Abstract
The p53 tumor suppressor responds to certain cellular stresses by inducing transcriptional programs that can lead to growth arrest or apoptosis. However, the molecular mechanisms responsible for choosing between these two cell fates are not well understood. Previous studies have suggested that p53 selectively activates proarrest target genes, due to the higher affinity of p53 for their promoters compared with proapoptotic genes. Here we show using microarray and chromatin immunoprecipitation that p53 binds to and transcriptionally activates both its proarrest and proapoptotic target genes proportionally to induced p53 expression levels. Further, we provide evidence that to trigger apoptosis, cells must overcome an apoptotic threshold, whose height is determined by expression levels of p53 and its targets, the duration of their expression and the cellular context. We demonstrate in multiple cells lines that below this threshold, expression levels of p53 and its targets were sufficient to induce arrest but not apoptosis. Above this threshold, p53 and its targets triggered extensive apoptosis. Moreover, lowering this threshold with inhibitors of antiapoptotic Bcl-2 family proteins sensitized cells to p53-induced apoptosis. These findings argue that agents that lower the apoptotic threshold should increase the efficacy of p53-mediated cancer therapy.
Keywords: affinity, apoptosis, cell fate decisions, p53, threshold
The p53 transcription factor responds to genotoxic, oncogenic and other stress signals by inducing antiproliferative transcriptional programs that can lead to growth arrest or apoptosis.1, 2 P53 mediates arrest by transcriptionally activating genes, including p21 (CDKN1A), 14-3-3σ (SFN) and GADD45α, whereas p53-dependent apoptosis is triggered by transactivation of proapoptotic genes, such as PIG3 (TP53I3), PUMA (BBC3), APAF1, FAS and BAX.3 The p53 pathway is compromised in most human cancers, either via inactivating mutations in the p53 gene or by inhibition of wild-type p53 function.4, 5, 6 Given that about half of human tumors retain wild-type p53, pharmacological restoration of its function has been a focus of intense study. In fact, the nutlin family of MDM2 antagonists that activate p53 by disrupting the p53–MDM2 interaction are in clinical trials.5, 6 Ideally, the aim of such p53-based therapy would be a preferential induction of apoptosis.7, 8 Therefore, understanding of the mechanisms that determine p53 cell fate decisions between arrest and apoptosis is paramount.
Previous studies suggested that p53-mediated cell fate decisions depend on the type of cell, tissue, stress, cofactors and other influences.9, 10 One mechanism that affects the outcome of p53 activation is the abundance of the p53 protein itself.11, 12 An ‘affinity model' proposed that low p53 levels preferentially bind to high-affinity p53 response elements (RE) in promoters of proarrest genes, whereas high p53 levels are necessary to bind to low-affinity p53 RE in proapoptotic promoters.9, 13 Studies investigating this model have yielded conflicting results. Chromatin immunoprecipitation (ChIP) analyses confirmed the presence of high- and low-affinity p53 RE within some proarrest (e.g. p21) and proapoptotic (e.g. BAX) genes, respectively.14, 15, 16 However, high-affinity p53 RE were also identified in proapoptotic promoters of genes such as PUMA, suggesting that not all proapoptotic genes bind p53 with low affinity.14 Similarly, although microarray analyses identified distinct sets of genes activated during p53-dependent arrest or apoptosis, some proapoptotic genes like PUMA or PIG3 were activated during either cell fate.17, 18, 19, 20, 21
A potential limitation of these studies is that p53-dependent arrest and apoptosis were usually caused by stimuli that trigger DNA damage.14, 15, 16, 17, 18 Thus, observed variations in p53 binding may reflect not only increased p53 levels but also a myriad of p53 post-translational modifications and p53 cofactors induced by different genotoxic stresses. Furthermore, genotoxic insults, which induce p53, also induce p53-independent pathways that may activate or repress p53 target genes.3, 18, 22 Studies that varied p53 levels using inducible systems did not achieve both arrest and apoptosis in the same cells unless they used super-physiological p53 levels or added a genotoxic insult to trigger apoptosis.19, 20, 21 These caveats have prevented a direct comparison of p53-induced arrest with p53-induced apoptosis within the same cellular context and via the same p53-activating signal. To achieve a comparison where the only variable is the level of p53, we established an inducible system that allows tight regulation of p53 expression. We show that low and high p53 expression within a physiologically relevant range triggers arrest and apoptosis, respectively. Using microarray and ChIP, we demonstrate that p53 directly activated proarrest and proapoptotic genes proportionally to its expression levels. Likewise, low and high p53 levels activated by nutlin-3 led to arrest or apoptosis in wild-type p53 cancer cells. Our results suggest a mechanism whereby the biological outcome of p53 activation is determined by different cellular thresholds for arrest and apoptosis. Lowering the apoptotic threshold was sufficient to switch the p53 cell fate from arrest to apoptosis, which has important implications for the effectiveness of p53-based cancer therapy.
Results
Characterization of p53-inducible B5/589 human mammary epithelial cells
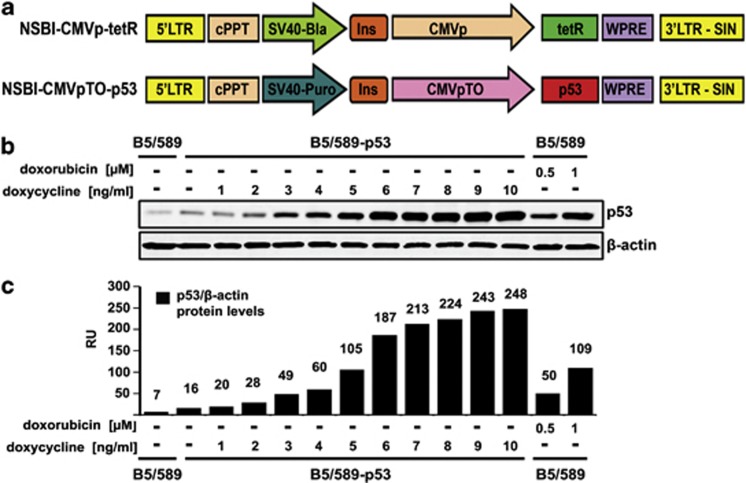
To study the effects of varying p53 expression levels in human epithelial cells, we established a doxycycline (dox)-inducible system in immortalized but non-tumorigenic wild-type p53 human mammary epithelial cells (HMECs) B5/589,23 designated B5/589-p53 (Figure 1a). We chose non-tumorigenic cells because cancer cells often contain defective signaling pathways that compromise their apoptotic response to p53 activation.24 To identify physiological levels of doxycycline-induced p53, we treated these cells with increasing doses of doxycycline (Figure 1b). P53 levels induced by 1–5 ng/ml doxycycline were comparable to endogenous p53 levels induced by the DNA-damaging agent doxorubicin (Figures 1b and c).25
Figure 1.
