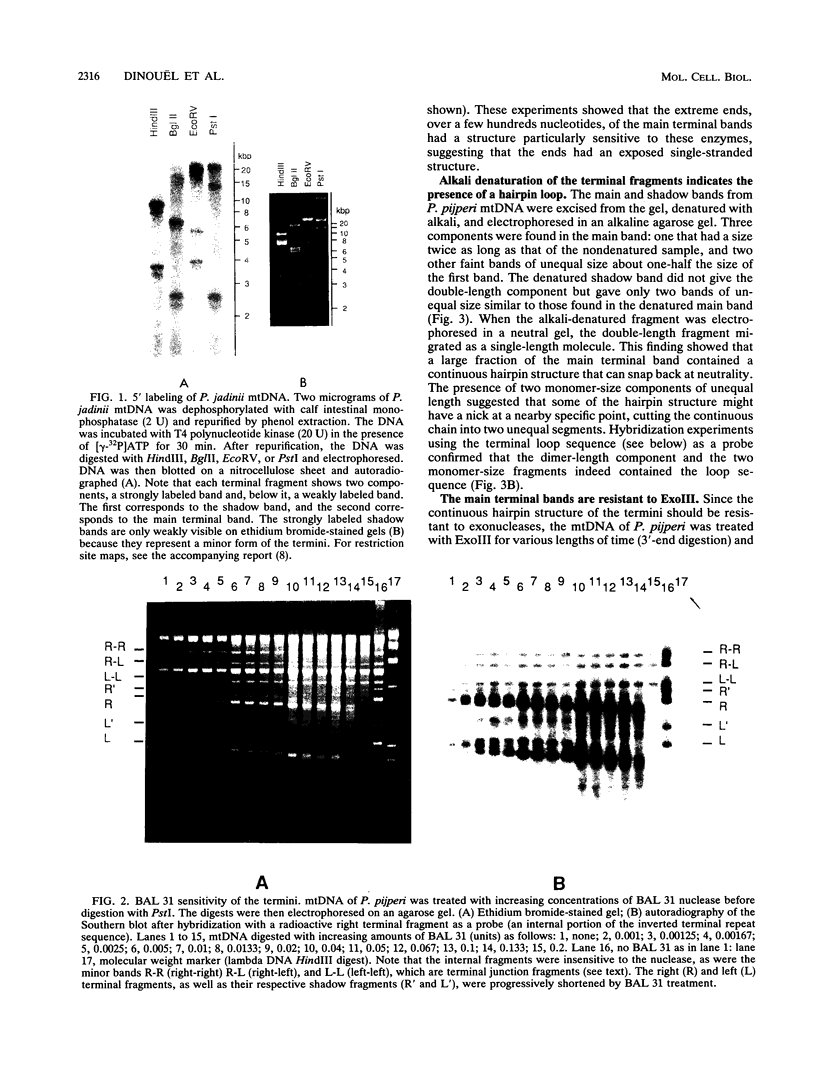
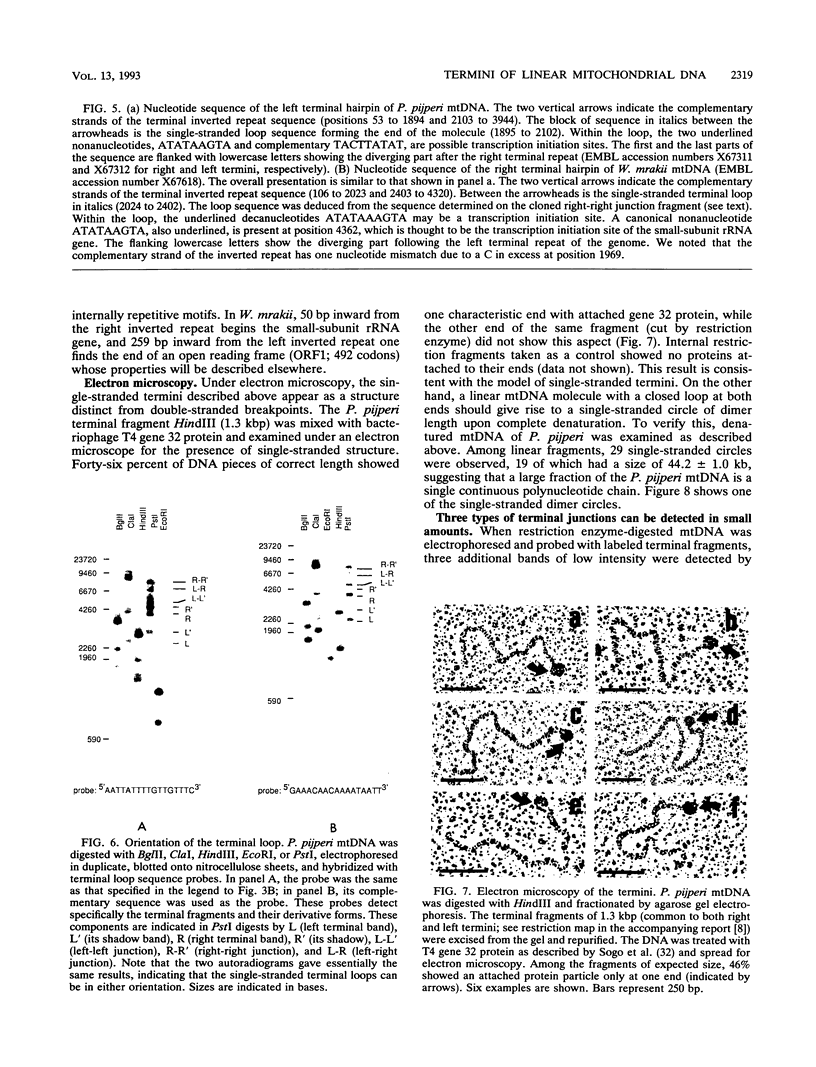
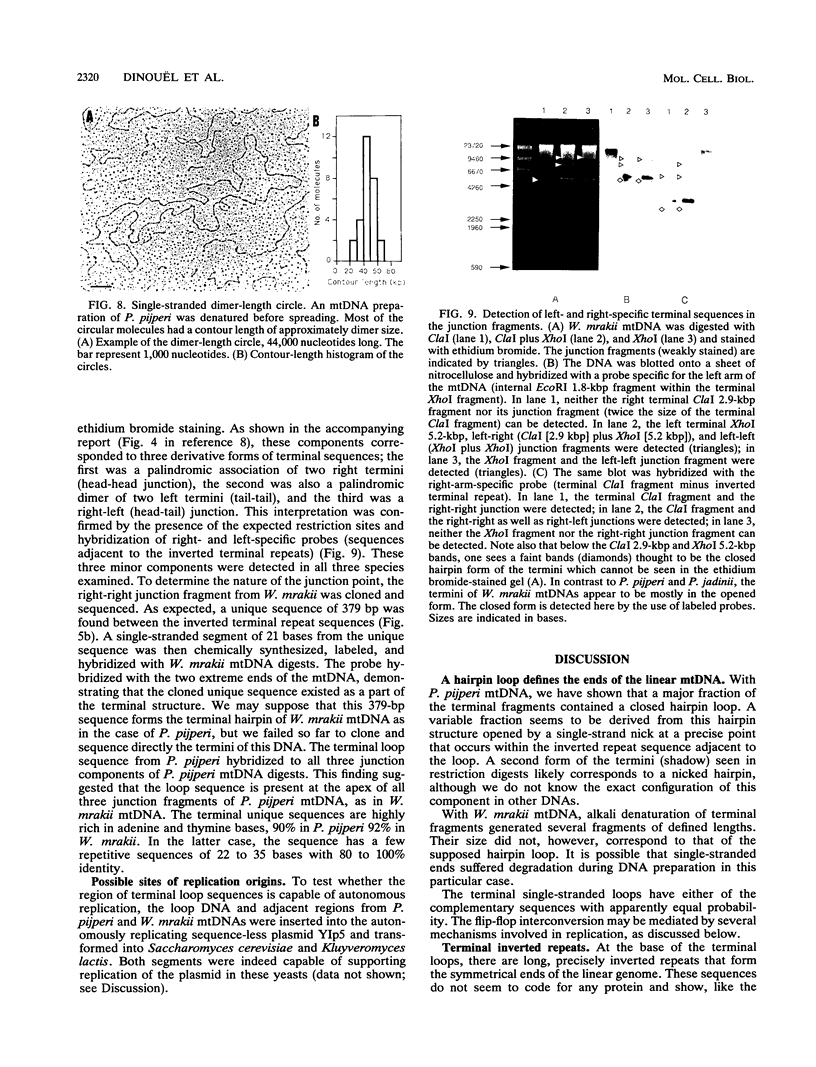
Abstract
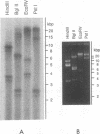
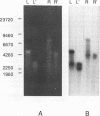
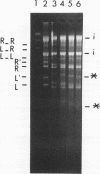
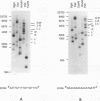
The terminal structure of the linear mitochondrial DNA (mtDNA) from three yeast species has been examined. By enzymatic digestion, alkali denaturation, and sequencing of cloned termini, it was shown that in Pichia pijperi and P. jadinii, both termini of the linear mtDNA were made of a single-stranded loop covalently joining the two strands, as in the case of vaccinia virus DNA. The left and right loop sequences were in either of two orientations, suggesting the existence of a flip-flop inversion mechanism. Contiguous to the terminal loops, inverted terminal repeats were present. The mtDNA from Williopsis mrakii seems to have an analogous structure, although terminal loops could not be directly demonstrated. Electron microscopy revealed the presence, among linear molecules, of a small number of circular DNAs, mostly of monomer length. Linear and circular models of replication are considered, and possible conversion mechanisms between linear and circular forms are discussed. A flip-flop inversion mechanism between the inverted repeat sequences within a circular intermediate may be involved in the generation of the linear form of mtDNA.
