Abstract
Despite the prominent pro-apoptotic role of p53, this protein has also been shown to promote cell survival in response to metabolic stress. However, the specific mechanism by which p53 protects cells from metabolic stress-induced death is unknown. Earlier we reported that carnitine palmitoyltransferase 1C (CPT1C), a brain-specific member of a family of mitochondria-associated enzymes that have a central role in fatty acid metabolism promotes cell survival and tumor growth. Unlike other members of the CPT family, the subcellular localization of CPT1C and its cellular function remains elusive. Here, we report that CPT1C is a novel p53-target gene with a bona fide p53-responsive element within the first intron. CPT1C is upregulated in vitro and in vivo in a p53-dependent manner. Interestingly, expression of CPT1C is induced by metabolic stress factors such as hypoxia and glucose deprivation in a p53 and AMP activated kinase-dependent manner. Furthermore, in a murine tumor model, depletion of Cpt1c leads to delayed tumor development and a striking increase in survival. Taken together, our results indicate that p53 protects cells from metabolic stress via induction of CPT1C and that CPT1C may have a crucial role in carcinogenesis. CPT1C may therefore represent an exciting new therapeutic target for the treatment of hypoxic and otherwise treatment-resistant tumors.
Keywords: carnitine palmitoyltransferase, carcinogenesis, p53, metabolism, AMPK
Hypoxia is an important chronic stress on tumor cell growth and has been shown to correlate with poor disease-free and reduced overall survival in a variety of carcinomas and sarcomas.1 To enhance survival in an altered environment such as hypoxia cancer cells undergo a so-called metabolic transformation.2, 3, 4 The best-known aspect of metabolic transformation is the Warburg effect, whereby cancer cells upregulate glycolysis to limit their energy consumption. However, there is increasing evidence that not only glucose metabolism, but also fatty acid oxidation (FAO) is involved in metabolic transformation. Although glucose seems to be the major energy source for tumor growth and survival, there is increasing evidence that alternative energy sources such as fatty acid metabolism are altered in cancer cells, even under hypoxic conditions. Indeed, fatty acid synthase has been found to be upregulated in many human cancers,5 and inhibitors of the fatty acid synthase show antitumor activity.6
As recently published, we identified carnitine palmitoyltransferase (CPT) 1C (CPT1C) as a potential novel p53-target gene.7 By their restriction of fatty acid import into mitochondria,4 the CPT 1 (CPT1) family of enzymes represent key regulatory factors of FAO. There are three tissue-specific isoforms of CPT1: CPT1A that is found in liver, CPT1B in muscle and CPT1C in brain and testes. Loss-of-function of CPT1C was generated in mouse embryonic stem cells (Cpt1cgt/gt ES cells). Importantly, Cpt1cgt/gt ES cells readily succumbed to cell death under hypoxic conditions, whereas control cells were resistant. ES cells deficient for CPT1C showed a spontaneous induction in cell death through the mitochondrial apoptosis pathway. Using transient knock-down models for Cpt1c, we reported that Cpt1c promotes tumor growth in response to metabolic stress.7 These results suggest that cells can use a novel mechanism involving CPT1C to protect against metabolic stress.
Cpt1c-deficient mice show a complex metabolic phenotype characterized by decreased food intake and lower body weight when fed a normal diet. However, the mice show a higher tendency to obesity on a high-fat diet when compared with wild-type mice.8, 9 Conversely, using a transgenic mouse model, Cpt1c gain-of-function results in postnatal microcephaly and when fed a high-fat diet, these mice are protected from weight gain.10 These data suggest a role for CPT1C in feeding behavior or metabolic sensing in the brain. Metabolic stress stimulates the activation of intracellular sensors, which mediate cellular adaptation in order to evade apoptosis. The tumor suppressor gene p53 is a well-studied transcriptional factor that is activated and stabilized by many cellular insults such as DNA damage, hypoxia, starvation and oncogenic activation. The AMP activated kinase (AMPK) is a cellular energy sensor activated by conditions of metabolic stress characterized by an increase in the intracellular AMP/ATP ratio.11 AMPK is now known to be activated by multiple factors, including AMP and ADP, as well as many nucleotide independent factors acting through upstream kinases.12 Starvation or low energy levels initiate activation of AMPK, which results in the induction of p53.13 Depending on the intensity and duration of the stress, p53 activation either leads to cell-cycle arrest, ROS clearance or survival signals or induces apoptosis and cell death signals. There is increasing evidence that p53, which has a key role in determining apoptotic cell fate, is involved in metabolic reprogramming, one of the key alterations in tumorigenesis.
Recent studies showed that CPT1C is localized both in the endoplasmic reticulum (ER) and mitochondria, but predominantly in ER.14 However, the exact subcellular localization of CPT1C and its cellular function remains unclear. Here, we show that CPT1C is upregulated in vitro and in vivo in a p53-dependent manner. We also demonstrate that CPT1C expression is induced by metabolic stress in an AMPK- and p53-dependent manner. Furthermore, we show that CPT1C can protect cells from cell death induced by hypoxia. Interestingly, CPT1C depletion increases survival and suppresses tumor development in the Nf1+/− : p53+/− tumor model. Analysis of these tumors confirms an activated AMPK/p53 signaling pathway. In addition, depletion of CPT1C leads to decreased proliferation. Our findings have implications for the cell survival effects of p53 under conditions of metabolic stress and might have a key role in carcinogenesis. Understanding the roles of CPT1C as a key downstream target in the AMPK/p53 pathway via regulation of metabolism may provide interesting potential targets for the development of new cancer therapies.
