survival in short-term apoptosis assays, we next investigated clonal cell survival after low-dose IR. First, we evaluated the impact of 10 Gy IR on cell survival over a period of 6 days, but noted no difference in cell numbers between wt or PIDD-deficient cells, whereas PARP-1-deficient cells showed initially decreased survival (Figure 6a). Next, we performed colony-forming assays using MEF from all three genotypes exposed to graded doses of IR and observed that neither genotype showed significant differences in long-term clonal survival (Figure 6b and c). Finally, to rule out the possibility that immortalisation by the SV40 large T antigen specifically impacted on the phenotype under these conditions, we also performed colony-forming assays with c-Myc/Ras-transformed MEF and exposed them to IR. However, these MEF seemed actually even more sensitive to IR than those immortalised by SV40 large T and died readily upon DNA damage independent of their genotype (not shown).
Figure 6.
Clonal survival of MEF does not depend on PIDD or PARP-1. (a) Cells (20 000/genotype) were plated in 3-cm dishes and the next day cells were either left untreated or irradiated with 10 Gy. The following days, cells were harvested by trypsin treatment and counted with a haemocytometer. The number of surviving cells was determined after normalisation to untreated cells. Data represent mean±S.E.M; n=4; *P<0.05 versus wt. (b) Fixed numbers of cells were plated and the following day irradiated with the indicated doses. After 6 days, resulting colonies were visualised by crystal violet staining, solubilised and the resulting OD was determined (c). Survival was normalised to untreated cells. Data represent mean±S.E.M.; n=5–8
Loss of PIDD does not affect clonal stem/progenitor survival after IR
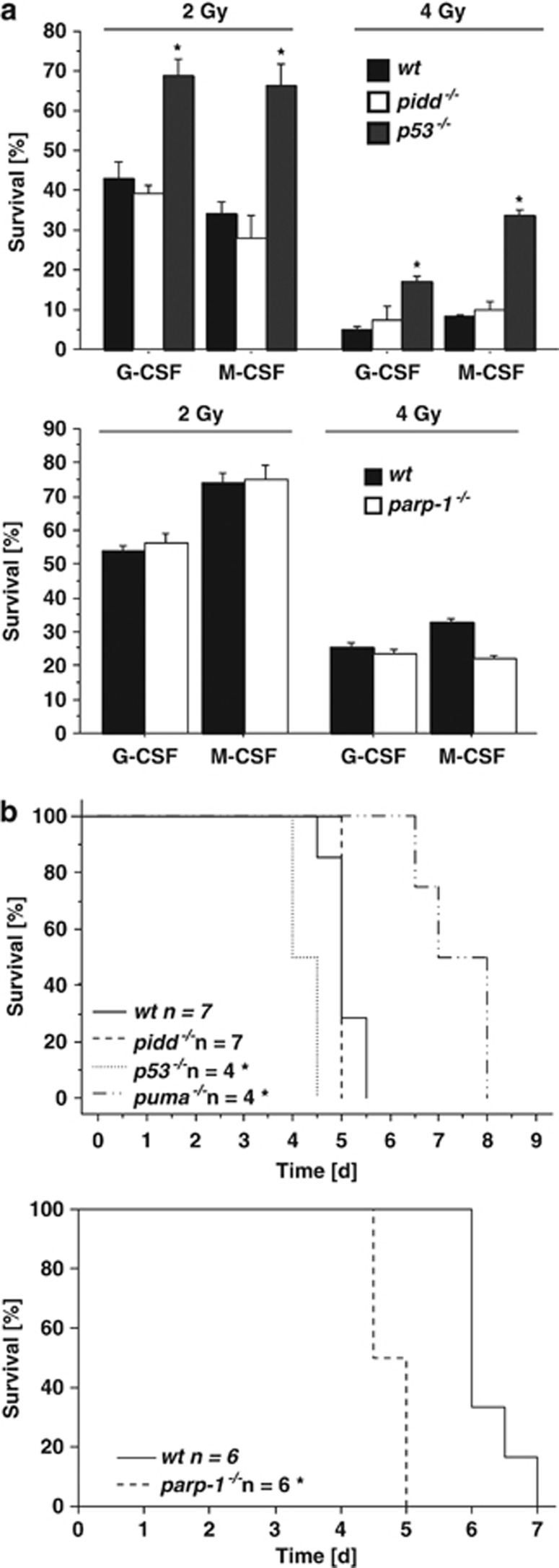
Haematopoietic stem/progenitor cells (HSPC) can escape IR-mediated DNA damage and apoptosis below a certain threshold because of successful DNA repair, allowing their clonal survival.28 Hence, we isolated bone marrow from wt and PIDD-deficient mice, exposed these cells to 2 or 4 Gy of IR or left them untreated and compared colony formation of myeloid progenitors in methylcellulose. Although the number of colonies was reduced after exposure to IR in a dose-dependent manner, we did not note a significant difference between both genotypes whereas p53 deficiency was highly protective (Figure 7a). In a different set of experiments, we also explored the role of PARP-1 deficiency under these conditions that also failed to sensitise HSPCs to IR (Figure 7a).
