Abstract
Lysyl oxidase (LO) catalyzes crosslink of collagen, elastin, and histone H1, stabilizing the extracellular matrix and cell nucleus. This enzyme displays dual functions for tumorigenesis, i.e., as a tumor suppressor inactivating the ras oncogene and as a tumor promoter enhancing malignant cell metastasis. To elucidate LO transcriptional regulation, we have cloned the 804 base pair region upstream of the translation start site (ATG) of the rat LO gene with the maximal promoter activity. Computer analysis indicated that at least four hypoxia-response element (HRE) consensuses (5′-ACGTG-3′) exist in the cloned LO promoter. Treatment of rat lung fibroblasts (RFL6) with CoCl2 (Co, 10–100 μM), a chemical hypoxia reagent, enhanced LO mRNA expression and promoter activities. Overexpression of LO was associated with upregulation of hypoxia-inducible factor (HIF)-1α at mRNA levels in cobalt (Co)–treated cells. Thus, LO is a hypoxia-responsive gene. Dominant negative-HIF-1α inhibited LO promoter activities stimulated by Co. Electrophoretic mobility shift, oligonucleotide competition, and in vitro translated HIF-1α binding assays indicated that only one HRE mapped at −387/−383 relative to ATG was functionally active among four consensuses. Site-directed mutation of this HRE significantly diminished the Co-induced and LO promoter-directed expression of the reporter gene. Cadmium (Cd), an inducer of reactive oxygen species, inhibited HIF-1α mRNA expression and HIF-1α binding to the LO gene in Co-treated cells as revealed by RT-PCR and ChIP assays, respectively. Thus, modulation of the HRE activity by Co and Cd plays a critical role in LO gene transactivation.
Key Words: lysyl oxidase, cobalt, cadmium, hypoxia, hypoxia-response element, hypoxia-inducible factor (HIF)-1α.
Lysyl oxidase (LO) (E.C.1.4.3.13), a copper-dependent enzyme, oxidizes peptidyl lysine residues in substrates, e.g., collagen, elastin, and histone H1, essential for organization and stabilization of the extracellular matrix (ECM) and the cell nucleus (Kagan and Li, 2003; Li et al., 2011). Of particular interest, considerable evidence supports LO as a tumor suppressor. This catalyst has been shown to antagonize transforming activity of ras, a proto-oncogene (Kenyon et al., 1991). Defected expression of LO was detected in a variety of spontaneous tumors in humans (Li et al., 2011). Thus, LO as an intra- and extracellular effector is implicated in various human pathological states such as organ fibrosis, atherosclerosis, emphysema, carcinogenesis, etc. (Kagan and Li, 2003; Li et al., 2011).
Hypoxia is a profibrotic stimulus, which enhances the expression of LO and its substrates such as collagen in different assay systems (Higgins et al., 2008; Manalo et al., 2005). Furthermore, high levels of LO were also detected in the hypoxia stage of some tumors, facilitating the metastasis (Erler et al., 2006). Hypoxia-inducible factor-1 (HIF-1), a transcription enhancer, plays a critical role in the cell response to oxygen deficiency (Kaluz et al., 2008). The HIF-1 is composed of HIF-1α and HIF-1β subunits. The HIF-1β is constitutively expressed, whereas HIF-1α is maintained at a low level in normoxic cells. Upon hypoxia, HIF-1α is upregulated and HIF-1 complex binds to the hypoxia-response element (HRE) of the gene promoter for transactivation.
Cobalt (Co), an essential metal for the formation of vitamin B12, mimics the hypoxia condition in the cell culture system activating the HIF-1 targeting genes (Kaluz et al., 2008). Cadmium (Cd) is a toxic carcinogen without any biological availability (Li et al., 2011). Cd exposure enhances cellular levels of reactive oxygen species (ROS) (Cuypers et al., 2010). To elucidate regulation of LO gene transcription under hypoxia conditions, we used the cloned 804 base pair rat LO promoter with the maximal promoter activity as a model (Gao et al., 2007) to characterize HRE status in the LO gene under Co exposure conditions. Furthermore, we also examined Co-activated LO promoter in response to Cd, a ROS inducer in the biological system. We found that only one HRE mapped at −387/−383 relative to ATG was functionally active in response to Co among four HRE consensuses existing in the cloned LO promoter, and Cd inhibited HIF-1α expression and its binding to the HRE for Co activation of the LO gene.
MATERIALS AND METHODS
Materials.
Co chloride and Cd chloride, each 99.9% pure, were from Aldrich Chemicals (Milwaukee, WI). Mouse IgG, anti-RNA polymerase II (RNA-Poly II), anti-HIF-1α, and anti-glyceradehyde 3-phosphate dehydrogenase (GAPDH) were from Santa Cruz Biotech (Santa Cruz, CA). Synthetic oligonucleotides containing the cis elements used for the electrophoretic mobility shift assay (EMSA) or oligonucleotide primers used for the PCR were purchased from Integrated DNA Technologies (Coralville, IA). All tissue culture products were from Invitrogen Co. (Carlsbad, CA).
Cell culture and metal exposure.
The rat fetal lung fibroblasts (RFL6) obtained from ATCC were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 and 95% air incubator as previously described (Zhao et al., 2006). Stock cultures were derived from the frozen cell line and passaged every 4 days. A total of six passages were used for experiments. To obtain growth-arrested cultures, cells were incubated in 0.3% FBS/DMEM for 3 days, changed to fresh medium, and used for experiments. Growth-arrested cells were exposed to Co, Cd, or both at indicated concentrations for 24h. To identify effects of chronic Cd exposure on cell phenotype changes, we have isolated Cd-resistant (CdR) RFL6 cells by incubation of cells with graded concentrations of Cd. Different degrees of CdR cells used in this study such as those resistant to 20, 40, and 80µM Cd were referred to as CdR20, CdR40, and CdR80 cells (Zhao et al., 2006). Growth-arrested CdR cells were exposed to Co at indicated concentrations for 24h.
Reverse transcription-PCR analysis.
Total RNA was extracted from control and treated cells using TRIzol reagent (Invitrogen). The first-strand cDNA was synthesized with 1 µg of the total RNA using the SuperScript first-strand synthesis system for reverse transcription (RT)-PCR (Invitrogen). Using one-twentieth of the cDNAs as a template, the PCR was carried out under conditions as described (Chen et al., 2005). The primer pairs used for DNA amplification by the PCR are 5′-GACTGGATCCATGGAGGGCGCCGGCGGCGAGAA-3′ and 5′-GACTCTCGAGTTAACTTGATCCAAAGCTCTG-3′ for HIF-1α, 5′-GACTAAGCTTATGGCGGCGACTACAGCTAACC-3′ and 5′-GACTC TCGAGTATTCAGAAAAAGGGGGAAACA-3′ for HIF-1β, and 5′-GA CTGGATCCATGCGAAGCAAAGAGTCTGAAG-3′ and 5′-GACTCTCGA GAGCTTGTCGAAGAGGCAGCTC-3′ for dominant negative form of HIF-1α (DN-HIF-1α). The primer pairs used for PCR amplification of the LO and GAPDH genes are the same as those described previously (Chen et al., 2005).
Plasmid constructions.
The cDNA fragments of wild-type HIF-1α, DN-HIF-1α, and wild-type HIF-1β with the full coding region were amplified by RT-PCR and inserted into the BamHI-XhoI, HindIII-XhoI, and BamHI-XhoI sites of pcDNA3.1/v5-his expression plasmid, respectively, (Invitrogen) to create expression constructs of pcDNA3.1-HIF-1α, pcDNA3.1-DN-HIF-1α, and pcDNA3.1-HIF-1β as described (Gao et al., 2007).
Transient transfection and luciferase assay.