Full text
PDF


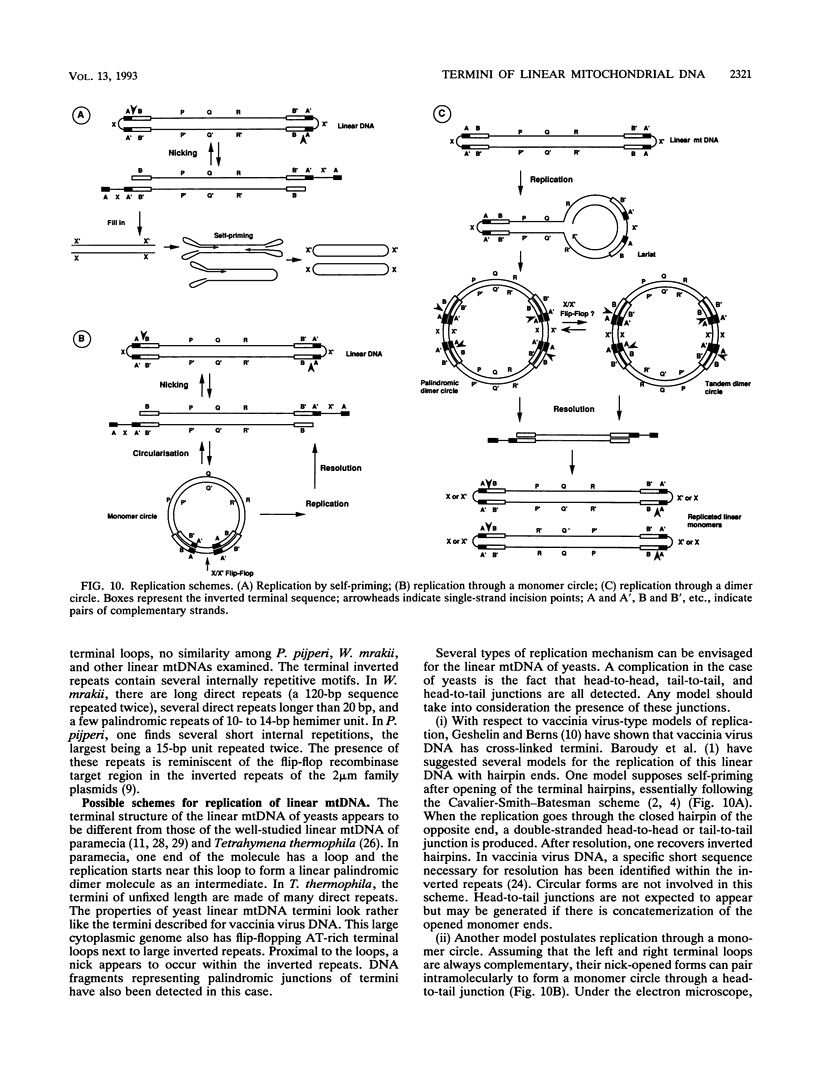


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Bateman A. J. Letter: Simplification of palindromic telomere theory. Nature. 1975 Jan 31;253(5490):379–380. doi: 10.1038/253379a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974 Aug 9;250(5466):467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Partial duplication of the large ribosomal RNA sequence in an inverted repeat in circular mitochondrial DNA from Kloeckera africana. Implications for mechanisms of the petite mutation. J Mol Biol. 1981 Apr 15;147(3):399–415. doi: 10.1016/0022-2836(81)90492-7. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Futcher A. B. The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast. 1988 Mar;4(1):27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- Geshelin P., Berns K. I. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J Mol Biol. 1974 Oct 5;88(4):785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- Goddard J. M., Cummings D. J. Structure and replication of mitochondrial DNA from Paramecium aurelia. J Mol Biol. 1975 Oct 5;97(4):593–609. doi: 10.1016/s0022-2836(75)80061-1. [DOI] [PubMed] [Google Scholar]
- Grant D., Chiang K. S. Physical mapping and characterization of Chlamydomonas mitochondrial DNA molecules: their unique ends, sequence homogeneity, and conservation. Plasmid. 1980 Jul;4(1):82–96. doi: 10.1016/0147-619x(80)90085-2. [DOI] [PubMed] [Google Scholar]
- Gray M. W. Origin and evolution of mitochondrial DNA. Annu Rev Cell Biol. 1989;5:25–50. doi: 10.1146/annurev.cb.05.110189.000325. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Bradford C. J., Grossman L. I. Organization of Achlya mtDNA: a population with two orientations and a large inverted repeat containing the rRNA genes. Proc Natl Acad Sci U S A. 1983 Jan;80(1):142–146. doi: 10.1073/pnas.80.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. P., Mahendran R., Spottswood M. R., Yang Y. C., Miller D. L. Mitochondrial DNA of Physarum polycephalum: physical mapping, cloning and transcription mapping. Curr Genet. 1990 Apr;17(4):331–337. doi: 10.1007/BF00314881. [DOI] [PubMed] [Google Scholar]
- Kiss G. B., Amin A. A., Pearlman R. E. Two separate regions of the extrachromosomal ribosomal deoxyribonucleic acid of Tetrahymena thermophila enable autonomous replication of plasmids in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):535–543. doi: 10.1128/mcb.1.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Inverted repeats in chloroplast DNA from higher plants. Proc Natl Acad Sci U S A. 1979 Jan;76(1):41–45. doi: 10.1073/pnas.76.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovác L., Lazowska J., Slonimski P. P. A yeast with linear molecules of mitochondrial DNA. Mol Gen Genet. 