Figure 7.
Loss of PIDD does not sensitise stem/progenitor cells to IR. (a) Bone marrow from mice of the indicated genotypes was irradiated with 2 or 4 Gy and 40 000 cells were plated in duplicates in Methylcellulose containing G-CSF or M-CSF. Colonies were scored after 10 days and survival was normalised to untreated controls. Data represent mean±S.E.M.; n=3; *P<0.05 versus wt. (b) Mice of the indicated genotypes were exposed to whole-body IR of 15 Gy and survival was monitored over time. *P<0.05 versus wt (log-rank test)
High-dose IR elicits gastrointestinal (GI) syndrome due to massive GI epithelial cell death and stem cell failure, a phenomenon reportedly aggravated by loss of the NF-κB subunit p50.29 Therefore, we lethally irradiated wt, pidd−/− and parp-1−/− mice with 15 Gy and monitored their survival. Mice lacking p53 or pro-apoptotic Puma were included as controls in this analysis. Whereas both wt and PIDD-deficient mice succumbed to GI syndrome within the same period of time, mice lacking the pro-apoptotic BH3-only protein Puma showed increased survival, and mice lacking p53 or PARP-1 showed shortened survival after high-dose IR (Figure 7b), as reported earlier.30, 31, 32
Together, this indicates that impaired NF-κB activation due to loss of PIDD or PARP-1 does not impinge on the clonal survival of MEF or HSPC after IR, whereas GI stem cells and/or their progeny benefit from p53- or PARP-1-mediated protective mechanisms that spare them from Puma-mediated apoptosis, an effect that yet may not directly relate to NF-κB activation.
Loss of NF-κB activation downstream of PIDD or PARP-1 does not affect IR-driven transformation
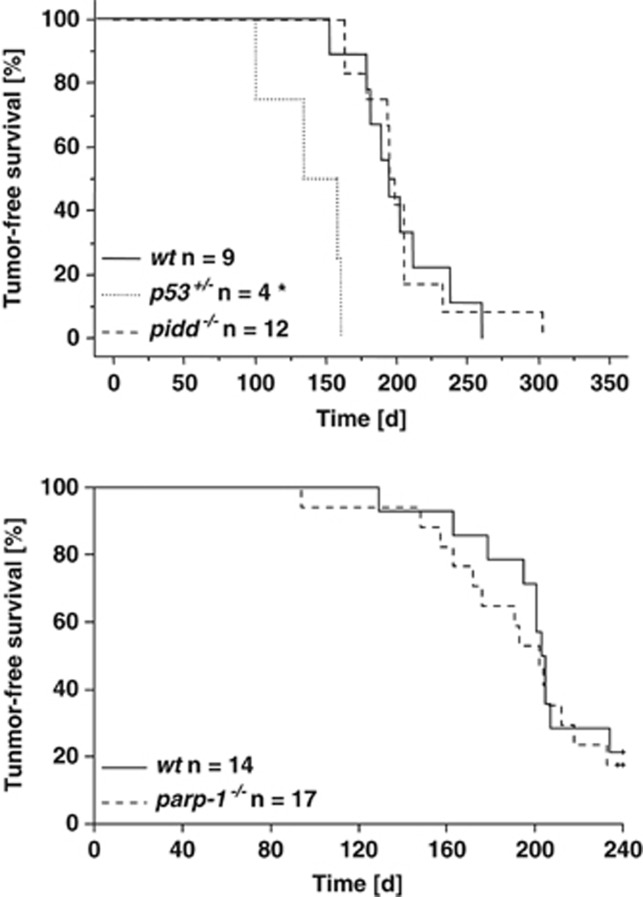
We next explored whether loss of NF-κB activity may influence tumour formation triggered by IR-mediated DNA damage, as compromised repair efficiency should facilitate malignant disease, whereas increased cell death rates of damaged cells should limit tumorigenesis. Therefore, we exposed wt, PIDD- and PARP-1-deficient mice to split-dose IR that triggers the formation of thymic lymphomas.33 As demonstrated before, loss of one allele of p53 accelerated tumour formation in this model, but neither loss of PIDD nor PARP-1 had any effect (Figure 8). These experiments may suggest that the lack of NF-κB activation after acute DNA damage or in response to subsequent oncogenic stress in pre-malignant cells does not have an impact on thymic lymphomagenesis.
Figure 8.