Cell transfection and assays for reporter gene products were carried out as described previously (Gao et al., 2007). Briefly, RFL6 cells were plated at 2.5 × 105 cells per 35-mm well containing 2ml of 10% FBS/DMEM. After 24-h incubation, cells were cotransfected with LO promoter-luciferase constructs prepared by this lab (1 µg) (Gao et al., 2007) and phRL-TK expression vectors (0.5 µg; Promega, Madison, WI), the latter used as an internal control to monitor the transfection efficiency, by using lipofectamine reagent (Invitrogen). Following 6h posttransfection incubation, cells were immersed in 10% FBS/DMEM for 18h, washed, and then changed to 0.3% FBS/DMEM for 6 h. Such prepared cells were finally treated with or without different concentrations of Co for an additional 24h. The luciferase activity in the cell lysates was determined using dual luciferase assay reagents (Promega). The luciferase activities were normalized by the internal control values and represented as the mean ± SD for the three wells. For the transient overexpression assays in RFL6 cells, 0.5 µg of each expression vector was cotransfected together with the luciferase reporter vectors. After incubation for 18h, the medium was replaced with a 0.3% FBS/DMEM, and 6h later, these cells were treated with or without Co at indicated concentrations for an additional 24h.
Nuclear extract preparation and EMSA.
Nuclear extracts were prepared from control and Co-treated cells using Nuclear and Cytoplasmic Extraction kit (Pierce, Rockford, IL). Protein concentrations were determined by the BCA protein assay reagents (Pierce). For the EMSA (Gao et al., 2007), synthetic oligonucleotides were end labeled with [γ-32P]ATP (Perkin Elmer, Boston, MA) by T4 polynucleotide kinase (New England Biolabs, Boston, MA) and annealed to their complements. A total volume of 20 µl reaction mixture containing 20 µg of nuclear protein or bovine serum albumin, a negative control, 1 µg of poly(dI-dC).poly(dI-dC) (Sigma, St Louis, MO), and 10,000 to 20,000 cpm of labeled probes was incubated for 20min at room temperature. For competition experiments, cold human erythropoietin (Epo) HRE oligonucleotides as described (Mukhopadhyay et al., 2000) at 100-fold molecular excess were added 10min prior to addition of the radiolabeled probe. After reaction, samples were subjected to native 6% polyacrylamide gel electrophoresis and visualized by exposure of the dried gel to Kodak film. Supershift reactions were run as competition assays as described above with the exception that 2 µg of a specific antibody against HIF-1α (Santa Cruz Biotech) instead of cold probes was added.
In vitro transcription/translation.
The rat HIF-1α, HIF-1β, and DN-HIF-1α proteins were expressed by PCR based on the methodology of in vitro transcription/translation in rabbit reticulocyte lysates (Promega) according to the manufacturer’s instruction. The primer pairs are 5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGGAGGGCG CCGGCGGCGAGAA-3′ and 5′-TTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTAGTTAACTTGA TCCAAAGCTCTG-3′ for HIF-1α, 5′-GGATCCTAATACGACTCA CTA TAGGGAACAGCCA CCATGGCGGCGACTACAGCTAACC-3′ and 5′-TTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTATTCAGAAAAAGGGGGAAACA-3′ for HIF-1β, and 5′-GGATCCTAATACGACTCACTA AGG GAACAGCCACCAT GCGAAGCAAAGAGTCTGAAG-3′ and 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTAAAGCTT GTC GAA GAGGCAGCTC-3′ for DN-HIF-1α.
Site-directed mutagenesis.
The mutated HRE in the LO promoter-reporter construct was created by site-directed mutagenesis using the QuickChangeTM Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The pLOProm 804-reporter construct was used as a parental plasmid containing the wild-type HRE (labeled with the lower bold case), i.e., 5′-GCTGTCCGCCTTGcacgtTTCCAATCGCAT-3′ as described (Gao et al., 2007). Primers used for the point mutation is 5′-GGAGCTGTCCGCCTTGcgattTTCCAATCGCATTACG-3′, where the HRE and mutated nucleotides were labeled with the lower bold case and underline, respectively.
Chromatin immunoprecipitation assay.
To determine cellular HIF-1 binding to the LO promoter region, the chromatin immunoprecipitation (ChIP) assay was performed as described (Gao et al., 2007) with the EpiQuik Chromatin Immunoprecipitation Kit based on the protocol provided by the supplier (Epigentek Group Inc., Brooklyn, NY). Cellular components were crosslinked by incubation of control and metal (Co, Cd, or both)-treated cells at the same number (2 × 106) with 1% formaldehyde. The crosslinking reaction was stopped by addition of glycine. Nuclei were extracted and then sonicated to shear DNA to lengths between 200 and 1000bp. After centrifugation, aliquots of DNA containing supernatants were removed out as “input” DNA. DNA samples were transferred into the strip wells precoated with the antibody against rat HIF-1α, (Santa Cruz Biotech), RNA-Poly II (a positive control provided by the kit supplier), or nonspecific rat IgG (a negative control from Santa Cruz Biotech). After incubation followed by successive washing, precipitated DNA-protein complexes and the “input” samples were treated with proteinase K and collected by the P-spin columns. Using purified DNA as a template, PCR was conducted under conditions as described (Gao et al., 2007). Primer pairs were used in PCR to characterize the HIF-1α binding to the LO gene and the RNA-Poly II binding to GAPDH gene as follows: 5′-ctccctgtgcaacgtgtct-3′ and 5′-tgcagttacacaagccgttc-3′ were used for amplification of the LO HRE fragment (152bp), and 5′-ttgcttggcttcttctttgg-3′ and 5′-gagacgaggctggtactcca-3′ were used for amplification of the RNA-Poly II binding region in the GAPDH promoter (160bp). PCR products were analyzed on a 2.2% agarose gel, stained with ethidium bromide and visualized on a UV transilluminator. Densities of DNA bands on the gel were measured as described (Chen et al., 2005).
Statistical analysis.
Data were expressed as mean ± SD of at least three independent experiments. Statistical differences between means were determined using one-way ANOVA followed by Bonferroni’s post hoc test or two-tailed Student’s t-test when appropriate. A p value < 0.05 was considered significant.
RESULTS
Co Enhanced LO mRNA Levels and Promoter Activities in Treated RFL6 Cells
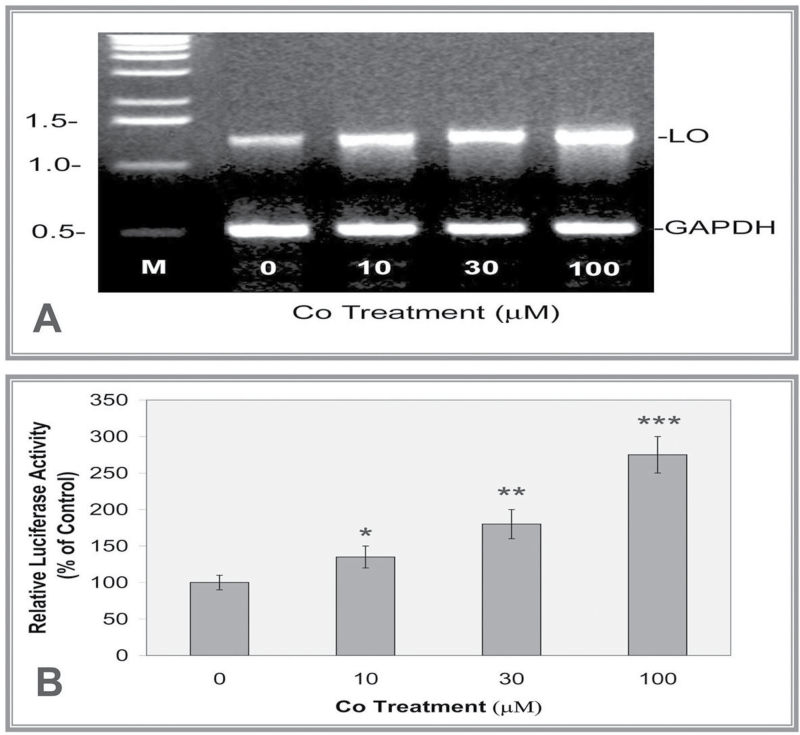
To assess hypoxia effects on the expression of the LO gene, we first examined LO mRNA levels in RFL6 cells in response to various dosages of Co, which mimics the hypoxia condition in the cell culture system (Kaluz et al., 2008). As shown in Figure 1A, exposure of cells to CoCl2 induced a dose-dependent enhancement of LO mRNA expression as determined by the RT-PCR reaching 144, 162, and 188% of the control in cells exposed to Co at 10, 30, and 100 µM concentrations, respectively. To answer the question whether enhancement of LO mRNA expression by Co is due to activation of the LO gene promoter, we have prepared a LO promoter-reporter construct containing a 804bp LO gene promoter fragment relative to the translation initiation codon ATG upstream of the luciferase gene in the pGL3-Basic vector (Promega) (pLOProm 804) (Gao et al., 2007). The pLOProm 804 construct and the phRL-TK vector, an internal control, were transiently cotransfected into RFL6 cells. Effects of CoCl2 at different concentrations on the reporter gene product expression were tested. As shown in Figure 1B, Co exposure increased luciferase activities amounting to 1.4-, 1.9-, and 2.9-fold of the control, respectively, in cells treated with Co at 10, 30, and 100 µM concentrations. Thus, LO is a Co/hypoxia-sensitive gene.