1984;197(3):420–424. doi: 10.1007/BF00329938. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Nucleotide sequence required for resolution of the concatemer junction of vaccinia virus DNA. J Virol. 1989 Oct;63(10):4354–4361. doi: 10.1128/jvi.63.10.4354-4361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis G., Vahrenholz C., Pratje E. Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol Gen Genet. 1990 Sep;223(2):211–216. doi: 10.1007/BF00265056. [DOI] [PubMed] [Google Scholar]
- Morin G. B., Cech T. R. The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell. 1986 Sep 12;46(6):873–883. doi: 10.1016/0092-8674(86)90069-3. [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Thompson W. F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982 Jun;29(2):537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E., Cummings D. J. Replication of linear mitochondrial DNA from Paramecium: sequence and structure of the initiation-end crosslink. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7341–7345. doi: 10.1073/pnas.78.12.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard A. E., Laping J. L., Seilhamer J. J., Cummings D. J. Inter-species sequence diversity in the replication initiation region of Paramecium mitochondrial DNA. J Mol Biol. 1983 Feb 15;164(1):1–15. doi: 10.1016/0022-2836(83)90084-0. [DOI] [PubMed] [Google Scholar]
- Ryan R., Grant D., Chiang K. S., Swift H. Isolation and characterization of mitochondrial DNA from Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3268–3272. doi: 10.1073/pnas.75.7.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumard D. S., Grossman L. I., Hudspeth M. E. Achlya mitochondrial DNA: gene localization and analysis of inverted repeats. Mol Gen Genet. 1986 Jan;202(1):16–23. doi: 10.1007/BF00330510. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Unequal excision of complementary strands is involved in the generation of palindromic repetitions of rho- mitochondrial DNA in yeast. Cell. 1983 Feb;32(2):391–396. doi: 10.1016/0092-8674(83)90458-0. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2486–2490. doi: 10.1073/pnas.87.7.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Troutman W. B., Riggsby W. S. Circular mitochondrial genome of Candida albicans contains a large inverted duplication. J Bacteriol. 1985 Oct;164(1):7–13. doi: 10.1128/jb.164.1.7-13.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Reddy P. M., Yu C. Y., Bastiani C., Higgs D., Stamatoyannopoulos G., Papayannopoulou T., Shen C. K. Transcriptional activation of human zeta 2 globin promoter by the alpha globin regulatory element (HS-40): functional role of specific nuclear factor-DNA complexes. Mol Cell Biol. 1993 Apr;13(4):2298–2308. doi: 10.1128/mcb.13.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]