Abstract
Light-stimulated adjustment of the circadian clock is an important adaptive physiological response that allows maintenance of behavioral synchrony with solar time. Our previous studies indicate that the aryl hydrocarbon receptor (AhR) agonist 2,3,7,8- tetrachlorodibenzo-p-dioxin attenuates light-induced phase resetting in early night. However, the mechanism of inhibition remains unclear. In this study, we showed that another potent AhR agonist—β-naphthoflavone (BNF)—significantly decreased light-induced phase shifts in wild-type (WT) mice, whereas AhR knockout mice had an enhanced response to light that was unaffected by BNF. Mechanistically, BNF blocked light induction of the Per1 transcript in suprachiasmatic nucleus and liver in WT mice, and BNF blocked forskolin (FSK)-induced Per1 transcripts in Hepa-1c1c7 (c7) cells. An E-box decoy did not affect BNF inhibition of FSK-induced Per1 transcripts in c7 cells. cAMP-response element (CRE)-dependent induction of Per1 promoter activity in response to FSK in combination with phorbol 12-tetradecanoate 13-acetate was suppressed in cells that expressed high levels of AhR (c7) compared with cells lacking functional AhR activity (c12). In addition, the inhibitory effect of BNF on FSK-induced Per1 was dependent on phosphorylation of JNK. Together, these results suggest that AhR activation inhibits light-induced phase resetting through the activation of JNK, negative regulation of CREs in the Per1 promoter, and suppression of Per1.
Key Words: aryl hydrocarbon receptor, Period1, phase shift, JNK, cAMP-response elements.
Mammals generate and maintain physiological and behavioral rhythms with approximately 24h periodicity due to molecular oscillations in the suprachiasmatic nucleus (SCN) of the hypothalamus (Bendova and Sumova, 2006; Meijer et al., 1998; Pittendrigh and Daan, 1974; Ralph et al., 1990). Interacting transcriptional-translational feedback loops engendered by a core group of clock genes provide the underlying molecular framework for this circadian rhythmicity (Ko and Takahashi, 2006; Lowrey and Takahashi, 2004; Reppert and Weaver, 2002). The core clock components CLOCK and BMAL1 form a heteromeric complex that binds to E-boxes in the promoters of the Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes, thereby driving their expression; Per and Cry proteins, in turn, enter the nucleus to inhibit the CLOCK-BMAL1-mediated gene transcription (Kume et al., 1999; Zheng et al., 2001).
On the other hand, the circadian clock period is not exactly 24h, so internal clocks require daily adjustments to reset appropriately to match with environmental time (Challet et al., 2003). Light is the most prominent zeitgeber used to synchronize the organism with the external environment. Light-induced phase resetting in the SCN is mediated primarily by glutamatergic neurotransmission in response to retinal light information (Tischkau et al., 2003). During the subjective night, a single light pulse is sufficient to cause phase resetting through increasing Ca2+-cAMP-mediated cAMP-response element (CRE)-binding protein (CREB) phosphorylation and activation of CRE-mediated transcription (Ding et al., 1997). Light-induced resetting of locomotor activity and glutamate-induced resetting of SCN firing rate rhythms can be blocked by mPer1 antisense oligonucleotides, and this response to light does not occur in mPer1-deficient mice (Akiyama et al., 1999; Albrecht et al., 2001). CRE elements in the Per1 promoter are required for this response. Thus, Per1 is essential both for conveying the light-entrainment signal and for generation of circadian rhythm.
Our previous study demonstrated that aryl hydrocarbon receptor (AhR) activation inhibits basal Per1 expression and alters the endogenous rhythm of Per1 through repressing CLOCK/BMAL1 activity at E-box elements (Xu et al., 2010). We have also demonstrated that 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) exposure significantly reduces phase shifting responses to light pulses during the early night in vivo (Mukai et al., 2008). This study was undertaken to determine the underlying mechanism by which AhR activation attenuates responsiveness to nocturnal light.