Doxycycline-inducible p53 expression in B5/589 HMECs. (a) Schematic representation of the binary lentiviral vector system. NSBI-CMVp-tetR vector constitutively expresses the tetracycline repressor (tetR) driven by the CMV promoter (CMVp). NSPI-CMVpTO-p53 vector inducibly expresses p53. cPPT, central polypurine tract; Ins, insulator; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; TO, tetracycline operator containing two repeats of the tetR operon downstream the CMV promoter; and SIN, self-inactivating. (b) Immunoblot analysis of p53 expression levels in B5/589-p53 cells induced with 1-10 ng/ml doxycycline for 48 h. Parental B5/589 cells were untreated or treated with 0.5 or 1 μℳ doxorubicin for 24 h. β-Actin was used as a loading control. (c) Quantification of the data shown in (b). Relative protein levels of p53 and β-actin were measured with the Odyssey Infrared Imaging System. RU, relative units
Activation of p53 with a genotoxic stimulus imposes a variety of p53 post-translational modifications, but their contribution to p53 function is unclear.4, 26 To ascertain whether doxycycline-induced p53 protein was post-translationally modified, we compared selected phosphorylation and acetylation sites between doxycycline-induced exogenous p53 and doxorubicin-activated endogenous p53 (Supplementary Figure 1a). We also examined phosphorylation of histone H2AX at serine 139 (γH2AX), a marker for DNA damage (Supplementary Figure 1b).27 As expected, doxorubicin treatment led to phosphorylation of endogenous p53 on serine 15 and 46, acetylation of lysine 382, and increased γH2AX levels. Doxycycline-induced p53 also exhibited phosphorylation and acetylation at these sites in a dose-dependent manner; however, no increase in γH2AX was evident. Endogenous p53 was phosphorylated on serine 15 and 46 even in the absence of doxorubicin, and addition of doxycycline did not influence this phosphorylation (Supplementary Figure 1c). Moreover, phosphorylation levels were similar on comparable levels of immunoprecipitated doxycycline-induced p53 (Supplementary Figure 1d). Non-phosphorylatable/acetylatable mutants of p53 on serine 15, 46 and lysine 382 demonstrated the specificity of the phospho/acetyl antibodies (Supplementary Figure 1e). Therefore, post-translational modifications on doxycycline-induced p53 do not result from increased DNA damage or doxycycline-dependent toxicity, but likely reflect increasing protein levels of an already modified p53. Presumably, these modifications arise as a consequence of low constitutive stress signals in the cultured cells.2
Expression levels of p53 determine cell fate decisions between G1 arrest and apoptosis
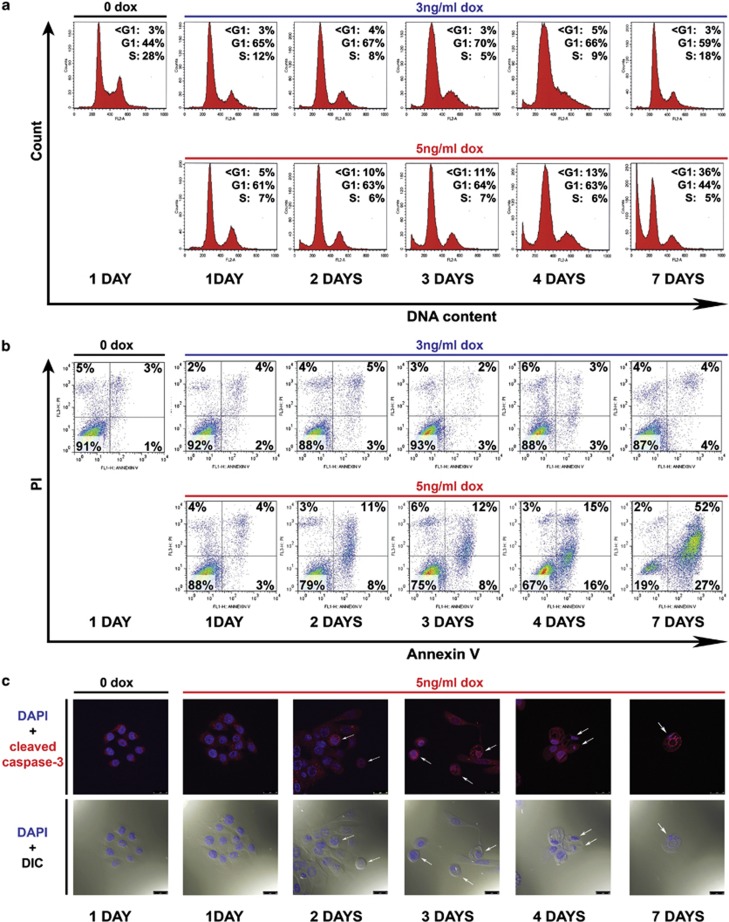
Next, we assessed the consequences of increasing p53 levels on cell fate decisions. Although p53 induced by a lower dose of 3 ng/ml doxycycline, defined as ‘low-p53', led to G1 arrest without concomitant apoptosis, p53 induced by a higher dose of 5 ng/ml doxycycline, defined as ‘high-p53', triggered apoptosis (Figure 2). Expression of both low- and high-p53 initially led to G1 arrest (Figure 2a). However, low-p53-mediated G1 arrest was resolved within 3-5 days, and the cells re-entered the cell cycle. By contrast, high-p53-mediated G1 arrest was followed by apoptosis within 2 days (Figures 2a and b). Of note, increased p53 expression to super-physiological levels positively correlated with the apoptotic rate, emphasizing the importance of p53 dose as a determinant of the intensity and kinetic of its response (Supplementary Figure 2). To further explore p53-mediated apoptosis in these cells, we performed confocal imaging of high-p53-induced B5/589-p53 cells. Following an apoptotic stimulus, cleaved caspase-3 has been shown to translocate to the nucleus.28 In agreement, we detected nuclear localization of cleaved caspase-3 in apoptotic cells within 2 days of high-p53 induction (Figure 2c). This was accompanied by morphological changes associated with apoptosis, including reduction in cell volume, appearance of cytoplasmic vacuoles and emergence of pyknotic and fragmented nuclei.
Figure 2.
The level of p53 in B5/589-p53 cells determines the cell fate decision between G1 cell cycle arrest and apoptosis. (a) Flow cytometry analysis of DNA content by PI staining of B5/589-p53 cells induced with 3 or 5 ng/ml doxycycline (dox) for 1, 2, 3, 4 and 7 days. Control cells were cultured without doxycycline (0 dox) for 1 day. (b) Flow cytometry analysis of apoptosis by Annexin V/PI staining of the same B5/589-p53 cells described in (a). (c) Confocal analysis of B5/589-p53 cells that were uninduced (0 dox) for 1 day or induced with 5 ng/ml dox for 1, 2, 3, 4 and 7 days. Blue, DAPI (4′,6-diamidino-2-phenylindole) nuclear staining. Red, cleaved caspase-3 staining. DIC (differential interference contrast microscopy) morphology was also photographed by the confocal microscope. White arrows point to examples of cell shrinkage (5 ng dox – 2 days; 5 ng dox – 3 days (left)), cytoplasm vacuolization (5ng dox – 3 days (middle and right)), pyknotic and fragmented nuclei (5 ng dox – 4 and 7 days). Magnification, × 63; scale bar, 25μm
Low- and high-p53 levels lead to transcriptional activation of both proarrest and proapoptotic p53 target genes
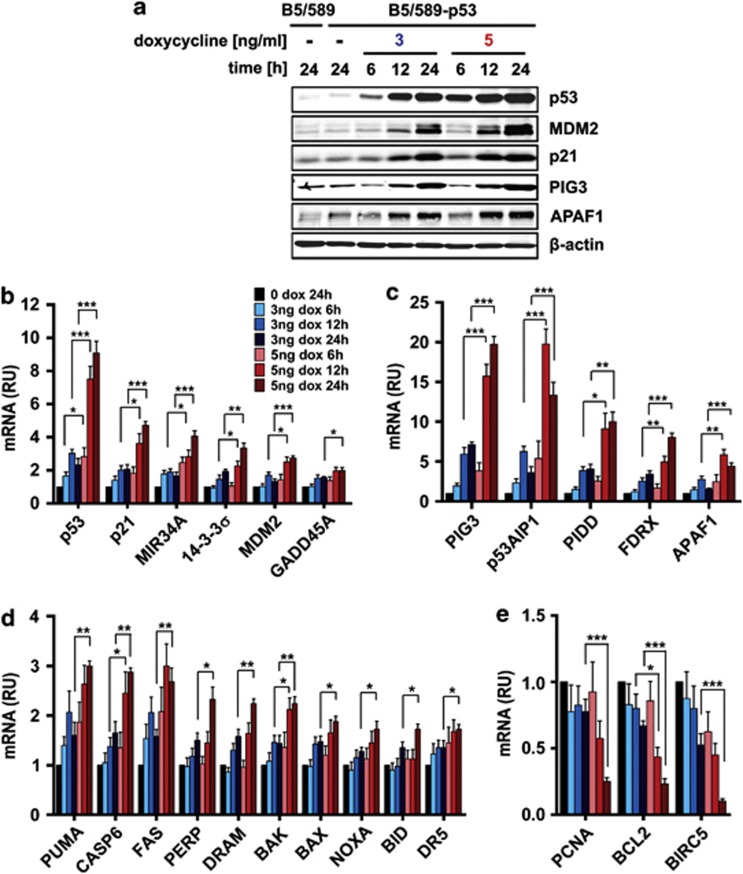
The ‘affinity model' asserts that low p53 levels preferentially transactivate its proarrest target genes, whereras high-p53 levels are necessary to transactivate proapoptotic targets. To test this, we examined selected proarrest and pro-apoptotic p53 target genes in B5/589-p53 cells induced with low- and high-p53. We focused on early time points of p53 induction to isolate p53 effects on its direct target genes. Protein levels of p53 targets MDM2, p21, PIG3 and APAF1 increased proportionally to p53 expression, irrespective of their function in arrest or apoptosis (Figure 3a). Similarly, mRNA expression levels of p53 target genes mainly involved in arrest (Figure 3b) or apoptosis (Figures 3c and d) were proportional to p53 mRNA expression. Furthermore, downregulation of mRNA expression of PCNA, BCL2 and survivin, reportedly repressed by p53,3, 29 was inversely proportional to p53 levels (Figure 3e). These findings were inconsistent with the ‘affinity model.'
Figure 3.