Fig. 1.
Co enhancement of LO mRNA levels (A) and promoter activities (B) in treated RFL6 cells. (A) Growth-arrested cells were incubated for 24h in 0.3% FBS/DMEM in the absence or presence of Co at indicated concentrations. Total RNA was extracted from cells using TRIzol reagent. LO mRNA levels in control and treated cells were determined by RT-PCR. GAPDH, an internal control; M, molecular ladder. Note: Densities of PCR-amplified gene fragments on the gel were measured with the 1D Scan software in this and below experiments. (B) Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and phRL-TK vector, an internal control, then exposed to Co at indicated concentrations for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control without Co treatment. *p < 0.05, **p < 0.01, and ***p < 0.001 relative to control.
Co Enhancement of the LO Promoter Activity via Stimulation by HIF-1
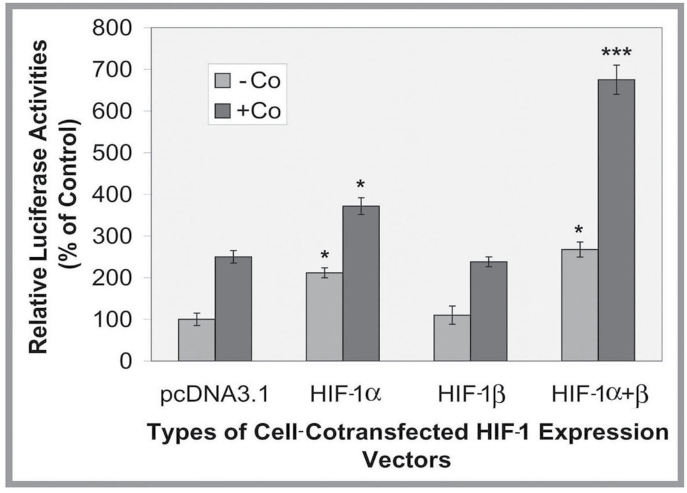
HIF-1 is a major transcription factor for the gene activation by hypoxia (Kaluz et al., 2008). To test whether the HIF-1 is involved in Co-induced LO gene expression in RFL6 cells, we examined effects of increased cellular level of HIF-1 by using gene expression vectors, i.e., the pcDNA3.1-HIF-1α, pcDNA3.1-HIF-1β, or both, on expression of the LO promoter-driven reporter gene. Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804), different HIF-1 expression vectors and the internal control phRL-TK vector, and incubated under normoxia and Co exposure conditions. As shown in Figure 2, overexpression of HIF-1α and HIF-1α + HIF-1β, but not HIF-1β alone, in the absence of 100µM Co, increased LO promoter-driven luciferase activity by 2.2- and 2.8-fold, respectively, compared with cells expressing the basic reporter only under normoxia. However, when cells were cotransfected with HIF-1α and HIF-1α + HIF-1β expression vectors in the presence of 100µM Co, LO promoter-driven reporter gene expression increased to 3.9- and 7.2-fold of the basal control, respectively. It should be noted that a 2.5-fold increase of the LO promoter-driven reporter gene expression in pcDNA3.1 co transfected cells in the presence of Co reflected effects of cell endogenous HIF-1 activation by this metal ion consistent with data as shown in Figure 1B. These data support that transactivation of the LO gene by Co is mediated by activation by HIF-1, and overexpression of HIF-1α plus HIF-1β had an additive effect on the maximal LO promoter activation in cells treated with Co or hypoxia conditions.
Fig. 2.
Co enhancement of the LO promoter activity stimulated by HIF-1 in treated RFL6 cells. Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and different HIF-1 expression vectors as shown, as well as the internal control phRL-TK vector, then treated without or with 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control in which cells were cotransfected with the pcDNA3.1 basic vector without the HIF-1 cDNA insert. *p < 0.05 and ***p < 0.001 relative to control cells cotransfected with the pcDNA3.1 basic vector.
DN-HIF-1α Inhibited LO Promoter Activity Stimulated by Co
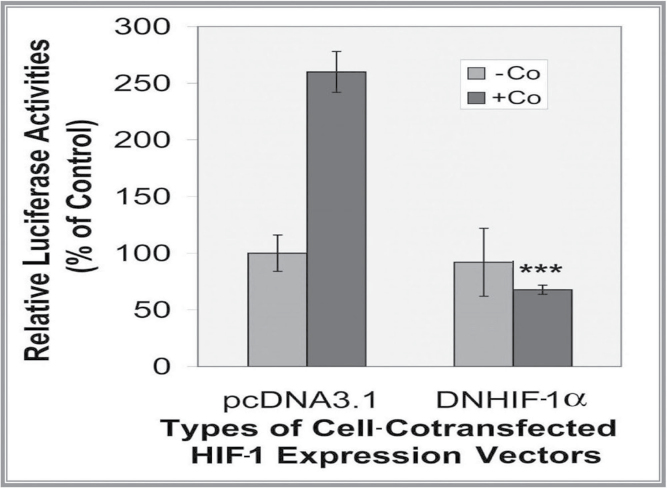
To illustrate the specificity of the HIF-1 in transactivating the LO promoter by Co, RFL6 cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and the DN-HIF-1α expression vector (Grosfeld et al., 2002) as well as the internal control phRL-TK vector. Transfected cells were then incubated in the presence or absence of Co at 100µM for 24h. The DN-HIF-1α mutant without the DNA binding domain can heterodimerize with HIF-1β to form an inactive dimer, thus inhibiting the activation of the HRE containing gene (Richard et al., 2000). As shown in Figure 3, overexpression of the DN-HIF-1α decreased luciferase activities to 25% of the control in cells treated with Co. These results further demonstrated the efficiency of the DN-HIF-1α in inhibition of LO promoter activity via the HIF-1 pathway in Co-treated RFL6 cells.
Fig. 3.
DN-HIF-1α inhibition of the LO promoter activity stimulated by Co in RFL6 cells. Cells were cotransfected with the LO promoter-reporter construct (pLOProm 804) and DN-HIF-1α expression vectors and the internal control phRL-TK vector, then treated without or with 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the control in which cells were cotransfected with the pcDNA3.1 basic vector without the DN-HIF-1α cDNA insert. ***p < 0.001 relative to control cells cotransfected with the pcDNA3.1 basic vector without the DN-HIF-1α cDNA insert and treated with Co.