Results
CPT1C is a p53-target gene
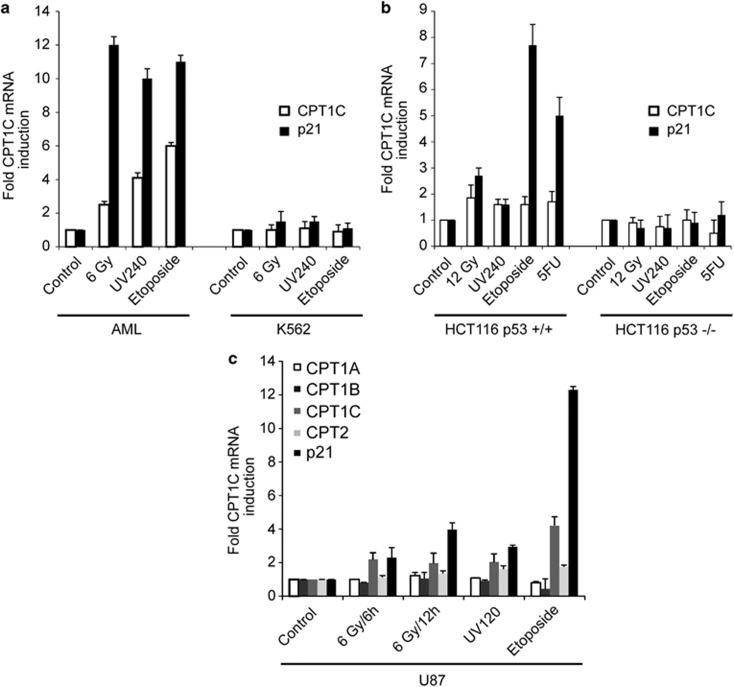
We previously reported on a cDNA microarray screen designed to identify novel p53 transcription targets and identified CPT1C as a potential novel p53-target.7 This screen employed Friend virus-transformed mouse erythroleukemia cells that lack endogenous p53 and express a temperature-sensitive form of p53 (DP16.1/p53ts cells). At the permissive temperature of 37 °C, mutated p53 protein is inactive and cells continue to proliferate. At the restrictive temperature of 32 °C, mutated p53 becomes active and cells are induced to undergo apoptosis. To identify genes differentially regulated upon p53 activation, we compared microarray mRNA expression patterns of DP16.1/p53ts cells cultured at 37 °C or 32 °C. Additionally, we recently published that upon p53 activation in DP16.1/p53ts cells, mRNA for EST AA050178.1, which represents a partial cDNA for Cpt1c, was increased 1.9-fold and 2.8-fold after 3 and 6 h at 32 °C, respectively.7 There were no significant changes in CPT1C mRNA in the parental DP16.1 cells (−1 and 1-fold change at 3 and 6 h after temperature shift, respectively (data not shown). We next examined whether CPT1C regulation was truly an effect of p53 activation. Real-time RT-PCR in a variety of cancer cell lines revealed that CPT1C is upregulated in multiple cell lines in a p53-dependent manner in response to several different stress stimuli such as ionizing radiation (6 or 12 Gray), ultraviolet (UV) radiation, etoposide and 5-fluorouracil (5-FU) (Figures 1a and b). Moreover, in U87 cells, CPT1C was the only CPT family member regulated by p53 (Figure 1c). Similar results were obtained using A549 and other human cancer cell lines (data not shown). Owing to the fact that the current available commercial antibodies were unable to detect endogenous level of human CPT1C in the tested cells, we could not confirm these results at the protein level.
Figure 1.
Induction of endogenous CPT1C by stress stimuli in a p53-dependent manner. (a and b) AML (p53 WT), K562 (p53 mutant) and p53 WT and mutant HCT116 cells were subjected to different stress stimuli known to activate p53 and RT-PCR was performed using SYBR Green to detect CPT1C mRNA. p21 served as a positive control for p53 activation. All values shown were normalized to GAPDH expression. Results shown are one trial representative of at least three experiments. (c) The U87 cell line was treated with the indicated DNA-damaging stimuli and real-time RT-PCR was performed to detect upregulation of expression of the indicated CPT family members. 6 Gy, 6 Gray of irradiation; UV240, 240 μJ/cm2 of UV; 5-FU, 50 μg/ml of Fluorouracil; Etoposide, 10 μM of Etoposide. p21 was used as positive control. All values shown were normalized to GAPDH and the response level was calculated relative to the untreated control. Results shown are one trial representative of at least three experiments
p53 directly activates CPT1C transcription
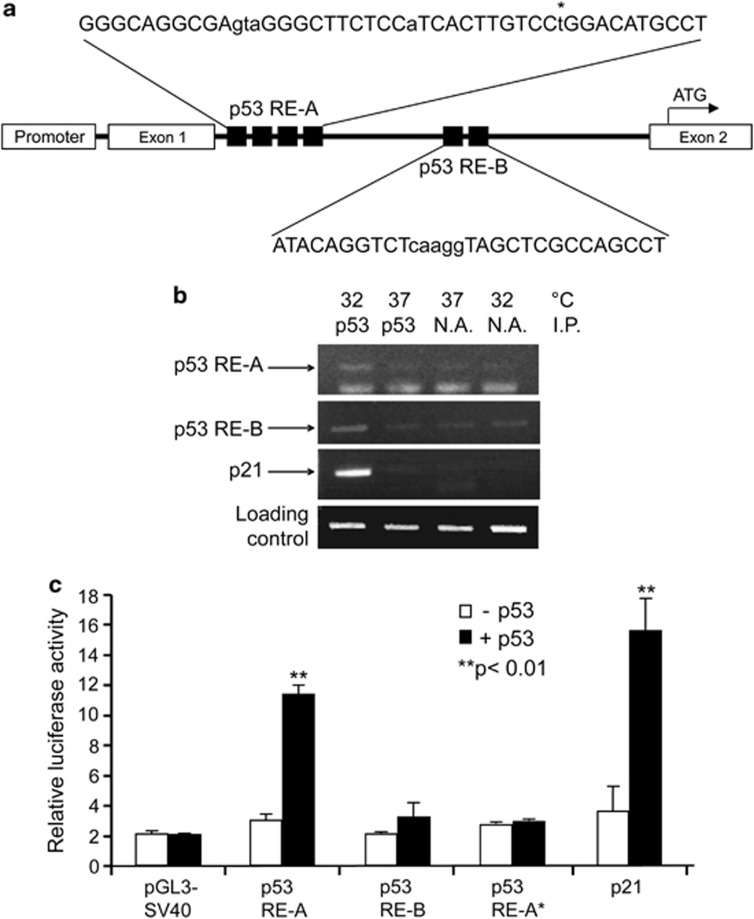
We analyzed the promoter of the murine CPT1C gene15 and identified two putative p53-responsive elements (p53-RE)16 in the first intron that were 330 bp apart: p53-RE-A, +174–219; p53-RE-B, +504–533 (Figure 2a). To investigate whether p53 could bind directly to either of these sites, we performed ChIP analyses on DP16.1/p53ts cells grown at 37 °C or 32 °C for 8 h. Using immunoprecipitation with anti-p53 antibody and PCR with primers specific for the two potential p53-binding sites, we observed a specific amplification of p53-RE-A at 32 °C under which p53 was activated (Figure 2b). It should be pointed out that the non-specific bands were also observed for p53-RE-B at all conditions. The proximity of p53-RE-A and p53-RE-B (330 bp) and the use of sonication, which allows analysis of 700 bp fragments make it difficult to clearly separate p53 binding. To further determine the specificity of these two binding sites, we cloned the p53-RE-A and p53-RE-B sequences into separate luciferase reporter vectors to test transcriptional activity. These constructs were co-transfected into p53−/− mouse embryonic fibroblasts (MEFs) with either WT p53 or p53 bearing a mutation in its DNA-binding domain. Only the luciferase vector containing p53-RE-A and not p53-RE-B showed increased luciferase activity in the presence of WT p53 (Figure 2c). This p53-dependent luciferase activity was blocked by a point mutation at position 42 (G->T) of p53-RE-A (p53-RE-A*) (Figure 2c). Co-transfection of p53-RE-A with the DNA-binding domain p53 mutant showed no increase in luciferase activity (data not shown), demonstrating that luciferase activation associated with p53-RE-A is dependent on the DNA-binding activity of p53. Taken together, these data suggest that the p53-consensus motif p53-RE-A is both sufficient and necessary to drive the p53-dependent transcription of CPT1C.