Expression levels of p53 target genes in low and high induced B5/589-p53 cells. (a) Immunoblot analysis of p53 and its transcriptional targets MDM2, p21, PIG3 and APAF1 in B5/589-p53 cells induced either with low (3 ng/ml) or high (5 ng/ml) levels of doxycycline (dox) for 6, 12 and 24 h. Control cells were cultured without dox for 24 h. β-Actin was used as a loading control. (b—d) Real-time PCR analysis of genes activated by p53 in B5/589-p53 cells treated as in (a) involved in cell cycle arrest (b) and apoptosis (c, d). (e) Real-time PCR analysis of genes repressed by p53 in B5/589-p53 cells treated as in (a). For all real-time PCR experiments, mRNA expression levels were normalized to 18S. Data are presented as the mean±S.E.M. from at least three independent experiments (n≥3). RU, relative units. Unpaired Student's t-test was conducted to determine whether mRNA expression of p53 transcriptional activation targets was higher (or lower for p53 transcriptional repression targets) in cells induced with 5 ng/ml compared with 3 ng/ml dox at the same time points. *P<0.05; **P<0.01; and ***P<0.001
Global changes in gene expression are proportional to p53 induction levels
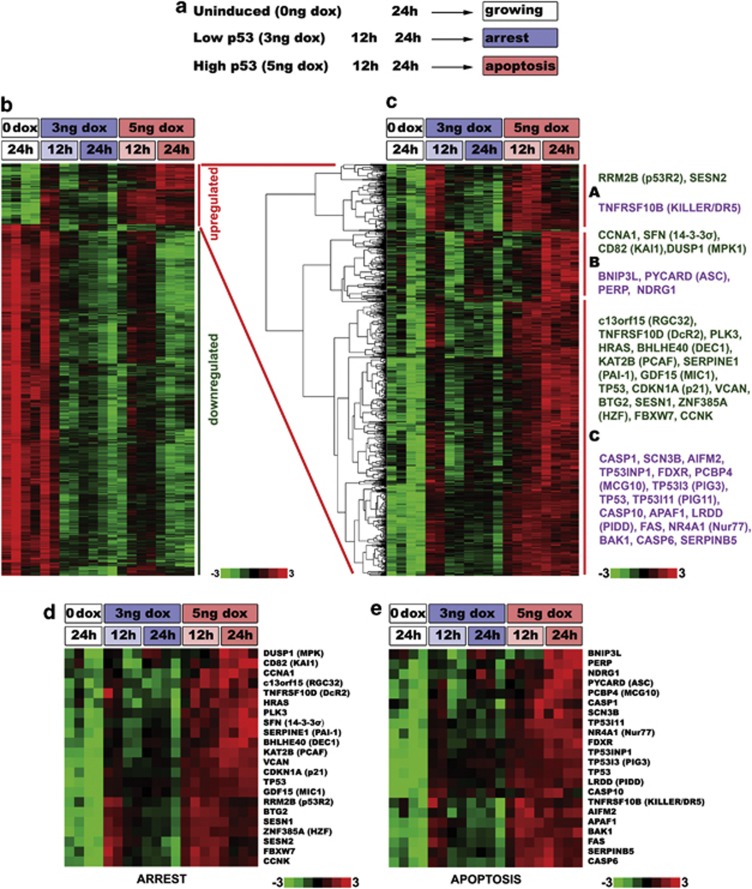
To determine whether other proarrest and proapoptotic p53 targets are also proportionally regulated by different p53 levels, we conducted a whole-transcript microarray in B5/589-p53 cells (Figure 4a). Using this approach, we detected 21 818 expressed genes, of which 947 were upregulated and 5467 were downregulated (Figure 4b). Such extensive p53-mediated repression has been reported previously.17, 21 p53 signaling pathway, pathways in cancer and apoptosis were among the highest ranked functional pathways in the group of 947 upregulated genes (Table 1 and Supplementary Table 1). Cell cycle, DNA replication and repair pathways were among those most significantly enriched in the downregulated group of 5467 genes (Table 1 and Supplementary Table 2).17, 29, 30, 31
Figure 4.
Microarray analysis of changes in gene expression upon low and high p53 expression. (a) Experimental design for the whole-transcript microarray analysis. B5/589-p53 cells were either uninduced (0 ng doxycycline (dox)) or induced with low- (3 ng/ml dox) and high- (5 ng/ml dox) p53 for 12 and 24 h. Four independent biological replicates were used for each of the experimental conditions. (b) Heat map of all differentially expressed genes upon p53 induction. In all, 947 upregulated genes are indicated by the red vertical bar, and 5467 downregulated genes are indicated by the green vertical bar. Variations in responses observed at 12 h with low-p53 may reflect small differences in kinetics among replicate arrays. (c) Cluster analysis of the 947 genes that were upregulated in at least one of the experimental conditions when compared with uninduced cells. These genes were grouped into three main clusters (A, B, C) according to the pattern of their expression. For each cluster, examples of proarrest (green) and proapoptotic (violet) genes are shown on the right. (d, e) Heat map of 21 proarrest (D) and 21 proapoptotic (e) genes. P53 gene was added for comparison to both analyses
Table 1. Up- and downregulated pathways upon p53 induction.
| Rank | KEGG ID | Upregulated pathways | Set size in pathwaya | Set size allb | P-valuec |
|---|---|---|---|---|---|
| 1 | hsa04115 | P53 signaling pathway | 19 | 74 | 0.00000 |
| 2 | hsa00330 | Arginine and proline metabolism | 10 | 45 | 0.00001 |
| 3 | hsa05200 | Pathways in cancer | 30 | 318 | 0.00004 |
| 4 | hsa04210 | Apoptosis | 12 | 87 | 0.00015 |
| 6 | hsa00590 | Arachidonic acid metabolism | 8 | 47 | 0.00027 |
| 8 | hsa01100 | Metabolic pathways | 63 | 982 | 0.00103 |
| 9 | hsa02010 | ABC transporters | 6 | 36 | 0.00115 |
| 10 | hsa04060 | Cytokine–cytokine receptor interaction | 18 | 196 | 0.00124 |
| 12 | hsa04662 | B-cell receptor signaling pathway | 9 | 71 | 0.00131 |
| 13 | hsa00480 | Glutathione metabolism | 6 | 40 | 0.00208 |
| 15 | hsa04512 | ECM–receptor interaction | 9 | 81 | 0.00335 |
| 16 | hsa04630 | Jak-STAT signaling pathway | 12 | 123 | 0.00339 |
| 18 | hsa04514 | Cell adhesion molecules (CAMs) | 10 | 104 | 0.00668 |
| 19 | hsa04640 | Hematopoietic cell lineage | 7 | 63 | 0.00703 |
| 20 | hsa04070 | Phosphatidylinositol signaling system | 8 | 78 | 0.00796 |
| 22 | hsa04360 | Axon guidance | 11 | 126 | 0.01033 |
| 23 | hsa04650 | Natural killer cell-mediated cytotoxicity | 10 | 112 | 0.01114 |
| 24 | hsa04142 | Lysosome | 10 | 113 | 0.01183 |
| 25 | hsa04660 | T-cell receptor signaling pathway | 9 | 98 | 0.01187 |
| 28 | hsa04622 | RIG-I-like receptor signaling pathway | 6 | 59 | 0.01581 |
| 29 | hsa04370 | VEGF signaling pathway | 7 | 74 | 0.01698 |
| 30 | hsa04664 | Fcɛ RI signaling pathway | 7 | 77 | 0.02091 |
| 33 | hsa04144 | Endocytosis | 13 | 189 | 0.03990 |
| 35 | hsa04010 | MAPK signaling pathway | 17 | 262 | 0.04097 |
| 36 | hsa04620 | Toll-like receptor signaling pathway | 7 | 90 | 0.04540 |
| |
|
Downregulated pathways |
|
|
|
| 1 | hsa04110 | Cell cycle | 86 | 136 | 0.00000 |
| 3 | hsa03030 | DNA replication | 32 | 40 | 0.00000 |
| 4 | hsa00240 | Pyrimidine metabolism | 51 | 93 | 0.00000 |
| 5 | hsa03010 | Ribosome | 58 | 116 | 0.00000 |
| 6 | hsa03410 | Base excision repair | 23 | 32 | 0.00000 |
| 7 | hsa03430 | Mismatch repair | 18 | 25 | 0.00003 |
| 8 | hsa03420 | Nucleotide excision repair | 25 | 44 | 0.00008 |
| 9 | hsa01100 | Metabolic pathways | 305 | 982 | 0.00009 |
| 10 | hsa03018 | RNA degradation | 30 | 58 | 0.00011 |
| 11 | hsa00230 | Purine metabolism | 56 | 138 | 0.00028 |
| 12 | hsa03440 | Homologous recombination | 18 | 31 | 0.00046 |
Pathways are truncated, see Supplementary Tables 1 and 2 for the complete tables including gene names
The analysis used the Comodus algorithm (A George, unpublished)
Number of probe sets in the pathway that were expressed in upregulated or downregulated experimental sets (total of 947 or 5467 probe sets, respectively)
Number of probe sets in the pathway that were expressed in all experimental sets (total of 21 818 probe sets)
Hypergeometric test P-value
Focusing on the 947 upregulated genes, we used hierarchical clustering to group genes based on their expression patterns. We identified three main clusters (Figure 4c; clusters A, B and C) and examined 21 representative proarrest and proapoptotic genes previously described as p53 targets. We reasoned that if p53 bound to the promoters of proapoptotic genes with a lower affinity than to the promoters of proarrest genes, we would expect proapoptotic and proarrest genes to show different kinetics in their expression profiles upon low-p53 and high-p53 induction. If true, proapoptotic and proarrest genes would group in separate clusters, indicative of their distinct expression profile signatures. However, all three clusters contained a mixture of both proapoptotic and proarrest genes (Figure 4c).