IR-induced tumorigenesis is not altered in PIDD or PARP-1-deficient mice. Mice of the indicated genotypes were treated with fractionated IR (4 × 1.75 Gy) in weekly intervals starting at the age of 4 weeks and tumour-free survival was monitored twice a week until disease onset. *P<0.05 versus wt (log-rank test)
Loss of PIDD leads to an impaired inflammatory response after DNA damage
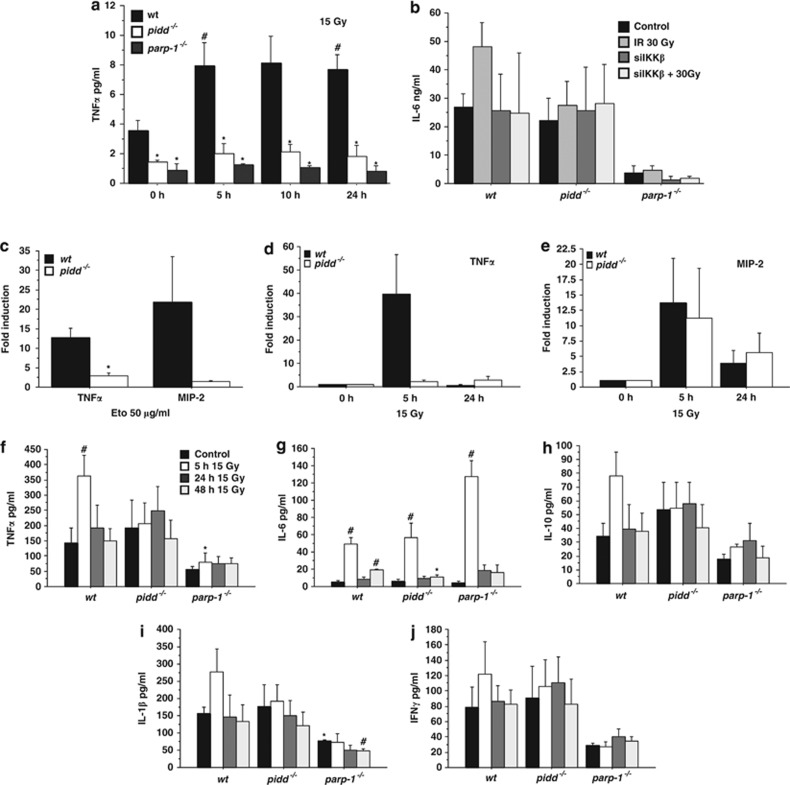
As many NF-κB target genes are inflammatory cytokines and as DNA damage was recently described to promote cytokine release,34 we wondered whether loss of PIDD may impact on production and/or secretion of such cytokines. Therefore, we monitored secretion of inflammatory cytokines in MEF and gene expression changes of cytokines in primary bone marrow-derived macrophages after DNA damage in tissue culture. We noted that the basal levels of TNFα detectable by ELISA in the supernatants of untreated MEF were already lower in the absence of PIDD or PARP-1, and that these cells also failed to induce TNFα secretion after IR-stress (Figure 9a). Similarly, IL-6 secretion into the supernatant increased slightly only in wt MEF cultures, whereas the levels did not change in cultures of PIDD-deficient cells (Figure 9b). Of note, PARP-1-deficient MEF showed very little IL-6 production, which was also not increased after IR. Consistently, knockdown of IKKβ prevented IR-triggered IL-6 secretion only in wt MEF, but had no effect on basal IL-6 levels. Assessment of MIP-2 secretion (IL-8 in humans) by bioplex analysis, however, failed to yield consistent results under these conditions (not shown). In support of our findings in MEF, we could also detect increased transcription of TNFα and MIP-2 mRNA in bone marrow-derived macrophages from wt mice exposed to etoposide. This effect was again less pronounced in the absence of PIDD (Figure 9c). Next, we analysed mRNA levels of inflammatory cytokines in whole intestines harvested at different times after exposure to IR. We noted a strong induction of TNFα in wt mice that was absent in PIDD-deficient mice (Figure 9d). MIP2, on the contrary, was comparably induced in both wt and PIDD-deficient mice (Figure 9e), whereas IL-6 mRNA analysis yielded inconsistent results (not shown).
Figure 9.
Attenuated cytokine release after DNA damage in the absence of PIDD or PARP-1. (a) MEF were irradiated with 15 Gy, cultured for additional 24 h and the supernatant was analysed by ELISA for TNFα. Data represent mean±S.E.M.; n=3–5; *P<0.05 versus wt; *P<0.05 versus control. (b) MEF transfected with siRNA against IKKß or mock controls were irradiated with 15 Gy and IL-6 levels in the supernatant were determined by ELISA after 24 h. (c) BMDM of wt and PIDD-deficient mice were treated with 50 μg/ml of etoposide for 5 h and mRNA levels of TNFα or MIP-2 were determined by qRT-PCR. Data represent mean±S.E.M.; n=3. *P<0.05 versus wt. (d, e) Mice were irradiated with 15 Gy and the intestine collected after the indicated time points. Induction of mRNA for TNFα and MIP-2 was determined by qRT-PCR. Data represent mean±S.D.; n=2. (f–j) Mice of the indicated genotypes were subjected to 15 Gy of whole-body IR and serum was collected from peripheral blood at the indicated time points. Cytokines were measured by Bioplex analysis according to the manufacturer's specifications. Data represent mean±S.E.M.; n=3–5. *P<0.05 versus wt; #P<0.05 versus control (Student's t-test)
To confirm the relevance of these observations in an in vivo setting, we quantified serum levels of different inflammation-associated cytokines after lethal body IR, triggering GI syndrome. We found increased serum levels of TNFα, IL-6, IL-10 and IL-1β 5 h after IR, followed by a subsequent decrease to basal levels after 24 h (Figure 9f to i). Notably, PIDD- and PARP-1-deficient mice showed a diminished or delayed release of TNFα and IL-1β, whereas IL-6 release was not impaired, contrasting with our results in MEF. Of note, serum IFNγ levels did not show any significant differences, while levels of anti-inflammatory IL-10 were also found unchanged in sera from PIDD or PARP-1 KO mice after IR (Figure 9h and j). Taken together, our data suggest that PIDD-dependent NF-κB activation might regulate mainly inflammatory responses, rather than having an impact on cell survival of primary cells suffering from acute IR damage.