Overexpression of LO mRNA Was Associated With Upregulation of Cellular HIF-1a mRNA in Co-Treated RFL6 Cells
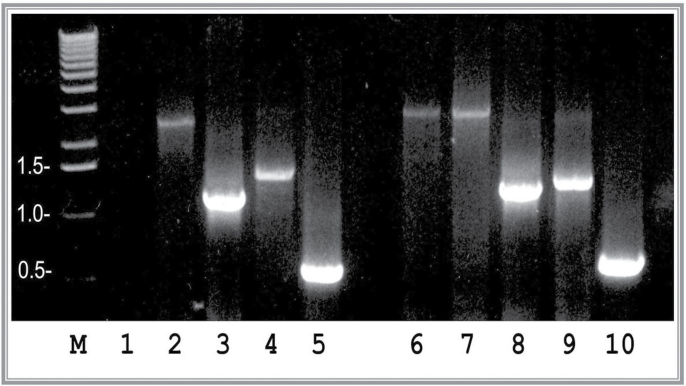
To establish the relationship between LO and HIF-1 expressions in Co-exposed cells, we examined their mRNA levels in control and Coexposed cells. Total RNA samples were prepared from RFL6 cells incubated in the presence or absence of 100µM Co. As shown in Figure 4, cellular HIF-1α mRNA was at a very low level with the density of 0.21 in the normoxia condition without Co treatment but had a higher level of expression in Co-treated condition reaching 51-fold of the control (compare lane 6 with lane 1). Concomitantly, LO mRNA was significantly increased in cells treated with 100µM Co amounting to 1.78-fold of the control under the normoxia condition (compare lane 9 with lane 4). However, HIF-1β mRNA did not display a noticeable change in Co treatment condition compared with normoxia condition. The DN-HIF-1α had a higher expression level either in normoxia or in Co treatment condition. There was no significant difference between both treatment conditions in GAPDH mRNA levels. These data suggest that HIF-1α was transactivated only upon Co treatment in association with upregulation of LO, whereas HIF-1β is constitutively expressed in both normoxia and Co treatment conditions. Higher expression of the DN-HIF-1α in both control and Co-treated cells suggests its potential regulatory effects on HRE activation or suppression.
Fig. 4.
Association of upregulation of HIF-1α and LO at mRNA levels in RFL6 cells treated with Co. Growth-arrested cells were incubated for 24h in 0.3% FBS/DMEM in the absence (lanes 1–5) or presence (lanes 6–10) of 100µM Co. Total RNA was extracted from cells using TRIzol reagent. Transcript levels of HIF-1α (lanes 1 and 6), HIF-1β (lanes 2 and 7), DN-HIF-1α (lanes 3 and 8), LO (lanes 4 and 9), and GAPDH (lanes 5 and 10), an internal control, in control (lanes 1–5) and treated cells (lanes 6–10) were determined by RT-PCR. M, molecular ladder.
Identification of HIF-1 Binding Sites in the 804bp LO Promoter Fragment
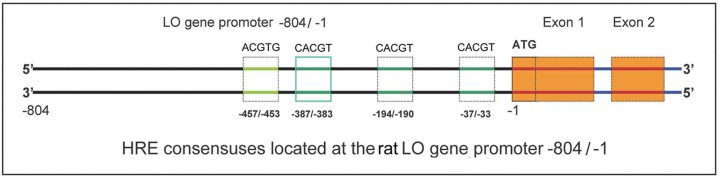
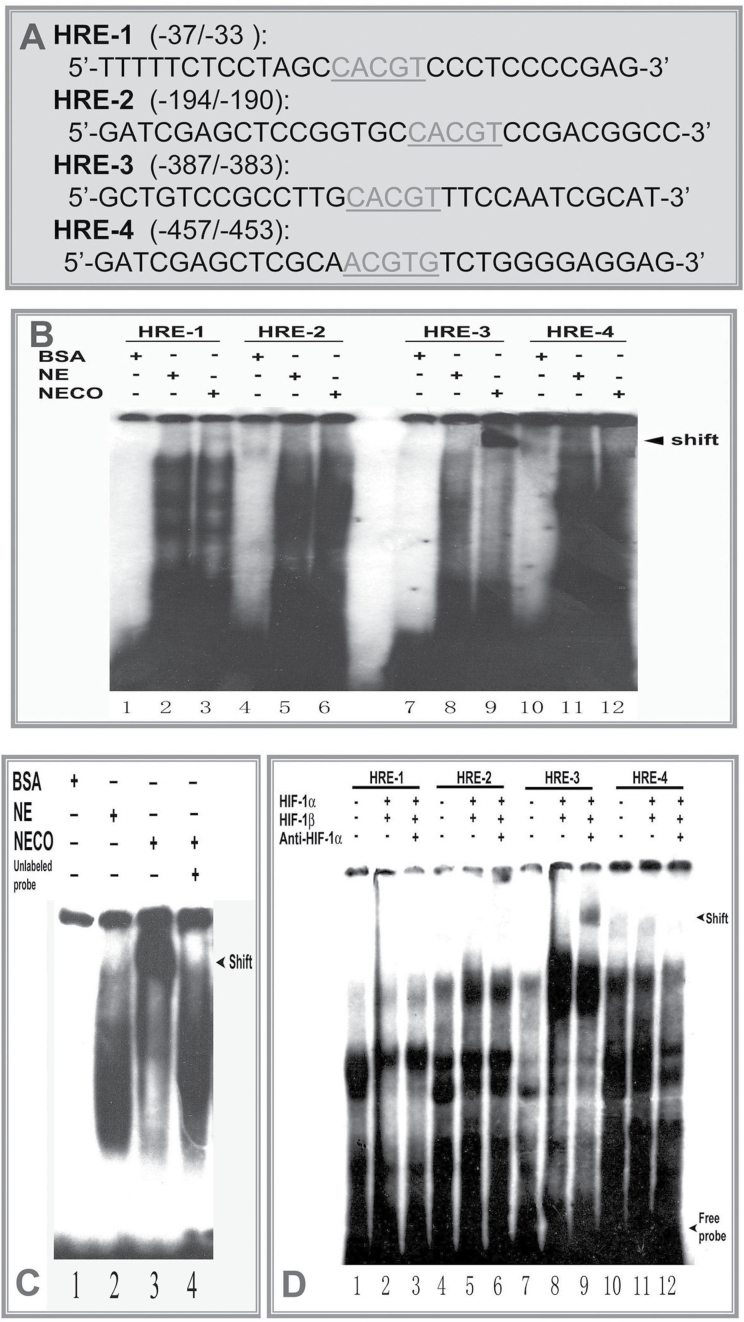
Sequence analysis reveals the presence of four 5′-RCGTG-3′ HIF-1 binding consensuses (HRE) within the 804bp LO promoter fragment upstream of ATG. Three putative HREs located at −33/−37, −190/−194, and −383/−387 are on the noncoding strand, and the fourth HRE located at −457/−453 is on the coding strand (Fig. 5). To determine whether HIF-1 binds to the functional HRE sequences in the LO gene, we performed EMSAs using four [32P]-labeled 30-bp probes encompassing each putative HRE sequence in the LO 804bp promoter (HRE-1, HRE-2, HRE-3, and HRE-4) (Fig. 6A). The shifted band was observed only using the radiolabeled HRE-3 probe in the presence of nuclear extracts from Co-treated cells (Fig. 6B, lane 9) but not that from the normoxic cells (Fig. 6B, lane 8), and no obvious shifted bands were observed using HRE-1, HRE-2, or HRE-4 probes under either normoxia or Co-treated condition (Fig. 6B). These results suggest that the HIF-1 complex binds to the HRE-3 sequence under Co chemical hypoxia condition. To confirm that the shifted band by the HRE-3 probe is a HIF-1 binding site, we performed a competition assay. As shown in Figure 6C, with a 100-fold molar excess of unlabeled human Epo HRE oligonucleotides (Mukhopadhyay et al., 2000), the binding of the radiolabeled HRE-3 probe with HIF-1 from nuclear extract of Co-treated cells was suppressed effectively (compare lane 4 with lane 3). This provides the evidence that the HRE-3 site is a target for HIF-1 binding under Co mimic hypoxia condition. Furthermore, we also assess whether HRE elements in the 804bp LO promoter region bind with the in vitro translated HIF-1 complex. For this purpose, two subunits of HIF-1, i.e., HIF-1α and HIF-1β, were synthesized in vitro by using the reticulocyte lysate system (Promega). EMSAs were performed with the proteins obtained in unprogrammed reticulocyte lysates, HIF-1α/HIF-1β-primed reticulocyte lysates and HIF-1α/HIF-1β-primed reticulocyte lysates plus anti-HIF-1α antibody. As shown in Figure 6D, a specific complex with retarded migration appeared exclusively when HIF-1α/HIF-1β-primed lysates with anti-HIF-1α antibody were incubated together with the labeled HRE-3 probe (lane 9). No complex was visualized when labeled HRE-1, HRE-2, or HRE-4 was used as the probe (Fig. 6D). These data further demonstrate that HIF-1 complex binds to the consensus HRE-3 located at the proximal promoter region of the LO gene.