To examine the expression patterns of these proarrest and proapoptotic genes in more detail, we performed a cluster analysis of the two groups separately. Although different expression patterns were apparent within each group, none were specific for either proarrest or proapoptotic groups (Figure 4d and e). In fact, cluster analysis of the representative proarrest and proapoptotic genes revealed that neither group exclusively clustered together. Of the seven clusters (A-G, Supplementary Figure 3) identified in this analysis, each contained both proarrest and proapoptotic genes. Thus, we found no evidence of differential upregulation of proarrest and proapoptotic genes by low- and high-p53. These results corroborate our mRNA expression data and provide further evidence that low and high levels of p53 transcriptionally activate both proarrest and proapoptotic target genes.
P53 is recruited to both proarrest and proapoptotic promoters
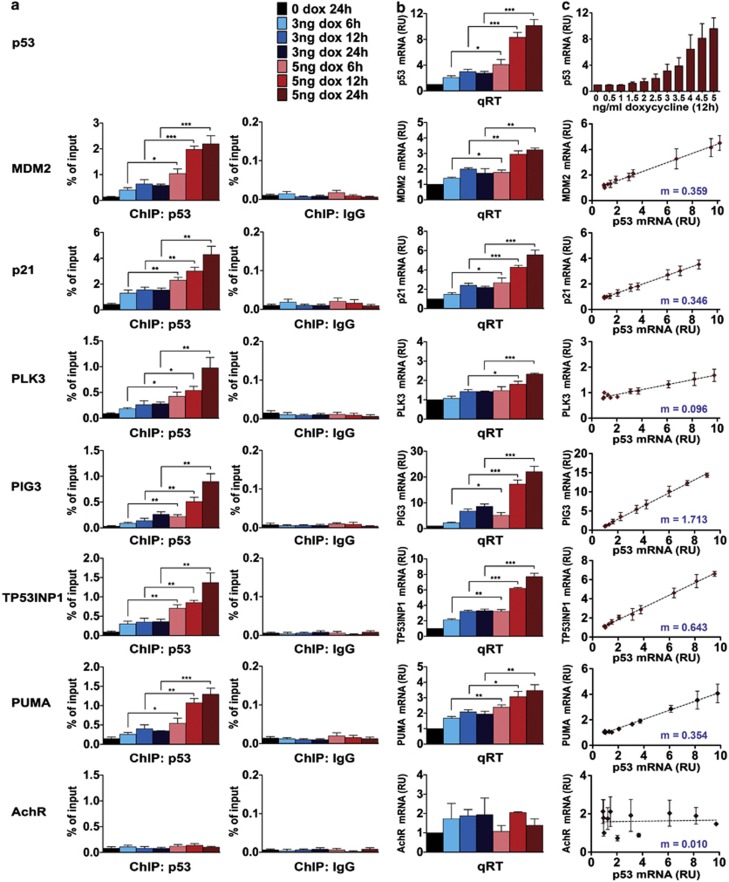
Next, we assayed p53 binding to selected proarrest and proapoptotic promoters in cells induced with low- and high-p53 by ChIP. We measured p53 occupancy of promoters in genes with well-defined p53 RE, including MDM2, proarrest p21 and polo-like kinase 3 (PLK3)16 and proapoptotic PIG3,32 TP53INP1 and PUMA. Acetylcholine receptor (AchR) served as a negative control.16 Concurrently, we analyzed the expression of the same genes via real-time PCR. Figures 5a and b show that the recruitment of p53 to both proarrest and proapoptotic promoters followed a similar pattern, which was proportional to p53 expression levels. Importantly, low levels of p53 were sufficient to occupy p53 RE on the proapoptotic promoters PIG3, TP53INP1 and PUMA. The pattern of p53 binding to all examined genes matched the pattern of their increased transcription (Figure 5b). These results indicate that p53 targets, whether proarrest or proapoptotic, are induced proportionally to the levels of p53, predicting a linear relationship between the expression levels of p53 targets and p53. To verify this, we titrated doxycycline within 0–5 ng/ml range in 0.5 ng/ml increments (Figure 5c, top panel) and measured mRNA levels of p53 and its targets MDM2, p21, PLK3, PIG3, TP53INP1 and PUMA by real-time PCR (Figure 5c). We observed a linear relationship between the expression levels of p53 and each of these proarrest and proapoptotic target genes. Of note, variations in slopes of different transcript levels following p53 induction may be indicative of the affinity of p53 for these targets. Even very low levels of p53 (3 ng/ml doxycycline and less) induced proapoptotic targets PIG3, TP53INP1 and PUMA. Thus, low levels of p53 are sufficient to bind and activate promoters associated with both arrest and apoptosis.
Figure 5.
The level of p53 binding to promoters of proarrest and proapoptotic genes is proportional to p53 expression levels. (a) ChIP analysis on the promoters of p53 target genes in B5/589-p53 cells. P53 was induced for 6, 12 and 24 h with either low (3 ng/ml) or high (5 ng/ml) levels of doxycycline (dox). ChIP was performed with p53 or control immunoglobulin G (IgG) antibody and quantified by real-time PCR on indicated promoters. (b) Real-time PCR analysis for mRNA levels of p53 and its indicated targets in B5/589-p53 cells treated as in (a). mRNA levels were normalized to 18S. (c) Real-time PCR analysis for mRNA levels of p53 and its indicated targets in B5/589-p53 cells. P53 was induced for 12 h with dox concentrations from 0.5 to 5 ng/ml in 0.5 ng/ml increments. m represents the slope, according to the formula of the linear equation y=mx+b. RU, relative units. Data are presented as the mean±S.E.M. from four independent experiments (n=4). Unpaired Student's t-test was conducted to determine whether promoter occupancy or mRNA expression was higher in cells induced with 5 ng/ml compared with 3 ng/ml dox at the same time points. *P<0.05; **P<0.01; and ***P<0.001
Prolonged, low-level expression of p53 and its proapoptotic targets is not sufficient to induce apoptosis
To address whether prolonged p53 expression resulted in a sustained and proportional upregulation of proarrest and proapoptotic targets, we measured protein levels of p53 and its targets MDM2, p21, PIG3 and APAF1 induced with low- or high-p53 over a 1-week period. As expected, expression levels of p53 targets were induced proportionally to p53 levels, whether they were involved in arrest (p21), apoptosis (PIG3 and APAF1) or p53 autoregulation (MDM2) (Figure 6a). The decrease in levels of all analyzed proteins (including β-actin) at 7 days of high-p53 coincided with extensive apoptosis (Figure 6b). Notably, even in the absence of apoptosis, protein levels of the proapoptotic PIG3 and APAF1 were increased in cells induced with low-p53, although to a lower level than in cells induced with high-p53.
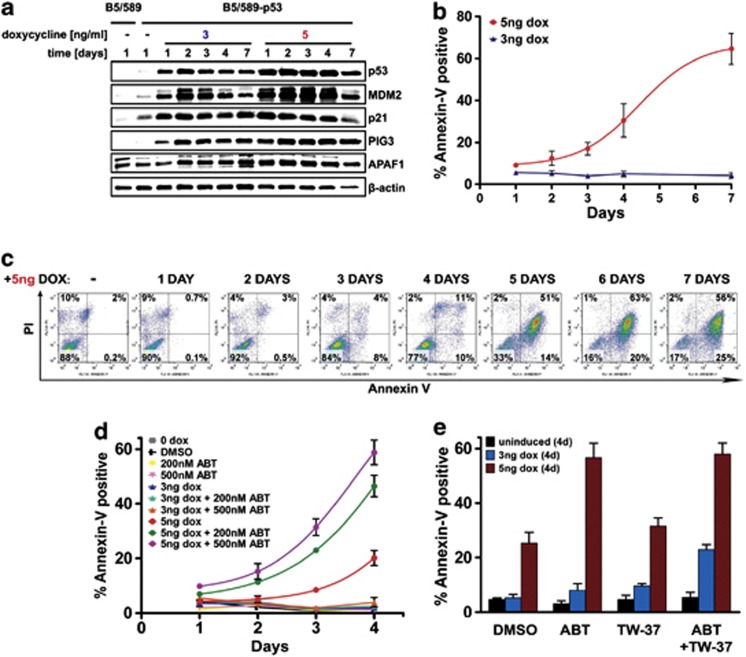
Figure 6.