Discussion
The events mediating activation of NF-κB signalling in response to DNA damage are still not fully defined. Given its implication in the drug resistance of tumours, understanding these mechanisms in detail is of high relevance. One factor linking nuclear lesions to the activation of the cytoplasmic IKK complex is PIDD. By using MEF derived from mice deficient for PIDD, we show that these cells fail to effectively activate NF-κB after DNA damage triggered by IR or etoposide treatment (Figures 1 and 2). This response is associated with a delayed transcription of pro-survival NF-κB target genes. Interestingly, PIDD deficiency does not completely abrogate this response to IR (Figure 2c). These findings are in concordance with studies using HEK293T cells where PIDD expression was reduced by RNAi, which also led to impaired phosphorylation of IκBα and impaired NF-κB target gene transcription.11 We assume that the residual/delayed gene transcription observed is due to PIDD-independent NF-κB activity or other factors inducing the investigated target genes with lower potency under these conditions, such as, for example, the signal transducer and activator of transcription 3 (STAT3).35 Alternatively, a second p53-dependent wave of NF-κB activation might occur with delayed kinetics, as described before,36, 37 and in line with a largely intact p53 response in PIDD-deficient cells (Figures 3 and 4).
In concordance with a previous study, loss of RIP1 in MEF phenocopied the defective NF-κB response observed in PIDD-deficient cells (Supplementary Figure 1), consistent with observations proposing the assembly of a nuclear RIP1/PIDD/NEMO complex facilitating sumoylation of NEMO.11, 24 However, in human HepG2 hepatocellular carcinoma cells this event appears to be independent of RIP1 upon DNA damage, pointing towards cell type or species-specific differences in these upstream signalling events.4 In another model, sumoylation of NEMO in the nucleus was found to depend on PARP-1, which builds a scaffold of polyADP-ribose, recruiting NEMO, ATM and PIASy into a nuclear signalling complex, the latter being the small ubiquitin-like modifier (SUMO) ligase for NEMO.4 Yet another model proposes that NEMO, ATM, Ubc13 and XIAP are required for K63-ubiquitination of ELKS, a modifier of the IKK complex, which in turn allows for the assembly of a TAK1/TAB2/3 complex activating IKK.5 Together, this suggests that all these pathways are either essential and non-redundant in MEF, or interconnected, possibly via PIDD, a hypothesis yet to be tested.
Surprisingly little is known about the possible pathophysiological consequences of impaired NF-κB activation after genotoxic stress, in particular in non-cancerous cells. Despite delayed NF-κB activation, we could not detect a survival benefit of wt MEF, leukocytes or HSPC in response to genotoxic stress over PIDD-deficient cells as well as numerous other tested death-inducing stimuli (Figures 5 and 6 and Manzl et al.21). Our observations are in line with findings in p50-deficient mice that show normal lymphocyte death upon IR, despite impaired NF-κB activation.29 Whether the observed late-phase activation of NF-κB target genes is sufficient to prevent enhanced apoptosis in PIDD-deficient cells remains unclear, as we also observed no differences in cell survival at earlier time points (Figure 5b and Manzl et al.21). Hence, we considered that PIDD-dependent NF-κB activation might serve different signalling outcomes than assisting DNA repair or survival. Interestingly, a recent paper suggested that excessive DNA damage leads to two waves of NF-κB activation, the first being orchestrated by NEMO leading to transcription of pro-survival genes but also autocrine production of TNFα. Although the role of PIDD has not been investigated in this study,34 our results suggest that PIDD might contribute to the first wave of NF-κB activation (Figures 1 and 2).