Fig. 5.
The schematic linear map of HRE consensuses in the cloned rat LO promoter. ATG, the translational start site; ACGTG or CACGT, the core HRE consensuses sequence; regions such as −387/−383 indicating the fragments between two nucleotide numbers using the first nucleotide preceding the ATG codon as −1. solid light blue line box, functionally active HRE consensuses; dash line boxes, nonfunction HRE consensuses; the green lines in the consensuses box, the HRE in the noncoding strand; and the light green lines in the coding strand; the red lines with orange boxes, coding regions.
Fig. 6.
EMSA to determine functionally active LO HRE and nuclear protein binding. (A) Synthetic oligonucleotides containing HREs in the LO promoter region from −804 to −1 relative to ATG. (B) EMSA to assess nuclear protein binding. Synthetic oligonucleotides were end labeled with [γ-32P]ATP and incubated with nuclear protein isolated from control (NE), Co-treated cells (NECO), or bovine serum albumin (BSA), a negative control, as described in Materials and Methods section. After reaction, samples were subjected to native 6% polyacrylamide gel electrophoresis and visualized by exposure of the dried gel to Kodak film. (C) Competition EMSA. Cold synthetic human Epo HRE oligonucleotide with high affinity for the HRE (see Materials and Methods section) at 100-fold molecular excess was added 10min prior to addition of the radiolabeled probe. After reaction, gels were run as described above in B. (D) Supershift EMSA to assess DNA probe binding with in vitro transcription/translation HIF-1 proteins. Reaction mixtures contain 2 µg in vitro transcription/translation proteins, HIF-1α and HIF-1β each. Supershift reactions were run as described above in B with the exception that 2 µg of a specific antibody against HIF-1α was added 10min prior to addition of labeled probes.
Mutational Analysis of the HRE-3 Within 804bp of the LO Promoter
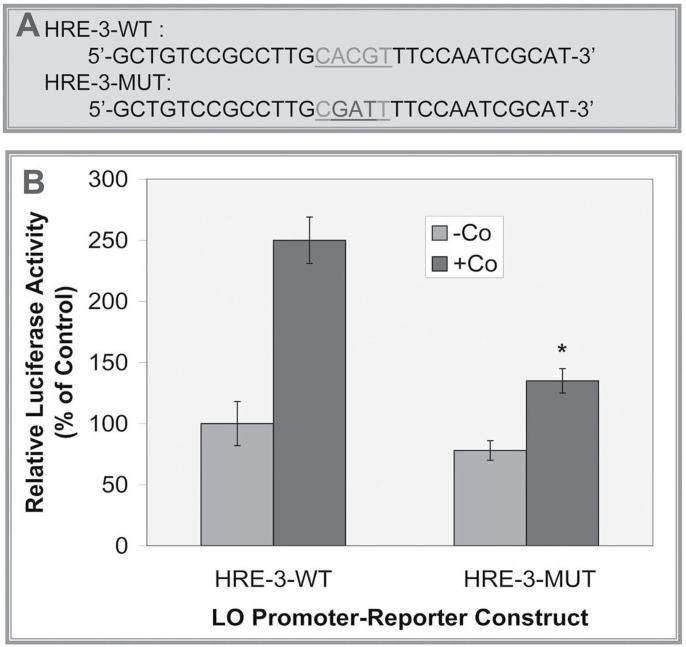
To assess the functional importance of the HRE-3 element in the proximal promoter region of the LO gene, this sequence was mutated in the pLOProm 804 reporter vector (Fig. 7A). The HRE wild-type or mutated construct and the pRL-TK vector, an internal control, were transiently cotransfected into RFL6 cells followed by treatment with or without 100µM Co. As shown in Figure 7, in the wild-type LO HRE promoter, the luciferase activity was increased to 2.5-fold of the control in the presence of Co. In contrast, in the HRE element–mutated LO promoter, the luciferase activity was reduced to 53.2% of the no mutation control in RFL6 cells incubated under same conditions. These results support the conclusion that the HRE-3 consensus sequence is required for hypoxia-mediated induction of LO promoter activity.
Fig. 7.
Determination of the functional HRE by mutagenesis. (A) Mutation of the LO HRE −387/−383. The site-directed mutation of the HRE −387/−383 was performed with the QuikChange mutagenesis kit using pLOProm 804 as a basic. The core HRE 5′-CACGT-3′ was mutated to 5′-CGATT-3′ labeled with gray color letters and underline. (B) Relative luciferase activities of LO promoter-reporter constructs. The wild or mutated LO promoter-reporter constructs each as shown above and the pRL-TK vector, an internal control, were transiently cotransfected into RFL6 cells. After transfection cells were treated with or without 100µM Co for 24h. Luciferase activity in each treatment was normalized to the internal control and expressed as % of the corresponding control. *p < 0.05 relative to the wild HRE vector–transfected control cells treated with Co.