High apoptotic threshold mediates p53-dependent decision between arrest and apoptosis in B5/589-p53 cells. (a) Immunoblot analysis of p53, MDM2, p21, PIG3 and APAF1 in B5/589-p53 cells induced with 3 or 5 ng/ml doxycycline (dox) for 1, 2, 3, 4 and 7 days. (b) Apoptosis in B5/589-p53 cells treated as in (a) measured by flow cytometry of Annexin V/PI staining. The percentages of Annexin V-positive cells are shown. In all, 5 ng dox data were fitted with a Boltzmann function. Data are presented as the mean±S.E.M. from five independent experiments (n=5). (c) Apoptosis in B5/589-p53 cells induced with 5 ng/ml dox for a number of days indicated by the ‘+5 ng DOX' legend, and subsequently de-induced for 4 days. Uninduced cells were grown for 4 days. (d, e) Apoptosis in B5/589-p53 cells measured by flow cytometry of Annexin V/PI staining. The percentages of Annexin V-positive cells are shown. (d) Cells were uninduced (0 dox) or induced with low (3 ng/ml) or high (5 ng/ml) levels of dox and treated with 200 or 500 nℳ ABT-263 for 4 days. Uninduced cells were treated with 0.05% dimethylsulfoxide (DMSO) as a control. Data were fitted with a Boltzmann function. (e) Cells were uninduced (0 dox) or induced with (3 ng/ml) or high (5 ng/ml) levels of dox and treated with 0.08% DMSO, 500 nℳ ABT-263 and/or 300 nℳ TW-37 for 4 days. Data are presented as the mean±S.E.M. from three independent experiments (n=3)
Prolonged, high-level expression of p53 is necessary to trigger apoptosis
It has been shown that cells within genetically identical populations can respond with different kinetics to the same death stimulus.33 We asked at which point of continued high-p53 expression the majority of B5/589-p53 cells irreversibly commit to apoptosis. Hence, we induced B5/589-p53 cells with 5 ng/ml doxycycline, followed by doxycycline removal at various times after induction. Cells were then further maintained without doxycycline for 4 days, and analyzed for apoptosis. As shown in Figures 6c, 3–4 days of sustained high-p53 resulted in a small but significant increase in the percentage of apoptotic cells after de-induction. However, 5 or more days of sustained high-p53 expression resulted in the majority of cells undergoing apoptosis after de-induction. These results illustrate that in the absence of additional stimuli, high and sustained expression of p53 is required to execute apoptosis in this system.
Lowering the apoptotic threshold sensitizes cells to p53-dependent apoptosis
The apoptotic threshold of a cell is determined by a ratio of proapoptotic and antiapoptotic signals, and apoptosis occurs when proapoptotic signals outweigh the counteracting antiapoptotic buffer.34 Antiapoptotic Bcl-2 family member proteins are known to influence the apoptotic threshold.35, 36, 37 Accordingly, we tested whether lowering the apoptotic threshold by inhibiting the functions of antiapoptotic proteins would facilitate the apoptotic program. We induced B5/589-p53 cells with low- and high-p53 in the presence or absence of ABT-263 (Navitoclax), an inhibitor of antiapoptotic Bcl-2 family members Bcl-2, Bcl-xL and Bcl-w.38 Although ABT-263 accelerated the onset of high-p53-induced apoptosis in a dose-dependent manner, it had no detectable apoptotic effect alone or in combination with low-p53 induction (Figure 6d). However, addition of TW-37, which also inhibits another Bcl-2 family member Mcl-1,39 was able to trigger apoptosis in approximately 20% cells induced with low-p53 (Figure 6e). This implies that although ABT-263 decreased the apoptotic threshold, it did so to a level insufficient to trigger apoptosis in low-p53-induced cells. However, lowering the threshold further by the addition of TW-37 allowed these cells to cross the apoptotic threshold and initiate apoptosis. Hence, neutralizing the activity of antiapoptotic Bcl-2 family proteins can promote the switch from arrest to apoptosis.
Nutlin-3-activated p53 determines cell fate decisions and synergizes with ABT-263 to induce apoptosis in RKO and LNCaP cancer cells
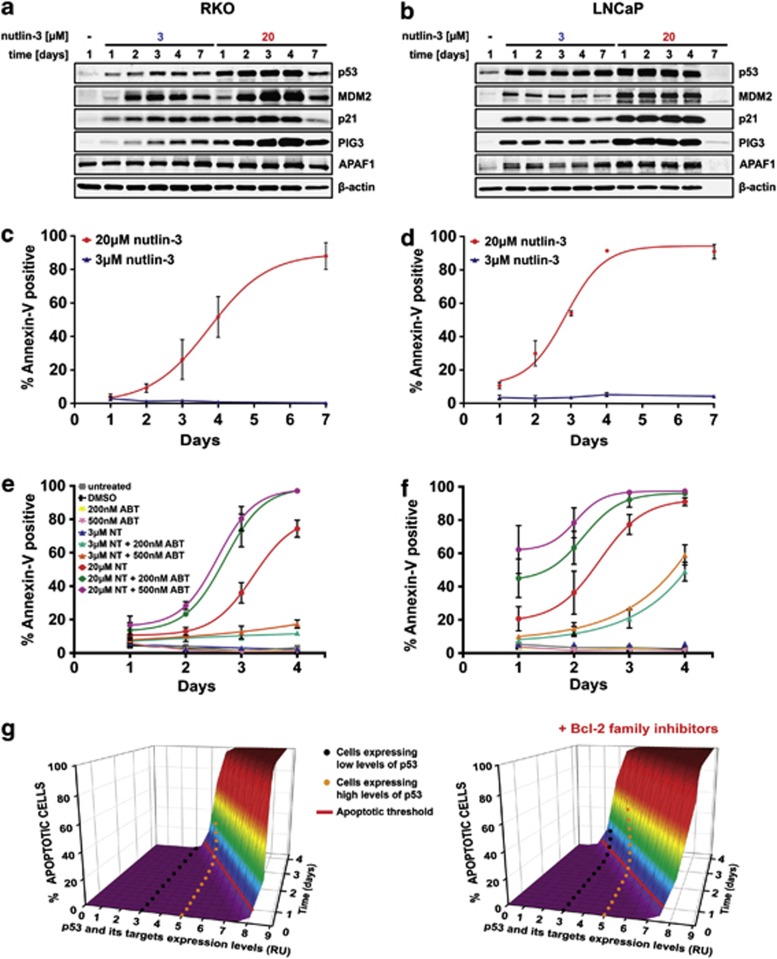
To validate our observations in another cell system, we examined the response to increasing p53 levels in wild-type p53 colon (RKO) and prostate (LNCaP) cancer cells. We used nutlin-3 to induce p53 to model therapeutic p53 activation in wild-type p53 tumors. Treatment with low doses of nutlin-3 (3 μℳ) led to growth arrest in both cell lines, whereas higher doses (20 μℳ) first triggered arrest, followed by apoptosis (Supplementary Figure 4). LNCaP cells were more sensitive to these nutlin-3 doses and exhibited sustained arrest and earlier onset of apoptosis than RKO cells, as reported previously.24 In both cell lines, increase in p53 protein matched the nutlin-3 dose, and the levels of p53 targets MDM2, p21, PIG3 and APAF1 increased proportionally to p53 expression (Figures 7a and b). The loss of all protein expression in LNCaP cells after 7 days of high-p53 (20 μℳ nutlin-3) activation is consistent with its higher apoptotic index compared with RKO cells (Figures 7c and d).24 As expected, lowering the apoptotic threshold with ABT-263 synergized with high-p53 activation in a dose-dependent manner to accelerate apoptosis in both RKO and LNCaP cells (Figures 7e and f). However, ABT-263 also synergized with low-p53 (3 μℳ nutlin-3) in a dose-dependent manner to trigger apoptosis in LNCaP cells, and to a lesser but reproducible extent in RKO cells. We did not observe this synergy in B5/589 cells (Supplementary Figure 5), suggesting that the RKO and LNCaP cancer cells are more sensitive to p53-induced apoptotic stimulus than the non-tumorigenic B5/589 HMEC. These results confirm that lowering the apoptotic threshold with ABT-263 significantly enhances the therapeutic impact of p53 activation.
Figure 7.