In an acute model of DNA damage leading to GI syndrome and early lethality, we observed reduced survival of mice lacking PARP-1 or p53, but not those lacking PIDD, whereas mice lacking the proapoptotic p53 effector Puma showed prolonged survival (Figure 7). This confirms that the cell type-dependent context can be critical, and that NF-κB activation involving PIDD or PARP-1 may lead to different signalling outcomes or have different additional roles, as demonstrated for PARP-1.38 For example, whereas PARP-1 activation may allow for more efficient repair of DNA lesions in GI epithelial stem cells enabling survival, PIDD and PARP-1-mediated NF-κB activation may serve a different purpose, such as the promotion of an inflammatory response (see below).
Whereas substantial progress has been made in understanding the role of NF-κB in inflammation-driven cancer, its influence on cancer arising in response to genotoxic stress has remained largely unexplored. Such analysis is clearly complicated because of the involvement of factors essential for NF-κB activation in other pathways important for tumour suppression, like the DNA damage response and DNA repair (e. g. ATM, PARP-1). However, as PIDD appearance is mainly required for activation of NF-κB in response to DNA damage but not for the viability of primary cells (Figure 520, Manzl et al.21), it may also be considered a suitable model to investigate the contribution of DNA damage-induced NF-κB activation to cancer progression. Surprisingly, we failed to observe any differences in cancer onset in PIDD-deficient mice in a model of DNA damage-induced lymphoma in two independent tumour cohorts (Figure 8 and Manzl et al.21). However, this cancer model seems largely unaffected by inflammation, as it progresses normally in MyD88 knockout mice,40 and hence it will be interesting to investigate how loss of PIDD affects cancers that are elicited by a combination of DNA damage and inflammation, such as diethylnitrosamine (DEN)-driven hepatocellular carcinogenesis. Of note, loss of PIDD delays c-Myc-driven lymphomagenesis, but the molecular mechanism remains to be identified; however, it appears to be unrelated to differences in tumour cell survival.39
Notably, PARP-1 also failed to suppress IR-driven lymphomagenesis (Figure 8), and was previously shown to limit tumorigenesis caused by nitrosamine41 or azoxymethane-driven DNA damage,42 but not in response to the DNA-damaging agent 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) that drives liver cancer.43 These findings also suggest a cell type and DNA damage type-dependent tumour suppressor function of PARP-1. Clearly, neither PARP-1-dependent DNA-repair nor NF-κB activation appears critical to limit lymphomagenesis driven by IR (Figure 8).
Our initial analysis of cytokine production suggests that PIDD and PARP-1 preferentially modulate inflammatory responses after genotoxic stress, contributing to sterile inflammation. This is supported by our observation that PIDD- or PARP-1-deficient MEF secrete less TNFα or IL-6 into the culture supernatant after IR, and that PIDD-defective macrophages showed impaired induction of TNFα and MIP-2 mRNA after etoposide treatment in culture (Figure 9). Inconsistencies in the pattern of cytokine release in vivo, however, may also be secondary and due to the impaired initial TNFα release or cell type-dependent requirements for PIDD in this process (Figure 9). The observation that parp-1−/− mice show comparable defects supports a similar role in NF-κB-dependent cytokine release in response to DNA damage. These observations are in line with a previous report on defective TNFα-regulated gene expression signatures in PARP-1-deficient heart endothelium cells.44
In summary, our study suggests that NF-κB activation driven by acute DNA damage in primary cells may serve primarily to launch a sterile inflammatory response to orchestrate the clearance of dying cells, for example, by recruitment of phagocytes to sites of lesions, rather than contributing to the survival of cells after DNA damage that may subsequently spread mutations, thereby facilitating transformation. This biological response depends in part on the NF-κB-activating activities of PIDD and PARP-1, and it will be interesting to explore the possibility that inhibition of PIDD function might reduce inflammation-associated side effects related to DNA damage-based anti-cancer therapy.
Materials and methods
Chemicals, cytokines and antibodies
The chemicals and cytokines used were mouse TNFα, mouse M-CSF and mouse GM-CSF (Peprotech, Vienna, Austria), caffeine, DAPI, propidium-iodide (all from Sigma, Vienna, Austria), AnnexinV-FITC (Biolegend, Fell, Germany) and etoposide (Alexis, Vienna, Austria). Antibodies from Cell Signaling: α-pIKKα/β (Ser176/180, #2697), α-pJNK (#9255). α-JNK (#9258), α-IκBα (#9242), α-pIκBα (Ser32, #9241), α-γH2A.X (#2577); from Santa Cruz Biotechnology: α-p65 (sc-372), α-p53 (sc-11311 and sc-6243), α-tubulin (sc-32293); others: α-53BP1 (Novus Biologicals NB100-305, Vienna, Austria), α-rabbit AlexaFluor 488 (Invitrogen, Vienna, Austria), goat α-rabbit HRP (DAKO, Vienna, Austria), goat α-mouse HRP (Dako).