Here, we show that a potent agonist of AhR, β-naphthoflavone (BNF), reduces the phase resetting induced by a light pulse in early night and attenuates Per1 gene induction in SCN, liver, and hepatoma cells. In addition, BNF increases JNK phosphorylation that inhibits CRE activity and CRE-mediated Per1 gene induction. These findings demonstrate a novel mechanism through which AhR activation by ubiquitous environmental pollutants or chemotherapy agents represses circadian clock resetting. Discordance between the internal clock and the external environment likely contributes to sleep disorders and fatigue.
MATERIALS AND METHODS
Cell culture.
Hepa-1c1c7 (c7) cells, and their c12 and c4 derivatives, were obtained from ATCC. Cells were cultured in Dulbecco’s Modified Eagle’s Medium Reduced Serum (DMEM-RS, HyClone) with 7.5% bovine growth serum (HyClone), penicillin/streptomycin/amphotericin (MP Biomedicals) at 37°C in a humidified 5% CO2 atmosphere. Cultures were treated with BNF (Sigma), forskolin (FSK, Fisher Scientific), phorbol 12-tetradecanoate 13- acetate (TPA, Sigma), ERK inhibitor (PD98059, Sigma), p38 inhibitor (SB203580, Calbiochem), and JNK inhibitor (SP600125, Calbiochem) in dimethyl sulfoxide (DMSO). Control cultures were treated with equivalent concentrations of the DMSO vehicle.
Animals and light exposure.
Six- to twelve-week-old male wild-type (WT) mice c57Bl/6J and AhR knockout (AhRKO) mice (from Richard Peterson, University of Wisconsin; Schmidt et al., 1996) were used in these experiments. Animal protocols were approved by the Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Except for those used in behavioral analysis (described subsequently), mice were housed 3–4 per cage in a light-tight chamber, provided with food and water ad libitum, and entrained for 1–2 weeks under controlled lighting (12L:12D, LD), temperature (22°C), and humidity (39%). After 1–2 weeks of entrainment in LD, animals were transferred into constant darkness (DD) for 7–10 days. At circadian time 0 (CT0 refers to the time lights were turned on in the previous light dark cycle, and CT12 refers to the time lights went off), mice were ip injected with BNF at 100 μg/kg body weight or vehicle (corn oil). One week later, a light pulse was performed in a separate chamber for 30min at CT16, following which mice were decapitated. Brains were fixed in 4% formaldehyde for in situ hybridization. Livers were snap frozen and stored at −80°C until use.
Wheel running and light pulse.
Detailed procedures for behavioral analysis of rhythms in the AhRKO mice have been described in our previous study (Mukai et al., 2008). In brief, WT and AhRKO mice were provided with 4.5-in. diameter activity wheels. After entrainment to a LD schedule for 7–14 days, the mice were placed in DD for 7–10 days. At CT0, mice were injected ip with 100 μg/kg BNF or vehicle, and 1 week later a 30-min (500 lx) light pulse was given at CT16 (where CT12 is defined as the time the animals start running on the wheel in DD). Wheel running activity data were not collected during the treatment. After 30min, the cage was returned back to DD, and collection of activity data was reinitiated for 7–10 days. Behavioral analysis of period before and after treatment was performed using Clocklab software (Actimetrics, Evanston, IL) as previously described (Mukai et al., 2008; Tischkau et al., 2000, 2003). Phase shifting of the circadian clock was assessed after re-establishment of a stable period by measuring the difference in activity onset on the day before the light pulse in comparison to the maximal phase delay at 2–3 days after the light pulse and restabilization of period.
qPCR.