Lowering the apoptotic threshold sensitizes RKO and LNCaP cancer cells to p53-mediated apoptosis. (a–b) Immunoblot analysis of p53, MDM2, p21, PIG3 and APAF1 in RKO (a) and LNCaP (b) cancer cells incubated with low (3 μℳ) or high (20 μℳ) doses of nutlin-3 for 1, 2, 3, 4 and 7 days. (c, d) Apoptosis in RKO (c) or LNCaP (d) cells treated as in (a, b) measured by flow cytometry of Annexin V/PI staining. The percentages of Annexin V-positive cells are shown. In all, 20 μℳ nutlin-3 data were fitted with a Boltzmann function. Data are presented as the mean±S.E.M. from three independent experiments (n=3). (e, f) Apoptosis in RKO (e) or LNCaP (f) cells measured by flow cytometry of Annexin V/PI staining. The percentages of Annexin V-positive cells are shown. Cells were incubated with low (3 μℳ) or high (20 μℳ) doses of nutlin-3 (NT) and treated with 200 or 500 nℳ ABT-263 for 4 days. Cells were also treated with 0.15% dimethylsulfoxide (DMSO) as a control. Data were fitted with a Boltzmann function. Data are presented as the mean±S.E.M. from three independent experiments (n=3). (g) A three-dimensional model of the apoptotic threshold. Apoptosis is triggered when the cell crosses the apoptotic threshold (solid red line). The rate of apoptosis initiation depends on the expression levels of p53 and its targets (x axis), on the duration of their expression (z axis) and on the activity of antiapoptotic Bcl-2 family proteins
Discussion
It is well established that in response to various stress signals, p53 acts as a barrier to tumor development by inducing growth arrest or apoptosis.1, 2 How p53 chooses between arrest and apoptosis is of great interest, especially because of its relevance to successful cancer therapy.8, 10, 11 Using a p53-inducible system, we demonstrated that p53 levels alone can determine the choice between arrest and apoptosis in B5/589 HMEC. Previous studies suggested that p53 regulated different biological outcomes by selectively activating distinct sets of target genes, due to p53 binding to promoters of proarrest genes with higher affinity than to promoters of proapoptotic genes.15, 16 By contrast, using real-time PCR, microarray and ChIP, we demonstrated that p53 can bind and transactivate the same sets of proarrest and proapoptotic genes proportionally to the level of p53 expression.
Although low levels of p53 proarrest targets could initiate growth arrest, low levels of proapoptotic targets failed to trigger apoptosis, even when expressed over an extended time period. Instead, prolonged higher levels of p53 and its proapoptotic targets were required for the onset of apoptosis. This behavior is analogous to the threshold mechanism described for receptor-induced apoptosis, whereby the rate of apoptosis increased with rising concentration of proapoptotic molecules, as long as their level was above a certain threshold.40, 41 Thus, we propose a model in which expression levels of both p53 proarrest and proapoptotic targets increase proportionally to p53 expression, and p53-mediated cell fate decision between arrest and apoptosis is determined by the higher execution threshold for the initiation of apoptosis than for arrest (Supplementary Figure 6).
The apoptotic threshold has been defined as the minimum ratio of proapoptotic to antiapoptotic signals necessary to induce apoptosis.40, 41 Cells expressing subthreshold concentrations of proapoptotic proteins do not undergo apoptosis, irrespective of the duration of their expression, due to their effective blocking by antiapoptotic proteins.41 The height of this threshold is not a fixed property of the cell; rather, it reflects a dynamic equilibrium between pro- and antiapoptotic signals.34, 35 By changing this equilibrium, it should be possible to lower the apoptotic threshold. In our study, apoptosis was triggered solely by p53. In vivo however, an apoptotic stimulus would activate p53-dependent as well as p53-independent pathways. For instance, DNA damage can stabilize p53 and the E2F1 transcription factors, which cooperate to increase the levels and the number of proapoptotic genes.18, 42 Equally, inhibition of antiapoptotic proteins should lower the apoptotic threshold. Accordingly, inhibitors of antiapoptotic Bcl-2 family proteins significantly accelerated apoptosis in multiple cell lines expressing high-p53 and triggered apoptosis in some cells expressing low-p53. Notably, wild-type p53 cancer cells, RKO and LNCaP, were more sensitive than the non-tumorigenic B5/589 cells to apoptosis triggered by p53 activation alone, or in combination with Bcl-2 inhibitors, consistent with evidence suggesting that some tumor cells are more sensitive to p53-dependent apoptotic stimuli than normal cells.5, 6
Based on these data, we modeled the apoptotic threshold (Figure 7g). Here, cells expressing low levels of p53 and its targets never cross the apoptotic threshold, whereas cells expressing high levels of p53 and its targets reach the apoptotic threshold after several days. However, lowering the threshold with Bcl-2 family protein inhibitors allows certain cells expressing low-p53 to reach the apoptotic threshold and undergo apoptosis. At the same time, cells expressing high p53 cross the apoptotic threshold faster, shown by increased apoptotic rate.
As a transcription factor, p53 regulates the expression of hundreds of genes.1, 3 This results in an activation and repression of complex signaling networks that need to be integrated into specific outcomes. From a systems biology perspective, threshold mechanisms present an elegant solution to the challenge of translating increases in stress severity to distinct biological responses.1, 43 A similar threshold model was reported for the Myc transcription factor, where different levels of Myc and its targets determined the cell fate decision between proliferation and apoptosis.44 Other investigators also demonstrated that increasing levels of Raf-1 or RasG12V could switch the outcome from proliferation to arrest or senescence.45, 46, 47 Thus, distinct execution thresholds may provide a common mechanism for determining cell fate decisions.
The fact that the p53 pathway is impaired in almost all human cancers makes the restoration of p53 function an attractive target for therapy.5, 6 The aim of these strategies is to reactivate p53 that would preferably trigger apoptosis to achieve maximum tumor regression.7, 8 Our results suggest that low-level p53 activation is an ineffective way to induce apoptosis. In fact, insufficient p53 protein induction has been shown as one of the causes of resistance to MDM2 inhibitors.48 Conversely, strong p53 activation could be toxic in normal tissues.26 Thus, lowering the apoptotic threshold has the potential to increase the efficacy of p53-based cancer therapies by preferentially inducing apoptosis, although careful titration of p53 activity would be necessary to optimize their therapeutic index.
Materials and Methods
Cell culture and reagents
The immortalized B5/589 HMECs23 were cultured in RPMI-1640 medium (Sigma, St. Louis, MO, USA; R8758) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma; F0926), 0.5% penicillin/streptomycin (P/S; Invitrogen, Carlsbad, CA, USA; 15140-122) and 5 ng/ml hEGF (Peprotech, Rocky Hill, NJ, USA; AF-100-15). RKO cells were grown in minimum essential medium (Invitrogen; 12571-063) supplemented with 1 × non-essential amino acids (Invitrogen; 11140-050), 100 μℳ sodium pyruvate (Sigma; S8636), 10% FBS and 0.5% P/S. LNCaP cells were maintained in RPMI-1640 supplemented with 10% FBS and 0.5% P/S. All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. Cells were seeded at exactly 0.8 × 105 cells/10 cm plate 12-24 h before treatment. Media with or without doxycycline (or other treatments) were replenished every 24 h. Doxycycline was purchased from Clontech (Mountain View, CA, USA; 631311), doxorubicin and polybrene from Sigma (D1515 and 107689, respectively), puromycin from Calbiochem (San Diego, CA, USA; 540222) and blasticidin from Invitrogen (R210-01). ABT-263 (ChemieTek, Indianapolis, IN, USA; CT-A263), TW-37 (Selleck Chemicals, Houston, TX, USA; S1121) and ±nutlin-3 (racemic; Cayman Chemical, Ann Arbor, MI, USA; 10004372) were dissolved in DMSO (Sigma; D2650). TrypLE Express Stable Trypsin Replacement Enzyme (Invitrogen; 12604) was used for the dissociation of attached cells.