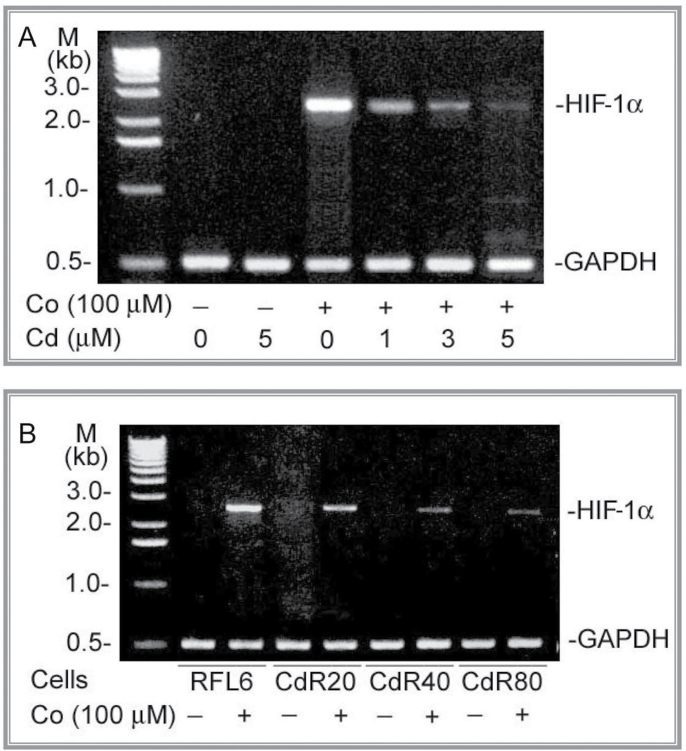
HIF-1α Binding to the LO HRE-387/-383 in RFL6 Cells in Response to Co and Cd
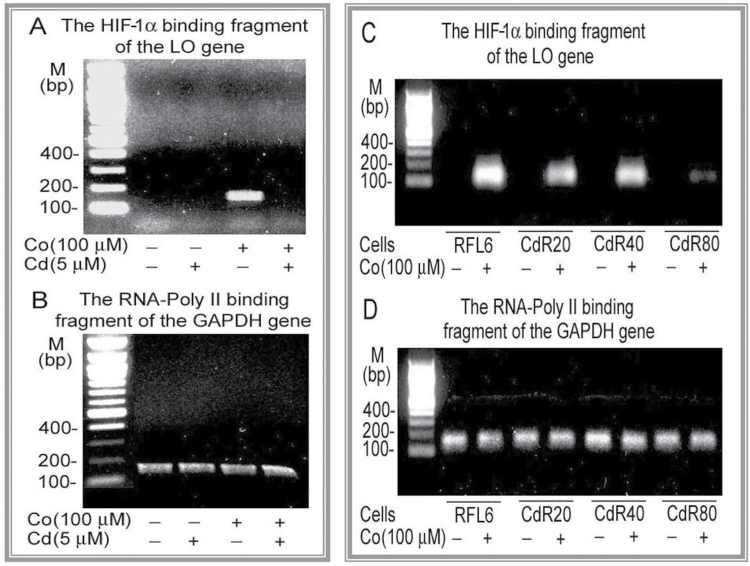
To identify effects of oxidative stress on Co activation of the LO HRE, we examined HIF-1α binding to the LO gene in Co-treated RFL6 cells in response to Cd, a ROS inducer (Cuypers et al., 2010). CdR cells (Zhao et al., 2006) were also used as a long-term Cd exposure model in this study. HIF-1α binding to the LO HRE-387/-383 was determined in cell models by ChIP assays as described (Gao et al., 2007). RNA-Poly II binding to the GAPDH gene was included as an internal control. As demonstrated above (Fig. 6), there was at least one functionally active HRE located at the regions −387/−383 in the LO promoter region −804/−1 that displayed the maximal promoter activity. As shown in Figure 8A, approximately, a 150-bp band was observed on the gel as the chromatin-protein crosslinked by formaldehyde fixation, immunoprecipitated with the HIF-1α antibody, and PCR-amplified using the primer pair that encompasses the HRE −387/−383 in cells treated with 100µM Co alone (lane 3). In contrast, no signal was detected in control cells without any treatment, cells treated with 5µM Cd alone, and cells treated with 100µM Co plus 5µM Cd. Cd at 5µM was used in this study because this concentration of Cd has been shown to inhibit LO at mRNA, protein, and catalytic levels (Li et al., 1995; Zhao et al., 2010). Furthermore, HIF-1α in the long-term Cd exposure model, CdR cells, exhibited a weak binding activity for the LO HRE in cells response to Co. The HIF-1α binding to the LO HRE levels was decreased to 72, 63, and 7% of the control in CdR20, CdR40, and CdR80 cells, respectively, in response to 100µM Co (Fig. 8C). In parallel, as an internal control, the RNA-Poly II binding to the GAPDH gene was not significantly changed in cells incubated under same corresponding conditions. No band was found in the gels using the samples immunoprecipitated by nonspecific IgG, and there was no significant difference in the yields of PCR products among groups using “input” (before immunoprecipitation) DNA as a template (data not shown). These results indicated that (1) Co stimulated HIF-1α binding to the LO HRE and (2) Cd inhibited LO HRE activation elicited by Co in RFL6 cells. Thus, the status of cellular HIF-1α binding to the LO gene −387/−383 is specific and highly sensitive to modification of Cd.
Fig. 8.
HIF-1α binding to cis-element −387/−383 in RFL6 cells treated with Co, Cd, or both revealed by the ChIP assay. Growth-arrested RFL6 cells were treated without or with 100µM Co, 5µM Cd, or both for 24h (A and B). Control and CdR cells were treated without or with 100µM Co (C and D) for the same time. Cells at 2 × 106 for each group were processed for chromatin immunoprecipitation with an antibody against HIF-1α (A and C) or RNA-Poly II, a positive control (B and D). Using immunoprecipitated DNA as a template, the PCR with primer pairs, as shown in Materials and Methods section, amplified LO gene fragments containing the HRE −387/−383 with 150bp (A and C) and the GAPDH promoter fragment with 160bp (B and D). PCR products were analyzed on 2.2% agarose gels. Various treatments were indicated at the bottom on each gel. The left lane on each gel shows the DNA molecular ladder.
Cd Effects on HIF-1α mRNA Expression Elicited by Co
Cells exposed to Co displayed upregulation of HIF-1α mRNA (Fig. 4). To further assess Cd effects on the Co-elicited HIF-1α expression, cells were treated with or without Co, Cd or both, or CdR cells with different degree of Cd resistance treated with Co alone. Then, total RNA were extracted from cell models, converted to the cDNA for each sample, and amplified by RT-PCR as described (Chen et al., 2005). As shown in Figure 9A, Cd induced a dose-dependent inhibition of Co-stimulated HIF-1α expression. Levels of HIF-1α mRNA were decreased to 47, 28, and 16% of the control, respectively, in 100µM Co-treated cells coincubated with 1, 3, and 5µM Cd. Furthermore, CdR20, CdR40, and CdR80 cells also exhibited a reduced HIF-1α mRNA levels in response to 100µM Co, amounting to 48, 20, and 14 of the RFL6 control, respectively (Fig. 9B). These results provide strong evidence for Cd inhibition of Co-stimulated LO HRE activation at the HIF-1α mRNA levels.
Fig. 9.
Cd inhibition of HIF-1α mRNA expression in RFL6 cells elicited by Co. Growth-arrested RFL6 cells were treated without or with 100µM Co in the presence of various concentration of Cd for 24h (A). Control and CdR cells were treated without or with 100µM Co for the same time (B). Total RNA was extracted from cells using TRIzol reagent. HIF-1α mRNA levels in control and treated cells were determined by RT-PCR. GAPDH, an internal control; M, molecular ladder.
DISCUSSION
O2 as a final electron acceptor is required for aerobic metabolism in mammalians. Reduced levels of the O2 concentration below the normal 40–60mm Hg range in mammalian cells are defined as hypoxia occurring under physiological and pathological conditions (Kaluz et al., 2008). Hypoxia-responsive genes contain the cis-acting element called the “HRE” with the core sequence 5′-RCGTG-3′ (R = purine), which in most cases is 5′-ACGTG-3′. Currently, functionally active HREs have been identified in the promoter region of more than 100 mammalian genes involved in erythropoiesis, glycolysis, angiogenesis, carcinogenesis, and other biological activities (Manalo et al., 2005; Semenza, 2010). Here, as reported by us, the rat LO gene is highly responsive to hypoxia as evidenced by increased mRNA expression in RFL6 cells exposed to Co, a chemical hypoxia reagent (Fig. 1A). Furthermore, Co exposure significantly enhanced LO promoter-driven reporter (luciferase) activities in cells cotransfected without (Fig. 1B) or with HIF-1 expression vectors (Fig. 2). The rat LO promoter region –804/–1 contains four putative HREs (Fig. 5). Using both the Co-treated cell nuclear extract (Fig. 6B) and the in vitro synthetic HIF-1 (Fig. 6D) as ligands, EMSA assays demonstrated that there is only one active HRE at –387/–383 for HIF-1 binding in the LO promoter −804/−1. Moreover, nonlabeled probe competition (Fig. 6C) and point mutation (Fig. 7) assays illustrated the specificity of this active site. These results suggest that the core HRE is important, but not unique, for the gene activation by hypoxia because several identical core HRE consensuses such as –37/–33, –194/–190, and –457/–453 in the rat LO promoter failed to bind with HIF-1 (Figs. 6B and D). In agreement with our findings, nonfunctional core HREs were also detected in other genes (Kaluz et al., 2008). Thus, it would seem reasonably to assume that in addition to the core HRE, the variable flanking sequences located at both sides of the core HRE (−387/−383) may also play a critical “helper” role in Co activation of the LO gene.