Mice
All mice used were on a C57BL/6 genetic background. The generation of PIDD-,21 Puma-45 and PARP-1-deficient mice30 has been described. Mice lacking p53 were kindly provided by Manuel Serrano (CNIO, Madrid). To induce thymic lymphomas, 4-week-old mice were exposed four times to 1.75 Gy of IR in weekly intervals and monitored three times a week for onset of disease.33 For total body IR, mice were irradiated with 15 Gy and monitored twice a day. Animal experiments were in accordance with the Austrian law and approved by the Austrian Ministry for Science and Education (BMWF-66.011/0020-II/3b/2011 and -66.011/0140-II/10b/2009) or by the Barcelona Biomedical Research Park Animal Care and Use Committee (JYL-10-1306 and TG-09-1223-JYL).
Statistical analysis
Estimation of statistical differences between groups was carried out using the unpaired Student's t-test, or, where indicated, by the paired Student's t-test. Comparison of tumour-free survival or survival after IR was performed using a log-rank test. P-values of <0.05 were considered to indicate statistically significant differences.
Cells and tissue culture
Wt, PIDD- and PARP-1-deficient MEF were obtained from day 13.5 embryos using standard procedures. Cells were cultured in DMEM containing 250 μM L-Gln (Invitrogen), penicillin/streptomycin (Sigma) and 10% FCS (PAA), and immortalised by transduction with retroviruses encoding for SV40 large T or c-Myc plus Ha-RasV12 in combination. RIP1-deficient and corresponding background matched wt SV40 MEF were obtained from J Silke. All experiments were performed with at least two independent bulk cultures derived from individual embryos, with the exception of PARP-1 and RIP1- deficient MEF.
Primary bone marrow-derived macrophages (BMDM) were generated by flushing out both femurs into KDS/BSS with 10% FCS and plating 10 millions on a petri dish in RPMI containing 250 μM L-Gln, penicillin/streptomycin, 10% FCS and 10 ng/ml M-CSF (PeproTech). After 3 days, fresh medium was added and cells were replated after 6 days (1 × 106 cells per 6-wells) for analysis.
Quantitative RT-PCR
RNA was isolated with Trizol (Invitrogen) and after DNAse digestion (Promega, Mannheim, Germany) reversely transcribed with Omniscript (Quiagen, Hilden, Germany) using random hexamer primers. cDNA was amplified with 5prime RealMasterMix SYBR ROX on an Eppendorf Mastercycler ep realplex2 cycler (40 cycles with 30 s 94 °C, 30 s 60 °C, 30 s 68 °C). Primers used were Bcl-xL (forward 5'-GAATGGAGC CACTGGCCA-3' reverse 5'-GCTGCCATGGGAATCACCT-3'), XIAP (forward 5′-TTGGAAGCCAAGTGAAGACC-3′ reverse 5′-CAAGGGTCTTCACTTGGCTT-3′), TNFα (forward 5′-CCAGTGTGGGAAGCTGTCTT-3′ reverse 5′ -AAGCAAAAGAGGAGGCAACA-3′), MIP-2 (forward 5′-AACATCCAGAGCTTGAGTGTG A-3′ reverse 5′-TTCAGGGTCAAGGCAAACTT-3′) and actin (forward 5'-ACTGGGACGACATGGAGAAG-3' reverse 5'-GGGGTGTTGAAGGTCTCAAA-3'). Fold induction compared to untreated controls was calculated by the delta–delta CT method. RNA from intestines was isolated from the small intestine of treated mice lysed in Trizol and processed as described above.
Survival, colony formation, ROS and ATP assay
MEF cells were plated at a density of 100 000 per 3-cm dish and exposed to IR or DNA-damaging agents the next day at the indicated doses. Cell viability was determined by staining with 5 μg/ml propidium iodide and AnnexinV-FITC (Biolegend) in AnnexinV-binding buffer (10 mM Hepes pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) and analysed on a FACSscan instrument (Beckton Dickinson). Cells negative for propidium iodide and AnnexinV were considered as alive. For subG1 analysis, cells were fixed in 70% EtOH/PBS, stained with 40 mg/ml propidium-iodide for 30 min at 37 °C and analysed on a FACSscan instrument.
For long-term survival assays, 20 000 MEF were plated in 3-cm dishes, left untreated or exposed to IR in a linear accelerator. Cell numbers were counted for 5 consecutive days and survival was normalised to untreated cells. For clonogenic survival assays, MEF immortalised with SV40 or Myc/Ras were plated at the stated density and irradiated the following day with the indicated doses. After 6 days colonies were visualised by staining with 0.2% crystal violet in 50% methanol and quantified by solubilisation in 1% SDS in PBS, and OD measurement was done at 595 nm.
The differentiation and colony formation potential of HSPC was analysed by isolating bone marrow from wt, PIDD-, PARP-1- and p53-deficient mice. Single-cell suspensions were exposed to graded doses of IR and 40 000 cells were plated in duplicates in methylcellulose (Stem Cell Technologies #3634, Grenoble, France), supplemented with either granulocyte colony-stimulating factor (G-CSF) or granulocyte-macrophage colony-stimulating factor (GM-CSF). Colonies were evaluated in a microscope and counted after 10 days.