Figure 2.
CPT1C is a p53-target gene. (a) p53-binding sites. Computational analysis revealed two putative p53-RE, p53-RE-A and p53-RE-B, located in intron 1 in the CPT1C promoter region as indicated. (b) p53 binding to p53-RE-A. ChIP analysis was performed on DP16.1/p53ts cells cultured at 37 or 32 °C. The p53-RE of p21 was used as positive control, unprecipitated genomic DNA was the loading control. NA: no p53 antibody. Results shown are one result representative of three trials. (c) p53-RE-A binds to p53 and activates transcription. The indicated luciferase reporter constructs were transfected into E1A/ras-transformed p53−/−MEFS, with or without co-transfection of WT p53. Relative luciferase activity was taken as the relative transcriptional activity. pGL3-SV40, vehicle control; p53-RE-A*, mutated p53-RE-A (G→T at position (42); p21 was used as positive control for p53 transcription
p53 upregulates CPT1C in vivo
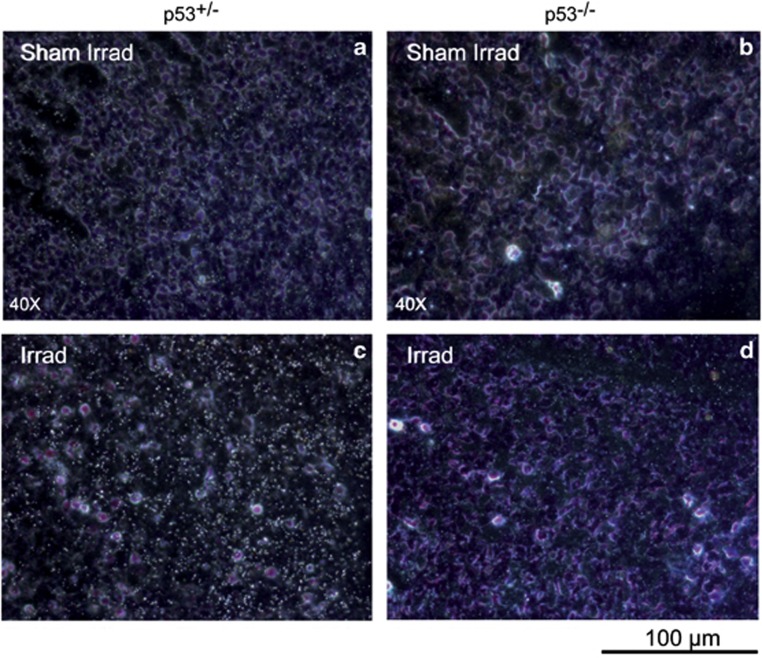
To determine whether CPT1C could be upregulated in response to p53 activation in vivo, we performed in situ hybridization to detect CPT1C mRNA in irradiated mouse embryos. At day 12.5 post coitum, embryos of C57BL/6 p53+/− and p53−/− mice were subjected in utero to 5 Gy irradiation. At 8 h post-irradiation, various tissues were excised and fixed for detection of CPT1C mRNA by in situ hybridization. Consistent with previous reports, the highest base levels of CPT1C mRNA were detected in neuronal tissues of non-irradiated embryos (Figure 3, midbrain). Irradiated p53+/− embryos showed a strong upregulation of CPT1C mRNA in most tissues examined, including the midbrain (Figure 3c) and heart (data not shown). This CPT1C upregulation was not detected in irradiated p53−/− embryonic midbrain (Figure 3d). These data indicate that CPT1C expression can be transcriptionally activated by p53 in vivo in response to DNA-damaging stimuli.
Figure 3.
p53 upregulates CPT1C in vivo. E12.5 C57BL/6 embryos from p53+/− and p53−/− mice were subjected to 5 Gy ionizing radiation (Irrad) in utero. Embryos were harvested and prepared for in situ hybridization at 8 h post irradiation. Incubation of midbrain sections with a CPT1C riboprobe showed that CPT1C mRNA was upregulated in irradiated p53+/− cells (c) but not in irradiated p53−/− cells (d) compared with sham-irradiated controls (a and b)
CPT1C expression is induced by hypoxia and glucose deprivation in a p53-dependent manner
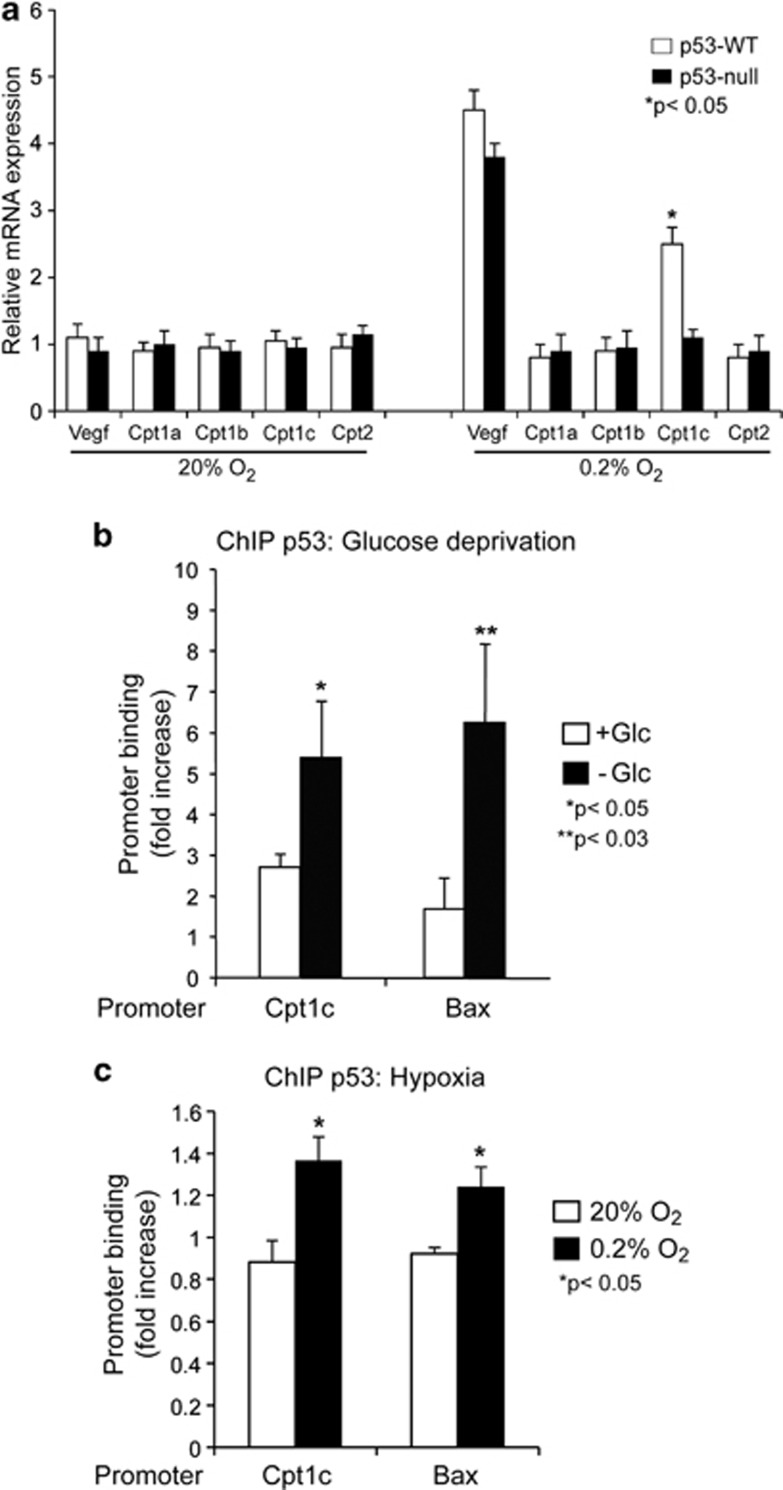
Key targets of p53 under hypoxic conditions are of special interest, as p53 is mutated in over 50% of all solid tumors. Rapidly growing cancer is often associated with hypoxia. Therefore, we investigated whether CPT1C is upregulated in response to hypoxia. We exposed transformed wild-type and mutant p53 MEFs to 0.2% O2 over 7 h and measured the expression levels of all CPT family members by real-time PCR. As shown in Figure 4a, CPT1C is the only family member upregulated in response to hypoxia in a p53-dependent manner.