Immunoblot analysis and immunoprecipitation
Cells were collected by centrifugation and harvested in lysis buffer (50 mℳ HEPES (pH 7.6), 150 mℳ NaCl, 0.5% NP-40, 5 mℳ EDTA (pH 8.0)) supplemented with protease and phosphatase inhibitors (Roche; 04693124001 and 04906845001). For γH2AX detection, cells were harvested in SDS lysis buffer (1% SDS, 10 mℳ EDTA (pH 8.0), 50 mℳ Tris (pH 8.0)) supplemented with protease and phosphatase inhibitors. Total cell protein (20 μg) was separated by 12% SDS-PAGE, transferred to Immobilon-FL PVDF membrane (EMD Millipore, Billerica, MA, USA; IPFL00010), incubated with primary antibodies overnight at 4 °C, followed by 40 min incubation with anti-mouse or anti-rabbit fluorescently tagged secondary antibodies. Bands were visualized and quantitated by the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). Primary antibodies used were as follows: p53 (1801; Mount Sinai School of Medicine Hybridoma Center, New York, NY, USA), MDM2 (Calbiochem; OP46), p21 (BD Biosciences, San Jose, CA, USA; 556431), PIG3 (Abcam, Cambridge, MA, USA; ab64798), APAF1 (R&D Systems, Minneapolis, MN, USA; MAB868), acetyl-p53 (Lys382) (Cell Signaling, Danvers, MA, USA; 2525), phospho-p53 (Ser15) (Cell Signaling; 9284), phospho-p53 (Ser46) (Cell Signaling; 2521), γH2AX (Abcam; ab26350) and β-actin (Sigma; A5441). Secondary antibodies used were: anti-mouse and anti-rabbit IgG antibodies conjugated to Alexa Fluor 680 or Alexa Fluor 750 (Invitrogen; A21057 or A21039). For immunoprecipitation, cells were harvested in lysis buffer (50 mℳ HEPES (pH 7.6), 150 mℳ NaCl, 0.5% NP-40, 5 mℳ EDTA (pH 8.0)) supplemented with protease and phosphatase inhibitors. In total, 200 or 800 μg proteins were incubated with 2 μg of p53 (DO-1) monoclonal antibody (Santa Cruz, Santa Cruz, CA, USA; sc-126) or 2 μg of normal IgG (Santa Cruz; sc-2025) overnight at 4 °C. In all, 20 μg of samples without primary antibody were saved as 10% input controls. Immunoprecipitated complexes were captured by 2 h incubation at 4 °C with Dynabeads Protein G (Invitrogen; 100.04D), followed by three washes in lysis buffer. Immunoprecipitated complexes were eluted by boiling for 10 min with 50 μl 2 × Laemmli buffer (100 mℳ Tris-Cl (pH 6.8), 20% glycerol, 4% SDS, 0.002% bromophenol blue, 2% 2-mercaptoethanol) and analyzed by immunoblotting with indicated primary antibodies and light-chain-specific, Alexa Fluor 647 anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, USA; 114-605-174 or 211-602-171) secondary antibodies. Bands were visualized by the Odyssey Infrared Imaging System (LI-COR Biosciences).
Flow cytometry analysis
Floating and adherent cells from 10-cm plates were combined and washed with PBS. Half were used for propidium iodide (PI) staining and the other half for Annexin V/PI staining. For PI analysis, cells were stained with PI using the CycleTEST PLUS DNA Reagent Kit (BD Biosciences; 340242). For Annexin V/PI analysis, cells were stained with Annexin V : FITC Apoptosis Detection Kit I (BD Biosciences; 556547). Cells were transferred to polystyrene tubes with cell-strainer caps (BD Biosciences; 352235). At least 10 000 cells were sorted by flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ, USA) and analyzed using Cell Quest 3.2 (Becton Dickinson) or FlowJo 7.6.5 (Tree Star Inc., Ashland, OR, USA) software.
Confocal microscopy
Cells grown on glass coverslips were washed twice with PBS and fixed in freshly prepared 4% paraformaldehyde (Polysciences, Warrington, PA, USA; 18814) in PBS for 1 h at room temperature. Fixed cells were then washed twice with PBS and permeabilized with 0.1% Triton X-100 in PBS for 5 min, followed by blocking for 30 min with 3% normal goat serum (Vector Laboratories, Burlingame, CA, USA; S-1000), and incubated for 1 h with cleaved caspase-3 antibody (Cell Signaling; 9664), all at room temperature. Cells were then washed three times with PBS and incubated for 1 h in the dark with secondary anti-rabbit Alexa Fluor 594 (Invitrogen; A-21207) at room temperature, followed by three washes with PBS and two washes with water. Coverslips were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen; P-36931). Confocal imaging was performed with the Leica SP5 DM confocal microscope (Leica Microsystems Inc., Buffalo Grove, IL, USA) using the × 63 oil objective.
RNA extraction and real-time PCR
Total RNA was isolated using an RNAeasy Mini kit (Qiagen, Valencia, CA, USA; 74104). In all, 10 μg RNA was reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen; 18064) according to the manufacturer's protocol, except for using a mixture of random nonamers (GeneLink, Hawthorne, NY, USA; 26-4000-06) and oligo(dT) primers (Invitrogen; 18418012) at 10 μℳ/1 μℳ final concentration. Real-time PCR was performed with FastStartSYBR Green Master (Roche, Indianapolis, IN, USA; 04673492001) on Agilent MxPro3005 system (Agilent Technologies, Santa Clara, CA, USA), using 50 ng cDNA in a 15 μl reaction mixture. Results were normalized to 18S. All reactions were performed in duplicate with primers described in Supplementary Table 3.
Whole-transcript microarray data analysis
Total RNA was isolated with an RNAeasy kit (Qiagen) and amplified, labeled and hybridized to 20 Affymetrix Human Gene 1.0 ST Arrays using GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix, Santa Clara, CA, USA). Raw CEL file data from Affymetrix GeneChip Operating Software were background corrected, normalized and summarized using both gcrma and plier functions in parallel using the Affymetrix power tools package (http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx). The power tools package was used to estimate a P-value for expression of gene in each array according to the Detection Above Background (DABG) method. The data were imported into an R Bioconductor session. Genes that were not expressed at DABG P<10−8 in all replicates of a set were discarded. This analysis detected 21 818 expressed genes. Linear models of differential expression were fit to each of the transformed data sets using the R Bioconductor limma package, and contrasts were estimated between each group of p53-regulated samples and its corresponding set of controls. The genes were tested for significant differential expression between each pair of groups at a level of P≤0.05 after multiple testing correction according to Benjamini–Hochberg procedure. Genes significantly differentially expressed in both plier and gcrma sets were taken to be the set of all-changers and used for downstream analysis. We used the Cluster 3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) to organize sets of differentially expressed genes into hierarchical clusters and then visualized these using the Java Treeview software (http://jtreeview.sourceforge.net/). Pathway analysis was performed using Comodus algorithm (A George, unpublished). To derive hypergeometric test P-values, we modeled each population (e.g. up- or downregulated) as the set of genes present in that experimental condition across the set of all expressed genes.
Accession numbers
Microarray data from this publication have been submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE30753.
Chromatin immunoprecipitation
Average of 5 × 106 cells were collected by centrifugation, crosslinked for 10 min at room temperature with 1% formaldehyde in PBS and neutralized by the addition of 125 mℳ glycine for 5 min. Cells were washed twice in PBS and lysed with 0.6 ml SDS lysis buffer (1% SDS, 10 mℳ EDTA (pH 8.0), 50 mℳ Tris (pH 8.0)) supplemented with protease inhibitors (Roche; 04693124001). Samples were sonicated to generate an average of 500 bp DNA fragments using Bioruptor (Diagenode, Denville, NJ, USA) on high setting (30 s on and off). Insoluble material was removed by centrifugation. A total of 200 μg of cleared samples were diluted 10-fold in ChIP dilution buffer (16.7 mℳ Tris (pH 8.0), 1.2 mℳ EDTA (pH 8.0), 167 mℳ NaCl, 1.1% Triton X-100) and incubated with 10 μg of p53 (DO-1) monoclonal antibody (Santa Cruz; sc-126) or 10 μg of normal IgG (Santa Cruz; sc-2025) overnight at 4 °C. In all, 20 μg of samples without primary antibody were saved as 10% input controls. Immunoprecipitated complexes were captured by 2 h incubation at 4 °C with Dynabeads Protein A (Invitrogen; 100.02D), followed by 5 min washes: once with low salt wash buffer (150 mℳ NaCl, 50 mℳ Tris (pH 8.0), 5 mℳ EDTA (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), once with high salt wash buffer (500 mℳ NaCl, 50 mℳ Tris (pH 8.0), 5 mℳ EDTA (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), four times with LiCl wash buffer (500 mℳ LiCl, 50 mℳ Tris (pH 8.5), 1% NP-40, 0.5% sodium deoxycholate) and twice with TE buffer (100 mℳ Tris (pH 8.0), 100 mℳ EDTA (pH 8.0)). Immunoprecipitated complexes and 10% input controls were eluted in two sequential 10 min incubations with 75 μl fresh elution buffer (50 mℳ Tris (pH 8.0), 10 mℳ EDTA (pH 8.0), 1% SDS) at 65 °C. DNA-protein crosslinks were reversed by the addition of 200 mℳ NaCl and incubation at 65 °C overnight. DNA was purified with QIAquick PCR Purification Kit (Qiagen; 28104), resuspended in 50 μl H2O and quantified by real-time PCR with FastStartSYBR Green Master (Roche; 04673492001) on Stratagene MxPro3005 system, using 2 μl DNA in a 15 μl reaction mixture. All reactions were performed in duplicate with primers described in Supplementary Table 3. Promoter occupancy was calculated with the ‘percent input method', using the formula: % of input=100 × 2^[Ctadjusted input−Ctenriched], where input Ct is adjusted from 10 to 100% by: Ctadjusted input=Ct10% input−log2 10.