To quantify cellular ATP levels, CellTiter-Glo luminescent cell viability assay (Promega) was used. A total of 5000 cells were seeded per well in a 96-well plate and the following day cells were treated as indicated. After 24 h ATP levels were assessed as instructed by the manufacturer.
Cytochrome c release was monitored by FACS staining with a cytochrome c antibody as described before.21
To quantify ROS production, MEF were seeded in 96 wells and treated as indicated. After washing with PBS, 25 μM DCFDA in PBS was added, incubated at 37 °C for 10 min and fluorescence intensity determined in a microplate reader over 30 min at 485/638 nm.
Transfection of siRNA targeting IKKβ (Sequence: 5′-AAACCAGAAUCCAGGAAGATT-3′) was performed with Metafectene (Biontex, Martinsried, Germany). Cells were replated 2 days after transfection for further analysis.
Immunoblotting
Cells were washed with PBS, harvested by trypsinization, washed again in PBS and one half boiled at 95 °C for 5 minutes in SDS-lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, 0.01% w/v bromophenol blue), whereas the other half was lysed in RIPA buffer (150 mM NaCl, 50 mM Tris, 1% v/v NP40, 0.5% v/v sodium deoxycholate, 0.1% v/v SDS) and used for protein quantification by Bradford. After sonication of the SDS-boiled samples, ∼50 μg of the lysates were loaded and run on a 4–20% gradient gel (Lonza, Vienna, Austria) and subsequently transferred to Hybond ECL nitrocellulose membranes (GE Healthcare, Vienna, Austria) by electroblotting. After incubation with the indicated antibodies, proteins were visualised by chemoluminescence (Pierce, Vienna, Austria).
Immunofluorescence
MEF were seeded in six wells onto sterilised glass cover slips and left to adhere overnight. Then cells were treated as indicated and after various time points washed with PBS and fixed with 4% PFA in PBS for 20 min at room temperature. Following three washes with PBS, cells were concurrently permeabilised and unspecific binding blocked by 1 h incubation with PBS containing 0.1% Tween20, 1% BSA and 10% FCS. Next, cells were incubated with α-p65 antibody (1:100) in blocking solution at 4 °C overnight, washed again for three times with PBS and incubated with α-rabbit AlexaFluor 488 secondary antibody in blocking solution for 1 h at room temperature. Finally, cells were briefly washed again, incubated with DAPI (2 μg/ml) for 10 min and mounted on microscope slides with Vectashield. Nuclear localisation of p65 was evaluated by imaging cells under a Nikon Eclipse E800 fluorescence microscope equipped with a × 20/0.75 Plan Apo objective and then manually counting cells in which p65 fluorescence in the nucleus was equal to or exceeded that of the cytoplasm in a blinded fashion. For staining of p53 (1:100) the same protocol was applied, whereas for 53BP1 staining (1:50) cells were fixed with methanol for 4 min at −20 °C, permeabilised with acetone for 4 min at −20 °C and blocked using 8% BSA in PBS-Tween20. Images were acquired on a fluorescence microscope and 53BP1 foci per cell were evaluated using the Image J software (NIH, Bethesda, MF, USA).
Cytokine measurement
Serum levels of IL-1β, IL-6, IL-10, IFNγ and TNFα in peripheral blood were monitored using the BioRad Bioplex Pro Mouse Th17 or the MIP-2 Kit as described by the manufacturer. Briefly, proteins in 12.5 μl serum were coupled to beads on a Bioplex 96-well plate and were detected by adding 1.5 μl of group 1 or MIP-2 detection-antibodies and secondary strep-PE antibodies. Fluorescence intensities were measured using a Bio-Rad Bioplex Suspension Array System, and were compared to dilution signals of Bioplex Pro Mouse Cytokine Standard Group I and II.
ELISA
To determine cytokine levels in the supernatant of MEF, 800 000 cells were plated in 3-cm dishes and medium was changed the following day before treatment. Supernatant was collected at the indicated times and used for ELISA for TNFα or IL-6 according to the manufacturer's conditions (BioLegend TNFα ELISA#430901 or IL-6 ELISA #431301).
Acknowledgments
We are grateful to K Rossi, C Soratroi, R Pfeilschifter, K Götsch and I Gaggl for technical assistance, P Lukas and his team (LINAC1-4) from the Department of Radio-oncology for enabling IR experiments. We thank A Strasser and M Serrano for mice and reagents, M Kelliher and J Silke for RIP1-deficient MEF and G de Murcia for parp-1−/− mice. This work was supported by grants from the Austrian Science Fund-FWF (SFB021), the MCBO Graduate School (W_1101-B12) and the EU-FP06 Marie Curie Research Training Network ‘Apoptrain' to AV and FB as well as the Austrian Cancer Society/Tirol (Tiroler Krebshilfe) to FB, GK and CM; the Spanish Ministerio de Ciencia e Innovación (SAF2008-01572, HA2008-0010 and SAF2011-26900), Generalitat de Catalunya (2009/SGR/524) to JY and Juan de la Cierva fellowship program to LL.