Figure 4.
Cpt1c is induced by hypoxia and glucose deprivation in a p53-dependent manner. (a) Mouse embryo fibroblasts from p53+/+ and p53−/− mouse embryos at day 13.5 were treated with either normoxia (20% O2) or hypoxia (0.2% O2) for 7 h, then the cells were harvested for total RNA isolation. The CPT1A, B, C and CPT2 mRNA levels were measured using real-time PCR. RNA levels were normalized to GAPDH. VEGF was used as positive control. Using ChIP analysis and PCR with primers specific for the p53-binding sites of Cpt1c, amplification of p53-RE was tested in p53 WT MEFs treated with either glucose deprivation (b) or hypoxia (c). Bax was used as positive control
We reported earlier that CPT1C depletion confers sensitivity to metabolic stress including hypoxia and glucose withdrawal.7 To investigate whether p53 directly binds to the verified p53-RE of Cpt1c upon hypoxia or glucose withdrawal, we performed ChIP analyses on p53 wild-type MEFs treated with either glucose-free DMEM (Figure 4b) or low oxygen (Figure 4c) using anti-p53 antibodies. Using immunoprecipitation with anti-p53 antibody and PCR with primers specific for the p53-binding sites of Cpt1c, we observed a strong amplification of the Cpt1c p53-RE (p53-RE-A) under hypoxia and glucose withdrawal conditions (Figures 4b and c). Primers specific for the p53-RE in the Bax gene were used as a positive control for p53 activation.
CPT1C is induced by energetic stress in an AMPK- and p53-dependent manner
AMPK activation leads to increased catabolic metabolism, which can activate a p53-dependent cell-cycle checkpoint.13 Our observation that glucose withdrawal induces Cpt1c expression and regulates cell proliferation suggests that Cpt1c may be a target of AMPK, the major sensor of cellular energy levels.11 We treated control or AMPKα-deficient MEFs (AMPKα1−/− : α2fl/fl MEFs expressing Cre recombinase) with 1 mM Metformin and found that increased endogenous CPT1C levels parallel increased phosphorylation of AMPKα and its downstream targets (acetyl-CoA carboxylase (ACC) and p53) (Figure 5a). These results indicate that CPT1C is upregulated by Metformin in an AMPK-dependent manner. To verify whether upregulation is dependent on p53, we examined by western blotting the endogenous level of CPT1C in 3T3 MEFs wild-type and p53-deficient cells. Western blotting analysis revealed that CPT1C protein levels in p53−/− cells was much lower than that in p53+/+ cells (Figure 5b, left). The CPT1C level was significantly elevated after Metformin treatment, where AMPK was activated in p53+/+ cells, but not in p53−/− cells. (Figure 5b, right). These data strongly support the observation that CPT1C is upregulated in a p53 and AMPK-dependent manner. We previously showed that CPT1C depletion in mouse ES cells leads to activation of the intrinsic mitochondrial apoptosis.7 Moreover, AMPK-deficient cells display increased sensitivity to apoptosis induced by metabolic stress.17 Interestingly, the ectopic expression of CPT1C protects AMPK-deficient cells from apoptosis induced by the metabolic stressor 2-deoxyglucose (Figures 5c and d). These data suggest that CPT1C is a key downstream mediator of AMPK signaling, important for mediating cell survival in response to metabolic stress.
Figure 5.
Cpt1c is induced by energetic stress in an AMPK and p53 dependent manner. (a) AMPKα1−/−, α2fl/fl MEFs +/−Cre cells were treated for 16 h in 1 mM Metformin. Cells were lysed with CHAPS buffer, and immunoblotted with antibodies indicated. (b) p53+/+ and p53−/− 3T3 MEF cells were treated for 16 h in 1 mM Metformin, Cells were lysed with CHAPS buffer and immunoblotted with antibodies indicated. (c) Cell lines stably expressing CPT1C protein were generated in control (Cre−) and AMPKα-deficient (Cre+) MEFs. Clones of each cell type were immunoblotted with antibodies indicated. (d) A PI-exclusion apoptosis assay was performed with AMPKα1−/−, α2fl/fl MEFs (Cre−, open bar; Cre+, closed bar) expressing either Flag-CPT1C (+) or control vector (−) MEFs were treated with 2-deoxyglucose. Data expressed in Mean±S.E.M.