Lentiviral constructs
All lentiviral vectors were generated using standard cloning procedures. To construct NSBI-CMVp-tetR lentiviral expression vector, a cassette containing puromycin selection marker in NSPI-CMVp-MCS49 was replaced with a cassette containing blasticidin selection marker (excised from pBabe-bla). tetR sequence was then excised from pcDNA6/TR vector (Invitrogen) and inserted into the vector downstream of CMVp. To engineer NSPI-CMVpTO-p53, we replaced the CMVp in NSPI-CMVp-MCS-Myc-His vector49 with a CMVp containing a tetracycline operator (referred to as CMVpTO) from the pcDNA 4/TO vector (Invitrogen). p53 gene (Arg72 variant) was PCR amplified with a forward primer containing a BamHI restriction enzyme site and a reverse primer containing three stop codons, followed by AgeI restriction enzyme site (forward: 5′-ATACTCGGATCCGCCACCATG-3′ reverse: 5′-ATCACCGGTCTAGTTATTCAGTCTGAGTCAGGCCCTTCTGTC-3′) and inserted into the vector between BamHI and AgeI sites. S15A, S46A, K120R and K382R p53 mutants within the NSPI-CMVpTO-p53 vector were generated with QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies; 210518) according to the manufacturer's protocol and primers listed in Supplementary Table 3.
Lentiviral production and transduction
Production of lentiviruses was carried out as described previously,49 with the following modifications. The lentivirus-containing supernatants were cleared by centrifugation at 3000 r.p.m. for 5 min at 4 °C, and in some cases concentrated by centrifugation at 6000 r.p.m. for 6–16 h at 4 °C. For transduction, cells were seeded on a six-well plate (50 000 cells/well) and transduced 24 h later with complete medium containing recombinant lentiviruses in the presence of 8 μg/ml polybrene. Following 24 h of incubation, cells were washed with PBS and left to recover in complete medium. To generate B5/589-p53 mass population, B5/589 cells were first transduced with NSBI-CMVp-tetR lentivirus constitutively expressing tetR and selected with 5 μg/ml blasticidin for 3 days. These cells were then transduced with NSPI-CMVpTO-p53 lentivirus containing a tetracycline-inducible (Tet-On) promoter-driving p53 expression. These cells were allowed to undergo negative selection for 1–2 weeks, which eliminated any cells with incomplete suppression of p53 because of its growth inhibitory effects, and then selected with 2 μg/ml puromycin for 3 days.
Statistics
Statistical analysis was performed using the GraphPad Prism 5.01 Software (La Jolla, CA, USA) using the unpaired, one-tailed Student's t-test. P-values of <0.05 were considered statistically significant.
Acknowledgments
We are grateful to Dr. Bo Zhao for his help with cloning procedures. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine Microscopy Shared Resources Facility, supported with funding from NIH-NCI shared resources (Grant no. 5R24CA095823-04), National Science Foundation Major Research Instrumentation (Grant no. DBI-9724504) and NIH shared instrumentation (Grant no. 1 S10RR0 9145-01). We are also grateful to Dr. Huei-Chi Wen for his help with the confocal imaging analysis. We thank Luis Carvajal and Dr. Pierre-Jacques Hamard from Dr. James J Manfredi's lab for sharing their ChIP assay expertise and providing us with the ChIP MDM2, p21 and PUMA primer sequences, Dr. Greg Khitrov in the Departmental Core Life Sciences Technology Laboratory of the Mount Sinai School of Medicine for performing the hybridization and processing of our Gene 1.1 ST Arrays, and Dr. Sam W Lee for helpful discussion. This study was supported by grants from the Breast Cancer Research Foundation, the Empire State Stem Cell Fund through New York State Department of Health Contract no. C024313 (SAA) and by P01 CA080058 from the National Cancer Institute (SAA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Empire State Stem Cell Board, the New York State Department of Health, the State of New York, the National Cancer Institute or the National Institutes of Health. Supplementary Information is available at Cell Death and Differentiation's website.
Glossary
- ChIP
chromatin immunoprecipitation
- dox
doxycycline
- HMEC
human mammary epithelial cells
- γH2AX
phosphorylated form of H2AX
- PI
propidium iodide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- Millar AW, Lynch KP. Rethinking clinical trials for cytostatic drugs. Nat Rev Cancer. 2003;3:540–545. doi: 10.1038/nrc1124. [DOI] [PubMed] [Google Scholar]
- Jackson JG, Post SM, Lozano G. Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol. 2010;223:127–137. doi: 10.1002/path.2783. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol. 2003;23:8576–8585. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA. Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol. 2001;21:3375–3386. doi: 10.1128/MCB.21.10.3375-3386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JG, Pereira-Smith OM. P53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res. 2006;66:8356–8360. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- Kho PS, Wang Z, Zhuang L, Li Y, Chew JL, Ng HH, et al. P53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J Biol Chem. 2004;279:21183–21192. doi: 10.1074/jbc.M311912200. [DOI] [PubMed] [Google Scholar]
- Hernández-Vargas H, Ballestar E, Carmona-Saez P, von Kobbe C, Bañón-Rodríguez I, et al. Transcriptional profiling of MCF7 breast cancer cells in response to 5-fluorouracil: relationship with cell cycle changes and apoptosis, and identification of novel targets of p53. Int J Cancer. 2006;119:1164–1175. doi: 10.1002/ijc.21938. [DOI] [PubMed] [Google Scholar]
- Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M, et al. Global genechip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinoma cells. Cancer Biol Ther. 2003;2:406–415. doi: 10.4161/cbt.2.4.437. [DOI] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- Xia M, Knezevic D, Vassilev LT. P21 does not protect cancer cells from apoptosis induced by nongenotoxic p53 activation. Oncogene. 2011;30:346–355. doi: 10.1038/onc.2010.413. [DOI] [PubMed] [Google Scholar]
- Stampfer MR, Bartley JC. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci USA. 1985;82:2394–2398. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minemoto Y, Gannon J, Masutani M, Nakagama H, Sasagawa T, Inoue M, et al. Characterization of Adriamycin-Induced G2 Arrest and Its Abrogation by Caffeine in FL-Amnion Cells with or without p53. Experimental Cell Research. 2001;262:37–48. doi: 10.1006/excr.2000.5072. [DOI] [PubMed] [Google Scholar]
- Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S. [gamma]H2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J Biol Chem. 2005;280:857–860. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Mercer WE, Shields MT, Lin D, Appella E, Ullrich SJ. Growth suppression induced by wild-type p53 protein is accompanied by selective down-regulation of proliferating-cell nuclear antigen expression. Proc Natl Acad Sci. 1991;88:1958–1962. doi: 10.1073/pnas.88.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Jiang S-W, Couch FJ. P53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J Biol Chem. 2003;278:15652–15660. doi: 10.1074/jbc.M211297200. [DOI] [PubMed] [Google Scholar]
- Lazaro-Trueba I, Arias C, Silva A. Double bolt regulation of Rad51 by p53: a role for transcriptional repression. Cell Cycle. 2006;5:1062–1065. doi: 10.4161/cc.5.10.2764. [DOI] [PubMed] [Google Scholar]
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Lowe SW. The p53–Bcl-2 connection. Cell Death Differ. 2006;13:1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinkevich VV, Nikulenkov F, Shi Y, Enge M, Bao W, Maljukova A, et al. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell. 2009;15:441–453. doi: 10.1016/j.ccr.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Pujals A, Renouf B, Robert A, Chelouah S, Hollville E, Wiels J. Treatment with a BH3 mimetic overcomes the resistance of latency III EBV (+) cells to p53-mediated apoptosis. Cell Death Dis. 2011;2:e184. doi: 10.1038/cddis.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Wang G, Nikolovska-Coleska Z, Yang CY, Wang R, Tang G, Guo J, et al. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J Med Chem. 2006;49:6139–6142. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- Spencer Sabrina L, Sorger Peter K. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, et al. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol. 2004;166:839–851. doi: 10.1083/jcb.200404158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. P53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. P53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acutemyelogenous leukemia. Blood. 2010;116:71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiri G, Cherian MM, Vijayakumar S, Liu G, Bafico A, Aaronson SA. Wnt pathway aberrations including autocrine Wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene. 2009;28:2163–2172. doi: 10.1038/onc.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.