Glossary
- PIDD
p53-induced protein with a death domain
- RAIDD
RIP1-associated ICH-1/CED-3-homologous protein with death domain
- NF-κB
nuclear factor of kappa light chain gene enhancer in B-cells
- NEMO
NF-κB essential modifier
- TRAF6
TNFα receptor-associated factor 6
- IKK
inhibitor of NF-κB kinase
- IκBα
kappa light polypeptide gene enhancer in B-cell inhibitor
- RIP1
α receptor interacting protein kinase 1
- PARP-1
poly (ADP-ribose) polymerase 1
- ATM
ataxia telangiectasia-mutated kinase
- PIASy
protein inhibitor of activated STAT4
- TNF
tumour necrosis factor
- TAK1
TGF β-activated kinase 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by A Ashkenazi
Supplementary Material
References
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced I kappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, et al. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40:75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. Embo J. 2011;30:3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma W, Pidd BenchimolS. A new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, LRDD LinLL. A novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- Bock FJ, Peintner L, Tanzer M, Manzl C, Villunger A.P53-induced protein with a death domain (PIDD): master of puppets Oncogene 2012. Jan 23;314733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, et al. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol. 2011;31:2774–2786. doi: 10.1128/MCB.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Kim IR, Murakami K, Chen NJ, Saibil SD, Matysiak-Zablocki E, Elford AR, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, et al. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol Cell. 2012;47:681–693. doi: 10.1016/j.molcel.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen's view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur GM, Lewis J, Yang Q, Lin Y, Nakano H, Nedospasov S, et al. The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes Dev. 2003;17:873–882. doi: 10.1101/gad.1062403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck WT, Mo YY, Bhat UG. Cytotoxic signalling by inhibitors of DNA topoisomerase II. Biochem Soc Trans. 2001;29 (Pt 6:702–703. doi: 10.1042/0300-5127:0290702. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA Damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meng A, Lang H, Brown SA, Konopa JL, Kindy MS, et al. Activation of nuclear factor kappaB in vivo selectively protects the murine small intestine against ionizing radiation-induced damage. Cancer Res. 2004;64:6240–6246. doi: 10.1158/0008-5472.CAN-04-0591. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova EA, Kondratov RV, Wang K, Christov K, Golovkina TV, Goldblum JR, et al. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23:3265–3271. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- Qiu W, Carson-Walter EB, Liu H, Epperly M, Greenberger JS, Zambetti GP, et al. PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell. 2008;2:576–583. doi: 10.1016/j.stem.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J, et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev. 2010;24:1602–1607. doi: 10.1101/gad.1940210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton S, NEMO AshkenaziA. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Barry SP, Townsend PA, Knight RA, Scarabelli TM, Latchman DS, Stephanou A. STAT3 modulates the DNA damage response pathway. Int J Exp Pathol. 2010;91:506–514. doi: 10.1111/j.1365-2613.2010.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohuslav J, Chen LF, Kwon H, Mu Y, Greene WC. p53 induces NF-kappaB activation by an IkappaB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J Biol Chem. 2004;279:26115–26125. doi: 10.1074/jbc.M313509200. [DOI] [PubMed] [Google Scholar]
- Strozyk E, Poppelmann B, Schwarz T, Kulms D. Differential effects of NF-kappaB on apoptosis induced by DNA-damaging agents: the type of DNA damage determines the final outcome. Oncogene. 2006;25:6239–6251. doi: 10.1038/sj.onc.1209655. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, et al. PIDDosome-independent tumor suppression by caspase-2. Cell Death Differ. 2012;19:1722–1732. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge AR, Wu L, Scott CL, Adams JM, et al. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev. 2010;24:1608–1613. doi: 10.1101/gad.1940110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi M, Masutani M, Nozaki T, Kusuoka O, Tsujiuchi T, Nakagama H, et al. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout mice to nitrosamine carcinogenicity. Carcinogenesis. 2001;22:1–3. doi: 10.1093/carcin/22.1.1. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Fujihara H, Watanabe M, Tsutsumi M, Nakamoto K, Kusuoka O, et al. Parp-1 deficiency implicated in colon and liver tumorigenesis induced by azoxymethane. Cancer Sci. 2003;94:497–500. doi: 10.1111/j.1349-7006.2003.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Masutani M, Kato K, Tang M, Kamada N, Suzuki H, et al. Parp-1 deficiency does not enhance liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline in mice. Cancer Lett. 2006;236:32–38. doi: 10.1016/j.canlet.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Carrillo A, Monreal Y, Ramirez P, Marin L, Parrilla P, Oliver FJ, et al. Transcription regulation of TNF-alpha-early response genes by poly(ADP-ribose) polymerase-1 in murine heart endothelial cells. Nucleic Acids Res. 2004;32:757–766. doi: 10.1093/nar/gkh239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.