CPT1C depletion increases survival and suppresses tumor development in the Nf1+/− : p53+/− tumor model
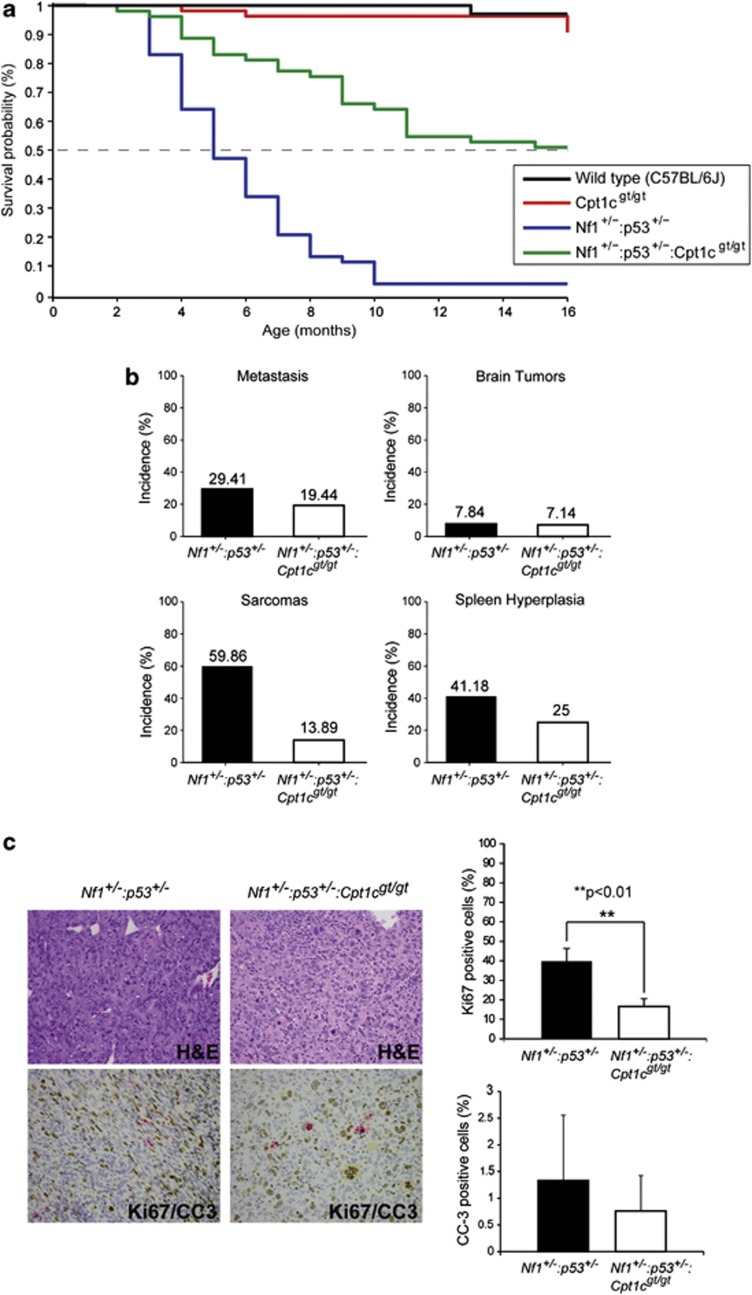
Based on our results that show upregulation of Cpt1c after activation of the tumor suppressor p53, we hypothesize that CPT1C has a crucial role in carcinogenesis by protecting tumor cells from hypoxic and metabolic stresses. To verify this hypothesis, we used the neurofibromatosis type I tumor model.18, 19 Nf1+/− : p53+/− mice, which are prone to develop soft tissue sarcomas, were crossed with Cpt1cgt/gt mice to generate Nf1+/− : p53+/− : Cpt1cgt/gt mice in a C57BL/6 background. Survival as well as tumor incidence was compared with C57BL/6, Cpt1cgt/gt and Nf1+/− : p53+/− mice (Figure 6a). Similar to previous reports,19, 20 we observed that Nf1+/− : p53+/− mice developed soft tissue sarcomas of the limbs and abdomen as well as lymphomas at around 3–6 months of age and with a penetrance of over 70%. CPT1C depletion in this murine tumor model highly increases the median survival time from 5–15 months (P<0.0001). Sarcomas developed in 59.9% of Nf1+/− : p53+/− mice, and metastases in 29.4%. In contrast, depletion of Cpt1c in Nf1+/− : p53+/− mice significantly decreased the incidence of sarcomas and metastases to 13.89% and 19.44%, respectively, (Figure 6b). Similarly, splenic hyperplasia was also significantly less in Nf1+/− : p53+/− : Cpt1cgt/gt mice (25%) compared with that in Nf1+/− : p53+/− mice (41.2%). In our cohort, there was no obvious difference in the onset of brain tumors. The reason why our Nf1+/− : p53+/− mice developed fewer brain tumors than previously reported is probably owing to the different C57BL/6 background.18
Figure 6.
Cpt1c depletion in the murine neurofibromatosis type I tumor model significantly increases their survival rate and suppresses tumor transformation and metastasis. (a) Kaplan–Meier survival curve. The survival of the Nf1+/− : p53+/− : Cpt1cgt/gt mice (green, n=53) was compared with the Nf1+/− : p53+/−mice (blue, n=53), the Cpt1cgt/gt mice (red, n=54) and a wild-type control (C57BL/6 strain, black, n=33). The survival of the mice was plotted on a Kaplan–Meier curve for individual genotypes against the animal age in months. (b) Nf1+/− : p53+/− : Cpt1cgt/gt mice showed less metastasis cases and fewer cases of spleen hyperplasia and sarcomas. There is no variation in the brain tumor cases when compared with Nf1+/− : p53+/− mice. (c) Histological analysis of tumor phenotypes in Nf1+/− : p53+/− and Nf1+/− : p53+/− : Cpt1cgt/gt mice. Nf1+/− : p53+/− (n=6) and Nf1+/− : p53+/− : Cpt1cgt/gt (n=4) sarcomas were analyzed using immunohistochemistry staining as indicated. Paraffin sections were stained with Ki67 (brown) and cleaved caspase-3 (CC-3, red) to analyze the proliferation and the apoptotic rate respectively. Data expressed is Mean±S.D.
Histological analysis reveals less proliferation in tumors from Nf1+/− : p53+/− : Cpt1cgt/gt mice
It has been shown that hypoxia in solid tumors is associated with rapid disease progression and poor outcome. As CPT1C protects tumor cells from apoptotic cell death, we hypothesize that CPT1C depletion in our murine tumor model reduces markers of tumor aggressiveness such as proliferation and apoptosis. In order to analyze the proliferative and apoptotic rate in Nf1+/− : p53+/− and Nf1+/− : p53+/− : Cpt1cgt/gt tumors, we performed immunohistochemistry with Ki67 and cleaved caspase-3 in paraffin-embedded tumor samples. As shown in Figure 6c, positive staining for Ki67 was significantly reduced in Nf1+/− : p53+/− : Cpt1cgt/gt tumor samples compared with Nf1+/− : p53+/−. There is also an apparent tendency towards reduced cleaved caspase-3-positive staining in Nf1+/− : p53+/− : Cpt1cgt/gt tumors when compared with Nf1+/− : p53+/−, though no significant difference was observed between those two groups. Taken together, Cpt1c depletion significantly decreases the incidence of sarcomas and metastases in the murine neurofibromatosis type I tumor model Nf1+/− : p53+/−, which might be caused by the cumulative effects of altered metabolism, an increase in sensitivity to hypoxia, and downregulation of proliferation.