The HIF-1 is a major HRE binding protein composed of HIF-1α and HIF-1β subunits, of which HIF-1α acts as a key sensor for hypoxia (Kaluz et al., 2008). The HIF-1α contains a unique O2-dependent degradation domain (ODDD). Under normoxic conditions, enzymatic hydroxylation of two prolines within ODDD leads to degradation of the HIF-1α by the proteasome. The proline hydroxylation is catalyzed by prolyl hydroxylase domain proteins (PHDs) including PHD1, PHD2, and PHD3 (Bruick and McKnight, 2001). In the reaction, PHDs using Fe(II) and ascorbate as cofactors split molecular oxygen and transfer one oxygen atom to the proline residue (Ke and Costa, 2006). Thus, PHDs function as intracellular oxygen sensors providing the molecular basis for regulation of HIF-1α protein expression by cellular O2 partial pressure. Upon hypoxia, inactivation of PHDs results in cellular accumulation of HIF-1α, which is then translocated into the nucleus, coupled with HIF-1β, and bound to HREs for transactivating hypoxia-inducible genes. The HIF family contains three α subunits (1α, 2α, and 3α) and two β units (ARNT and ARNT2) (Nakayama, 2009). In addition, several alternatively spliced variants have also been identified. The DN-HIF-1α variant such as lacking exons 11 and 12 displayed severely damaged transcriptional activity (Chun et al., 2002). Note that in this study, the DN-HIF-1α was used as a tool to identify the specificity of the activation of the LO HRE by Co-induced hypoxia (Fig. 3).
The direct link and overlap of gene expressions and signal transcriptional patterns between hypoxia and Co exposure have led to use this metal ion as the mimic of hypoxia in the biological research (Epstein et al., 2001). Co ion as a chelator can compete with the nonhemo iron for binding to the active site of PHDs and deplete ascorbate, two cofactors of PHDs, thus inhibiting PHD catalytic activity and subsequently stabilizing the HIF-1α protein (Epstein et al., 2001; Salnikow et al., 2004). It should be noted that in the RFL6 cell model as shown in this study, cobaltous ion not only increased HIF-1α protein levels in nuclear extracts (compare lane 9 with lane 8 in Fig. 6B) but also enhanced HIF-1α gene expression at mRNA levels (Fig. 9). Co-enhanced expression of the HIF-1α gene has been proposed by activation of the PI3-kinase/Akt pathway (Ardyanto et al., 2006). Importantly, Co stimulation of the HIF-1α expression at protein and mRNA levels (Figs. 6B and 9) was associated with upregulation of LO (Fig. 4). Moreover, Co enhanced the HIF-1α binding to the LO HRE −387/−383 in RFL6 cells (Fig. 8). These results strongly support the conclusion that the activation of the HRE by HIF-1 plays a critical role in Co upregulation of LO (Fig. 1A).
Co is an essential metal, but overexposure of human to this metal compounds induces diseases in different organs (EPA, 2000). Co-elicited hypoxia response is implicated in lung toxicity and pathology (Cugell, 1992; Saini et al., 2010). DNA microarray studies established the causal links between hypoxia and fibrosis. Hypoxia activated a striking number of genes relevant to the fibrogenic pathogenesis including collagens and its modulating enzymes in pulmonary endothelial cells (Manalo et al., 2005). In this study, we further identified Co activation of the LO gene in rat lung fibroblasts via the HIF-1 pathway. LO is an amine oxidase critical for lung morphogenesis and tissue repair. It initiates the covalent crosslinking of collagen and elastin by oxidizing specific lysine residues in these proteins stabilizing the lung ECM (Li et al., 2011). As hypoxia mimics, Co compounds have been evaluated as a Group 2A or a Group 2B human carcinogen by IARC (Rousseau et al., 2005). Hypoxia is associated with lung solid tumors such as non-small cell lung cancer. Patients with high tumoral LO expression had significantly lower survival compared with those with low-expressing tumors (Le et al., 2007). Upregulation of LO facilitated hypoxia-induced epithelial-mesenchymal transition in different tumor cell lines (Sahlgren et al., 2008) and enhanced hypoxia-induced tumor cell migration and invasion (Erler et al., 2006). Thus, findings that Co activated the LO gene through the HIF-1 pathway provide a critical basis for understanding mechanisms for Co-induced lung fibrosis and cancer progression.
Cd is a toxic metal, but still widely used in industries. In addition to occupational exposure, cigarette smoke constitutes a major source of Cd exposure for humans (IARC, 1993). The lung is a major Cd-target organ with a biological half-life of 9.4 years (IARC, 1993). Long-term exposure to Cd resulted in emphysema (Davision et al., 1988). Pulmonary Cd levels in smokers with severe emphysema pathology reached 7.5-fold greater than those in nonsmokers (Pääkkö et al., 1989). Thus, Cd was listed by the U.S. Environmental Protection Agency as one of the 126 priority pollutants (National Toxicology Program, 2000). Our previous studies indicated that Cd exposure induced downregulation of LO and its substrates (collagen and elastin) in Cd-pulsed cells and in CdR cells (Zhao et al., 2006). Furthermore, such relationship of Cd exposure with downregulation of LO, collagen, and elastin was also confirmed in emphysematous lungs of the rat animal model (Zhao et al., 2010). Cd is a ROS inducer (Cuypers et al., 2010), whereas HIF-1α is a hypoxia sensor transcription factor (Kaluz et al., 2008). Cd inhibition of HIF-1 expression and gene targeting has been assessed in other genes such as EPO in response to hypoxia or Co (Horiguchi et al., 2000; Obara et al., 2003). Cd blocks the expression of hypoxia-responsive genes by enhancement of proteasome-dependent degradation of HIF-1α (Chun et al., 2000). In contrast, the contradictory report indicated Cd elevation of HIF-1 expression through ROS, ERK, and AKT pathways inducing malignant transformation of human bronchial epithelial cells (Jing et al., 2012). Hypoxia is an important complication associated with lung diseases and tumors (Weissmann, 2008). Reduced expression of HIF-1α has been determined in the emphysema lung tissues in severe COPD patients (Yasuo et al., 2011), suggesting that deregulation of HRE activities may occur in HIF-1α targeting genes such as LO under this condition. Cd-inhibited Co activation of the LO gene via modulation of the HIF-1α targeting in this study is consistent with the endpoint for Cd-induced downregulation of LO, which acts as a key mechanism for Cd emphysema pathology.
Briefly, in this study, we identified (1) a unique active HRE located at −387/−383 in the rat LO gene promoter using Co as a hypoxia mimic, (2) Co enhanced the expression of the LO gene through activation of the HIF-1 gene and its binding to the HRE in the LO gene in rat lung fibroblasts, and (3) Cd abolishment of Co biological activity toward HIF-1. These results are expected to enhance our understanding of mechanisms for Co-induced lung fibrosis and cancers and Cd-induced emphysema.
FUNDING
National Institutes of Health (R01-ES 11340).
ACKNOWLEDGMENTS
There are no conflicts of interest.