Cpt1c is overexpressed in Nf1+/− : p53+/− sarcomas
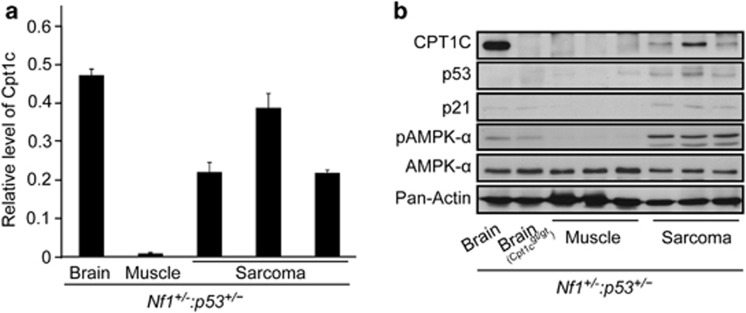
Previous studies19, 21 performed with the murine Nf1+/− :p53+/− tumor model revealed that the majority of sarcomas are malignant peripheral nerve sheath tumors and malignant triton tumors that arise within peripheral nerves. We next investigated whether Cpt1c is overexpressed in the tumors extracted from Nf1+/− : p53+/− mice. We performed real-time PCR and western blotting to analyze the expression level of CPT1C in normal muscle tissue and tumor samples from Nf1+/− : p53+/− mice that developed soft tissue sarcomas (Figure 7). Even though CPT1C is mainly expressed in brain,9, 14, 22, 23 we found high expression levels of Cpt1c in sarcomas extracted from Nf1+/− : p53+/− mice. In contrast, no Cpt1c expression was found in normal muscle tissues by performing real-time PCR and immunoblotting experiments. Motivated by our in vitro results that Cpt1c is regulated by p53 and AMPK, we next analyzed whether increased Cpt1c expression in tumors correlates with p53 and AMPK activation. p53 and p21 protein levels, and AMPK phosphorylation at T172 were highly upregulated in sarcomas, which showed elevated expression of Cpt1c (Figure 7b). It is worth noting that we observed p53 expression in all Nf1+/− : p53+/− sarcomas tested by western blotting (Figure 7b) and by PCR (data not shown) indicating that the Nf1+/− : p53+/− tumors did not undergo loss of heterozygosity of p53. These results imply that Cpt1c may give a p53-dependent growth advantage in tumors.
Figure 7.
CPT1C is upregulated in tumor tissues. Sarcomas and normal muscle tissues were isolated from Nf1+/− : p53+/−mice. RT-PCR analysis (a) and western blot (b) showed that Cpt1c is upregulated in sarcomas. Data represents at least three independent experiments. The brain tissues from Nf1+/− : p53+/− and Nf1+/− :p53+/− : Cpt1c−/− mice were used as Cpt1c positive and negative controls as indicated
Discussion
Here, we have shown that CPT1C is a bona fide p53 target and has a crucial role in sensitizing tumor cells to hypoxia and glucose withdrawal. In a murine tumor model, depletion of Cpt1c significantly reduces tumor development and increases survival of tumor bearing mice. These results suggest that CPT1C may act as an oncogene to promote cell survival in response to metabolic stress.
p53 is a tumor suppressor activated in response to a variety of cellular stresses.24, 25 Up to now, activation of p53 by hypoxia26, 27 has been commonly considered a death-inducing strategy of the cell because of its pro-apoptotic role in cancer cells. Alternatively, under acute cellular stress p53 is known to signal DNA repair, cell-cycle arrest or senescence to maintain the viability of the cell.28 Intriguingly, there is increasing evidence that p53 can promote cell survival by activating pathways of metabolic adaptation that seem to be crucial for successful cancer progression.29 Matoba et al.30 reported that p53 directly stimulates oxidative phosphorylation by activating the synthesis of cytochrome c oxidase 2 (SCO2). Interestingly, disrupting SCO2 in cancer cells with wild-type p53 leads to glycolytic metabolism in p53-deficient tumor cells. In addition, Bensaad et al.31 demonstrated that expression of TIGAR (TP53-induced glycolysis and apoptosis regulator) attenuates glycolysis. The ability of p53 to suppress glycolysis and to promote oxidative phosphorylation might help to prevent the unrestrained glycolytic flux that is associated with malignant cell growth, which represents another manifestation of the tumor suppressive activity of p53.32
Although alterations in glucose metabolism seem to represent a major source for metabolic transformation in cancer cells, there is increasing evidence that fatty acid metabolism has a crucial role. Fatty acid synthesis (FAS) is an energy-depleting process required for cell growth and proliferation, while FAO is an oxygen-dependent catabolic process that occurs in the lumen of mitochondria. Cytokines cause cells to activate FAS and concurrently reduce FAO.33 In conditions of ATP depletion, FAS is turned off in favor of FAO by AMPK-dependent inactivation of ACC.34 In light of energy and oxygen use implications, it is likely that the hypoxic response tightly regulates the balance between FAS and FAO. Thus far, the mechanism by which cells regulate a potential switch between FAS and FAO under hypoxic conditions has not been suggested. To date, no correlation has been found between CPT family members, which are key regulators of FAO, and metabolic adaption in tumor cells.
The CPT1 family (see review Bonnefont et al.35) consists of three members encoded by separate genes that appear to be expressed in a tissue-specific manner: CPT1A (liver isoform), CPT1B (muscle isoform) and CPT1C (brain isoform). CPT1A and CPT1B function to translocate free fatty acids to the lumen of mitochondria, where they can be degraded by beta-oxidation as source of energy. Although CPT1C appears to represent a more distant family member by homology,22 database searches suggest that CPT1C arose from a relatively recent gene duplication event.9 Indeed, unlike other CPT1 family members, two separate biochemical studies have failed to show palmitoyltransferase activity for CPT1C.9, 22 CPT1C has been recently demonstrated to be expressed in pyramidal neurons of hippocampus and is located in the ER. Sierra et al.14 demonstrated that CPT1C possessed CPT activity, while Carrasco et al.23 indicated that CPT1C may regulate ceramide levels in neurons. Nevertheless, it has been reported that CPT1C conserves the affinity for Malonyl-CoA, which inhibits all the CPT1 family members.36 However, it is still not clear whether the binding of Malonyl-CoA to CPT1C causes a decrease in food intake and weight loss. Current experiments in our laboratory are directed at elucidating the exact subcellular localization and molecular mechanism of CPT1C to better understand the functions of this protein.
The present study demonstrates that transcription of the mouse CPT1C gene emanates from a putative p53-RE in the first intron that is sufficient and necessary to drive the p53-dependent transcription of CPT1C. We were able to detect an increase of the levels of CPT1C mRNA in human cancer cells cultured under DNA damage conditions known to activate p53 (Figure 1). Interestingly, we found that CPT1C, but not p21, was induced by Staurosporin and UV240 stimuli in MCF7 cells (data not shown). This observation indicates that CPT1C is also regulated by certain unknown mechanisms apart from the p53 pathway. Unfortunately, it was not possible to confirm the increased CPT1C expression at the protein level owing to lack of a good quality antibody to detect endogenous levels of human CPT1C protein.