REFERENCES
- Ardyanto T. D., Osaki M., Tokuyasu N., Nagahama Y., Ito H. (2006). CoCl2-induced HIF-1alpha expression correlates with proliferation and apoptosis in MKN-1 cells: A possible role for the PI3K/Akt pathway. Int. J. Oncol. 29, 549–555 [PubMed] [Google Scholar]
- Bruick R. K., McKnight S. L. (2001). A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- Chen L. J., Zhao Y., Gao S., Chou I. N., Toselli P., Stone P., Li W. (2005). Downregulation of lysyl oxidase and upregulation of cellular thiols in rat fetal lung fibroblasts treated with cigarette smoke condensate. Toxicol. Sci. 83, 372–379 [DOI] [PubMed] [Google Scholar]
- Chun Y. S., Choi E., Kim G. T., Choi H., Kim C. H., Lee M. J., Kim M. S., Park J. W. (2000). Cadmium blocks hypoxia-inducible factor (HIF)-1-mediated response to hypoxia by stimulating the proteasome-dependent degradation of HIF-1alpha. Eur. J. Biochem. 267, 4198–4204 [DOI] [PubMed] [Google Scholar]
- Chun Y. S., Choi E., Kim T. Y., Kim M. S., Park J. W. (2002). A dominant-negative isoform lacking exons 11 and 12 of the human hypoxia-inducible factor-1alpha gene. Biochem. J. 362(Pt 1), 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugell D. W. (1992). The hard metal diseases. Clin. Chest Med. 13, 269–279 [PubMed] [Google Scholar]
- Cuypers A., Plusquin M., Remans T., Jozefczak M., Keunen E., Gielen H., Opdenakker K., Nair A. R., Munters E., Artois T. J, et al. (2010). Cadmium stress: An oxidative challenge. Biometals 23, 927–940 [DOI] [PubMed] [Google Scholar]
- Davision A. G., Newman-Taylor A. J., Darbyshire J., Chettle D. R., Guthrie C. J. G., O’Malley D., Mason H. J., Fayers P. M., Venables K. M., Pickering C. A. C, et al. (1988). Cadmium fume inhalation and emphysema Lancet 1, 663–667 [DOI] [PubMed] [Google Scholar]
- EPA (2000). Cobalt Compounds Available at: http://www.epa.gov/ttn/atw/hithef/cobalt.html Accessed on September 14, 2012.
- Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O’Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A, et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- Erler J. T., Bennewith K. L., Nicolau M., Dornhöfer N., Kong C., Le Q. T., Chi J. T., Jeffrey S. S., Giaccia A. J. (2006). Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 [DOI] [PubMed] [Google Scholar]
- Gao S., Zhao Y., Kong L., Toselli P., Chou I. N., Stone P., Li W. (2007). Cloning and characterization of the rat lysyl oxidase gene promoter: Identification of core promoter elements and functional nuclear factor I-binding sites. J. Biol. Chem. 282, 25322–25337 [DOI] [PubMed] [Google Scholar]
- Grosfeld A., Andre J., Hauguel-De Mouzon S., Berra E., Pouyssegur J., Guerre-Millo M. (2002). Hypoxia-inducible factor 1 transactivates the human leptin gene promoter. J. Biol. Chem. 277, 42953–42957 [DOI] [PubMed] [Google Scholar]
- Higgins D. F., Kimura K., Iwano M., Haase V. H. (2008). Hypoxia-inducible factor signaling in the development of tissue fibrosis. Cell Cycle 7, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi H., Kayama F., Oguma E., Willmore W. G., Hradecky P., Bunn H. F. (2000). Cadmium and platinum suppression of erythropoietin production in cell culture: Clinical implications. Blood 96, 3743–3747 [PubMed] [Google Scholar]
- IARC (1993). Beryllin, cadmium, mercury and exposures in the glass manufacturing industry. In Monographs on the Evaluation of Carcinogenic Risk to Humans, Vol. 58, pp. 119–236 IARC, Lyon, France: [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Liu L. Z., Jiang Y., Zhu Y., Guo N. L., Barnett J., Rojanasakul Y., Agani F., Jiang B. H. (2012). Cadmium increases HIF-1 and VEGF expression through ROS, ERK, and AKT signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol. Sci. 125, 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan H. M., Li W. (2003). Lysyl oxidase: Properties, specificity, and biological roles inside and outside of the cell. J. Cell. Biochem. 88, 660–672 [DOI] [PubMed] [Google Scholar]
- Kaluz S., Kaluzová M., Stanbridge E. J. (2008). Regulation of gene expression by hypoxia: Integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin. Chim. Acta 395, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Q., Costa M. (2006). Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 70, 1469–1480 [DOI] [PubMed] [Google Scholar]
- Kenyon K., Contente S., Trackman P. C., Tang J., Kagan H. M., Friedman R. M. (1991). Lysyl oxidase and rrg messenger RNA. Science 253, 802 [DOI] [PubMed] [Google Scholar]
- Le Q.-T., Erler J. T., Giaccia A. (2007). Tumor hypoxia and metastasis in non-small cell lung cancers: M02-M03. J. Thoracic Oncol. 2, S154–S155 [Google Scholar]
- Li W., Chou I. N., Boak A., Kagan H. M. (1995). Downregulation of lysyl oxidase in cadmium-resistant fibroblasts. Am. J. Respir. Cell Mol. Biol. 13, 418–425 [DOI] [PubMed] [Google Scholar]
- Li W., Zhou J., Chen L., Luo Z., Zhao Y. (2011). Lysyl oxidase, a critical intra- and extra-cellular target in the lung for cigarette smoke pathogenesis. Int. J. Environ. Res. Public Health 8, 161–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalo D. J., Rowan A., Lavoie T., Natarajan L., Kelly B. D., Ye S. Q., Garcia J. G., Semenza G. L. (2005). Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay C. K., Mazumder B., Fox P. L. (2000). Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J. Biol. Chem. 275, 21048–21054 [DOI] [PubMed] [Google Scholar]
- Nakayama K. (2009). Cellular signal transduction of the hypoxia response. J. Biochem. 146, 757–765 [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (2000). Ninth report on carcinogens. Department of Health and Human Services, Research Triangle Park, NC: [Google Scholar]
- Obara N., Imagawa S., Nakano Y., Suzuki N., Yamamoto M., Nagasawa T. (2003). Suppression of erythropoietin gene expression by cadmium depends on inhibition of HIF-1, not stimulation of GATA-2. Arch. Toxicol. 77, 267–273 [DOI] [PubMed] [Google Scholar]
- Pääkkö P., Kokkonen P., Anttila S., Kalliomäki P. L. (1989). Cadmium and chromium as markers of smoking in human lung tissue. Environ. Res. 49, 197–207 [DOI] [PubMed] [Google Scholar]
- Richard D. E., Berra E., Pouyssegur J. (2000). Nonhypoxic pathway mediates the induction of hypoxia-inducible factor 1alpha in vascular smooth muscle cells. J. Biol. Chem. 275, 26765–26771 [DOI] [PubMed] [Google Scholar]
- Rousseau M. C., Straif K., Siemiatycki J. (2005). IARC carcinogen update. Environ. Health Perspect. 113, A580–A581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlgren C., Gustafsson M. V., Jin S., Poellinger L., Lendahl U. (2008). Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. U.S.A. 105, 6392–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini Y., Kim K. Y., Lewandowski R., Bramble L. A., Harkema J. R., Lapres J. J. (2010). Role of hypoxia-inducible factor 1{alpha} in modulating cobalt-induced lung inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 298, L139–L147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K., Donald S. P., Bruick R. K., Zhitkovich A., Phang J. M., Kasprzak K. S. (2004). Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. J. Biol. Chem. 279, 40337–40344 [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2010). Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann N. (2008). Hypoxia-driven mechanisms in lung biology and disease: A new review series of the ERS Lung Science Conference. Eur. Respir. J. 31, 697–698 [DOI] [PubMed] [Google Scholar]
- Yasuo M., Mizuno S., Kraskauskas D., Bogaard H. J., Natarajan R., Cool C. D., Zamora M., Voelkel N. F. (2011). Hypoxia inducible factor-1α in human emphysema lung tissue. Eur. Respir. J. 37, 775–783 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chen L., Gao S., Toselli P., Stone P., Li W. (2010). The critical role of the cellular thiol homeostasis in cadmium perturbation of the lung extracellular matrix. Toxicology 267, 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Gao S., Chou I. N., Toselli P., Stone P., Li W. (2006). Inhibition of the expression of lysyl oxidase and its substrates in cadmium-resistant rat fetal lung fibroblasts. Toxicol. Sci. 90, 478–489 [DOI] [PubMed] [Google Scholar]