AMPK activation stimulates a number of biological pathways in order to conserve cellular energy. AMPK achieves this by two main mechanisms: (1) by limiting cellular energy usage through the inhibition of anabolic pathways such as mTOR-dependent mRNA translation or ACC-mediated FAS (2) or by activating pathways of catabolic metabolism to generate ATP. Recent results suggest that AMPK-dependent inhibition of anabolic growth has a key role mediating cell survival under nutrient limitation. Abrogating lipid synthesis by inhibiting ACC activity rescues AMPK-deficient cells from glucose deprivation.37 The data presented here suggest that AMPK- and p53-dependent activation of lipid catabolism via Cpt1c has an important role in this process as well. We show here that CPT1C protein levels accumulate in cells under energy stress conditions and its expression is dependent on AMPKα and p53 (Figure 5). Moreover, ectopic expression of Cpt1c is sufficient to rescue AMPKα-deficient cells from apoptosis induced by glycolytic inhibition. Together our data indicate that Cpt1c, the newest member of the CPT family, is a downstream target of the AMPK/p53 pathway, and provides a direct link between AMPK, p53 signaling and metabolic adaption in tumor cells.
The current literature together with our results suggests that CPT1C may have a unique function in the tumor milieu. Current targeting strategies against cancer mainly focus on specifically blocking molecular signals, which promote cell proliferation, hinder cell death, modulate the immune response or enhance neoangiogenesis. However, most of these signaling pathways are either redundant or essential in healthy tissue. A further strategy is to target the altered metabolism of cancer cells. The metabolic transformation that occurs in cancer cells and in response to hypoxia seems to represent an intrinsic part of carcinogenesis and might be altered by modulating CPT1C. Evidence that hypoxia-resistant tumors are highly aggressive and have a worse prognosis underscores that overcoming hypoxia is a major hurdle for viability in the tumor microenvironment.1
Material and Methods
cDNA microarray screen
Cell lines
DP16.1 and DP16.1/p53ts cell lines were maintained in α-modified Eagle's medium (α-MEM) containing 10% fetal calf serum (FCS). p53+/+ and p53−/− MEFs were derived from 14-day-old embryos, transformed with E1A/ras and cultured in a 5% CO2 atmosphere in Dulbecco's MEM containing 10% FCS. XL823, a gene trap ES cell line targeting CPT1C (BayGenomics, San Francisco, CA, USA), was maintained on 1% gelatin-coated dishes in DMEM supplemented with leukemia inhibitory factor, 15% FCS, ℒ-glutamine and β-mercaptoethanol. AMPKα1−/−, α2fl/fl/Cre+/− MEF cells were maintained in DMEM supplemented with 10% FCS, 100 IU penicillin, 50 μg/ml streptomycin, and transfected with flag-tagged CPT1C or vector control using Lipofectamine 2000 (Invitrogen, New York, NY, USA) as described previously.7
Prediction of promoter and p53-binding sites
Mouse genomic DNA sequence was obtained from National Center for Biotechnology Information Entrez Gene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene). Promoter sequence was predicted using WWW Promoter Scan program (http://www-bimas.cit.nih.gov/molbio/proscan/). Potential p53-RE were sought using TFBIND (http://tfbind.hgc.jp/).
ChIP analysis
Chromatin immunoprecipitation was carried out as previously described.39 Cells were cross-linked in formaldehyde and sonicated with 6 × 10 s pulses at 50 Watt, 50% max power (Vibra Cell TM, Sonics and Material Inc, Newtown, CT, USA). Extracts were subjected to ChIP assays using the acetyl-histone H3 ChIP assay kit (Upstate Biotechnology, New York, NY, USA) and anti-mouse p53 antibody (FL-393; Santa Cruz Biotechnology, Dallas, TX, USA). PCR amplification was performed using primers specific for the two regions in CPT1C intron 1 that contained consensus p53-binding sequences. The primers used were as follows: p53-RE-A, forward primer (GTACTAGTACCAGGTACAGGAGGGGC) and reverse primer (GAAGCACCCTACTGCGCATGCCC); p53-RE-B, forward primer (GCCTGGCAATTGGAAATGAACAG) and reverse primer (AGTTGGAGAGGGCTTTGGGACC).
Luciferase Assay
The two potential p53-binding sites in CPT1C intron 1 were individually PCR-amplified from murine E14K ES cells and cloned into a pGL3-promoter vector (Promega, Madison, WI, USA). These constructs were co-transfected with WT p53 or a DNA-binding mutant of p53 into p53−/− MEFs using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured in the presence or absence of p53 and normalized to β-galactosidase activity. A luciferase construct containing the p21 promoter region and a p53 construct with a mutation in the DNA-binding site were used as positive and negative controls, respectively.
Real-time PCR
Cells were treated with various stress stimuli (sham treatment, 12 Gy of irradiation, UV 240 μJ/cm2, 1 μM staurosporine for 8 h, 10 μM etoposide for 8 h, and 50 μg/ml 5-FU for 8 h). Total RNA was extracted using the Qiagen Mini Kit (Sigma, St Louis, MO, USA). RNA was reverse transcribed using Superscript (Invitrogen). Specific primers for mouse GAPDH, CPT1A, CPT1B, CPT1C, CPT2 and p21 were generated using either Oligo 5 or PrimerBank. Primer sequences are available upon request. Real-time PCR was performed using an SDS 7900 (Becton Dickinson, Franklin Lakes, NJ, USA) with SYBR green fluorescence (Applied Biosystems, Bedford, MA, USA). The samples were normalized to the stably expressed reference gene GAPDH.
From tumor samples RNA was isolated from frozen tumor samples using the Trizol method (Invitrogen) and the PureLink RNA Mini kit (Ambion, Austin, TX, USA). The RNA was quantified with NanoDrop (Thermo Scientific, Asheville, NC, USA). Two microgram of total RNA were reverse transcribed using the SuperScript III CellsDirect cDNA Synthesis kit (Invitrogen). Real-time PCR was performed using a LightCycler 480 System and SYBR Green I Master mix (Roche, Indianapolis, IN, USA). Raw Ct values were normalized against control housekeeping genes (GAPDH, beta actin and HPRT) and analyzed using the ΔΔCt method.
In situ hybridization
In situ hybridization was performed as previously described.40, 41 Briefly, E12.5 embryos of C57BL/6 p53+/− and p53−/− mice were sham-irradiated or subjected in utero to 5 Gy X-ray irradiation. At 8 h post-irradiation, recovered embryos were dissected, fixed in 4% paraformaldehyde, processed and embedded in paraffin. Tissue sections (4−6 mm) were cut, deparaffinized, acetylated and exposed to 33P-UTP-labeled riboprobes. The CPT1C cDNA template (from which the riboprobes were made) was a 700-bp fragment cloned into pBluescript SK (Invitrogen). The p21 cDNA template was a full-length fragment. Sense and antisense probes were synthesized from linearized templates using T3 or T7 RNA polymerase, labeled with [a33P]-UTP (Amersham, Arlington Heights, IL, USA), and processed as previously described.
Mouse models and animal care
Cis Nf1+/− : p53+/− mice18, 19 were kindly provided by K. Cichowski,21 Dana-Farber/Harvard Cancer Center, USA, and mice depleted in Cpt1c were generated using a gene trap approach as described (BayGenomics).7 Both mouse models were inbred in the C57BL/6 background. All mice were maintained within the Biologisches Zentrallabor barrier facility, University Hospital Zürich and all the experiments were approved by the Zurich Kantonales Veterinäramt (license number 161/2007, 15/2011).
We crossed Cpt1cgt/gt mice into the Nf1+/− : p53+/− background. The mice were monitored three times per week. As soon as the animals showed signs of tumors and distress, they were euthanized and the tumors were isolated.
Statistical analysis and survival studies
The survival of Nf1+/− :p53+/− : Cpt1cgt/gt mice was compared with Nf1+/− : p53+/− mice, Cpt1cgt/gt and wild-type control (C57BL/6). The number of mice per group is indicated in the legend. The survival of the mice was plotted on a Kaplan–Meier curve for individual genotypes against the animal age in months. The survival probabilities were calculated using a public survival calculator (http://www.hutchon.net/Kaplan-Meier.htm) and GraphPad Prism 5. The results were analyzed with the log-rank (P<0.0001) test using Graphpad Prism 5.
Histological analysis of tumors
Tumors were fixed in 4% formalin for 1 week at 4 °C. Then, they were embedded in paraffin, sectioned and stained with haematoxylin and eosin. Immunohistochemistry was performed to test for apoptosis and proliferation using a rabbit polyclonal anti-cleaved caspase-3 (Cell Signaling Technology, Beverly, MA, USA) and a rabbit monoclonal anti-Ki67 (clone SP6, Thermo Scientific). The samples were analyzed with Adobe Photoshop and ImageJ and the histological scores were obtained by calculating the ratio between the number of positive-stained cells and the total cell number/field.
Protein lysates and immunoblotting
Frozen tumor samples were thawed, washed in PBS and minced in extraction buffer containing 50 mM NaHCO3 pH 8.3, 0.25 M NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X100 and protease inhibitors. The tissues were then disrupted by Polytron homogenization and incubated on ice for 30 min. The homogenates were then centrifuged at 10 000 × g for 30 min at 4 °C to remove the cell debris. The supernatant was analyzed by western blotting. Fifty microgram of protein lysates we subjected to SDS-PAGE and immunoblotted with antibodies indicated. Antibody sources are as follow: mouse monoclonal anti-mouse CPT1C antibody (clone 1E11, generated in our Laboratory); mouse monoclonal anti-p53, rabbit polyclonal anti-Pan-actin, anti-AMPKα, anti-phospho-AMPKα (Thr172) and anti-phospho-ACC (Ser79) (Cell Signaling Technology); mouse monoclonal anti-p21 (Santa Cruz Biotechnology).
Acknowledgments
We thank members of the Mak laboratories for helpful comments and Sam Benchimol for helpful suggestions. This work was supported by grants from the Forschungskredit of the University of Zurich, Hartmann Müller Stiftung, Oncosuisse (OCS-02009-02-2007 and KLS 02569-02-2010) (KZ).
Glossary
- 5-FU
5-fluorouracil
- ACC
Acetyl-CoA carboxylase
- AMPK
AMP-activated kinase
- ChIP
chromatin immunoprecipitation
- CPT1
carnitine palmitoyltransferase 1
- CPT1C
carnitine palmitoyltransferase 1C
- ER
endoplasmic reticulum
- FAO
fatty acid oxidation
- FAS
fatty acid synthesis
- Gt
gene trap
- Irrad
ionizing radiation
- KO
knockout
- MEFs
mouse embryonic fibroblasts
- Nf1
neurofibromatosis type 1
- p53-RE
p53-responsive element
- UV
ultraviolet
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Edited by G Melino.
References
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Barger JF, Plas DR. Balancing biosynthesis and bioenergetics: metabolic programs in oncogenesis. Endocr Relat Cancer. 2010;17:R287–R304. doi: 10.1677/ERC-10-0106. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Cha SH, Millington DS, Cline G, Shulman GI, Suwa A, et al. Brain-specific carnitine palmitoyl-transferase-1c: role in CNS fatty acid metabolism, food intake, and body weight. J Neurochem. 2008;105:1550–1559. doi: 10.1111/j.1471-4159.2008.05255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Lane MD. Control of energy homeostasis: role of enzymes and intermediates of fatty acid metabolism in the central nervous system. Annu Rev Nutr. 2006;26:23–44. doi: 10.1146/annurev.nutr.25.050304.092532. [DOI] [PubMed] [Google Scholar]
- Reamy AA, Wolfgang MJ. Carnitine palmitoyltransferase-1c gain-of-function in the brain results in postnatal microencephaly. J Neurochem. 2011;118:388–398. doi: 10.1111/j.1471-4159.2011.07312.x. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Sierra AY, Gratacos E, Carrasco P, Clotet J, Urena J, Serra D, et al. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J Biol Chem. 2008;283:6878–6885. doi: 10.1074/jbc.M707965200. [DOI] [PubMed] [Google Scholar]
- Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchstaller J, McKeever PE, Morrison SJ. Tumorigenic cells are common in mouse MPNSTs but their frequency depends upon tumor genotype and assay conditions. Cancer Cell. 2012;21:240–252. doi: 10.1016/j.ccr.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- Price N, van der Leij F, Jackson V, Corstorphine C, Thomson R, Sorensen A, et al. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 2002;80:433–442. doi: 10.1006/geno.2002.6845. [DOI] [PubMed] [Google Scholar]
- Carrasco P, Sahun I, McDonald J, Ramirez S, Jacas J, Gratacos E, et al. Ceramide Levels Regulated by Carnitine Palmitoyltransferase 1C Control Dendritic Spine Maturation and Cognition. J Biol Chem. 2012;287:21224–21232. doi: 10.1074/jbc.M111.337493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase--development of the energy sensor concept. J Physiol. 2006;574 (Pt 1:7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Wolfgang MJ, Lane MD. Hypothalamic malonyl-CoA and CPT1c in the treatment of obesity. FEBS J. 2011;278:552–558. doi: 10.1111/j.1742-4658.2010.07978.x. [DOI] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihara K, Lapin V, Bakal C, Okada H, Brown L, Hirota-Tsuchihara M, et al. Ckap2 regulates aneuploidy, cell cycling, and cell death in a p53-dependent manner. Cancer Res. 2005;65:6685–6691. doi: 10.1158/0008-5472.CAN-04-4223. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider BF, Elia AJ, Gascoyne RD, Trumper LH, von Bonin F, Kapp U, et al. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2001;97:250–255. doi: 10.1182/blood.v97.1.250. [DOI] [PubMed] [Google Scholar]
- Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]