Total liver or cellular RNA was isolated using TRI-Reagent (MRC). Total RNA (2 μg) was added to 25 μl of reaction mixture with reverse transcriptase (Promega). Five microliters of diluted cDNA were then used in 20 μl of SYBR Green PCR mix (Quanta) containing 300nm Per1; Bmal1-specific primer pairs as previously published (Mukai and Tischkau, 2007); or 1× QuantiTect primer assay for Per2, clock, Cry1, Cry2, c-fos, or cyp1a1 gene expression analysis (Qiagen). Amplification was performed using a Smart Cycler rapid thermal cycler (Cepheid) according to the following protocol: an initial 10min denaturation step at 95°C followed by 40 cycles of denaturation (95°C) for 30 s, annealing (primer-optimized temperature) for 30 s, and extension (72°C) for 30 s. Detection of the fluorescent product was carried out during each 72°C extension period, and emission data were quantified using threshold cycle (Ct) values. Relative standard curves were created as previously described (Karman and Tischkau, 2006) and were used to calculate the relative amount of all genes. Relative gene expression fold change was calculated after normalization by actin as well as using standard curve. PCR product specificity from each primer pair was confirmed using melting curve analysis and subsequent agarose gel electrophoresis.
Immunoblot analysis.
Total cellular protein was extracted using Complete Lysis-M (Roche), and liver protein was isolated using T-PER (ThermoScientific). Protein concentrations were determined with a BCA kit (ThermoScientific), and 40–80 μg of protein was separated with SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (Bio-Rad). Blots were blocked with 5% nonfat milk for 1h and then incubated with a specific primary antibody overnight at 4°C. Blots were incubated with secondary antibody (LI-COR Bioscience) for 1h at room temperature following washes. Images were taken by LI-COR imaging system (LI-COR Bioscience). AhR antibody was from BIOMOL, β-actin antibody was from Sigma, and all other primary antibodies were from Cell Signaling. Densitometry was performed using Quantity One (Bio-Rad) software.
In situ hybridization.
The brains were fixed overnight at 4°C in 4% paraformaldehyde, followed by cryoprotection in 20% sucrose and sectioned at 10 µm on a cryostat. Hybridization was performed using digoxygenin-labeled riboprobes as described previously (Karman and Tischkau, 2006).
Luciferase assay.
mPer1 promoter regions and oligonucleotides corresponding to the CRE-mPer1 luciferase reporter were gifts from Dr Paolo Sassone-Corsi at the University of California at Irvine. Cotransfection experiments were performed with c7 and c12 cells. Cells were plated in 24-well plates in DMEM-RS, supplemented with 7.5% bovine growth serum. Transfection experiments were performed using lipofectamine (Invitrogen) according to the manufacturer’s protocol. Transfection mixtures generally contained 1 μg of mPer1-Luc or CRE-Luc, and the internal control pRL-TK vector (10ng). Transfected cells were cultured for an additional 24h, washed once with cold PBS, and resuspended in passive lysis buffer (Promega). The reporter gene firefly luciferase activity was initiated by mixing an aliquot of lysates (20 μl) with Luciferase Assay Reagent II (Promega). Then, Renilla activity was detected. Luciferase was normalized to the Renilla signal.
Decoy transfection.
Two hundred nanomoles of E-box sense sequence: TTTAGCCACGTGACAGTGTAACGACACGTGGGCCCTCAAGTCC‑ ACGTGCAGGGA and antisense sequence: TCCCTGCACGTGGACTT‑ GAGGGCCCACGTGTCGTTACACTGTCAC-GTGGCTAAA and corresponding mutant sense sequence: TTTAGCCTGGTGAC‑ AGTGTAAC-GA-CTGGTGGGCCCTCAAGTCCTGGTGCAGGGA and antisense sequence: TCCCTGCAC-CAGGACTTGAGGGCCCACCAGT CGTTACACTGTCCCAGGCTAAA were synthesized by Invitrogen. These sense and antisense sequences were mixed, then heated for 5min at 100°C, and slowly cooled down to room temperature for annealing. These oligonucleotides of 200nM were transfected using Lipofectamine (Invitrogen) and incubated overnight. These cells were treated with BNF, FSK, or BNF + FSK and harvested for total RNA.
Statistics.
Statistical analysis was performed by two-way or one-way ANOVA with Tukey’s post hoc comparison or two-sided Student’s t-test using SPSS 16.0. The p values were considered statistically significant at p < 0.05. All data are shown as mean ± SEM.
RESULTS
AhR Activation Reduced Light-Induced Phase Resetting
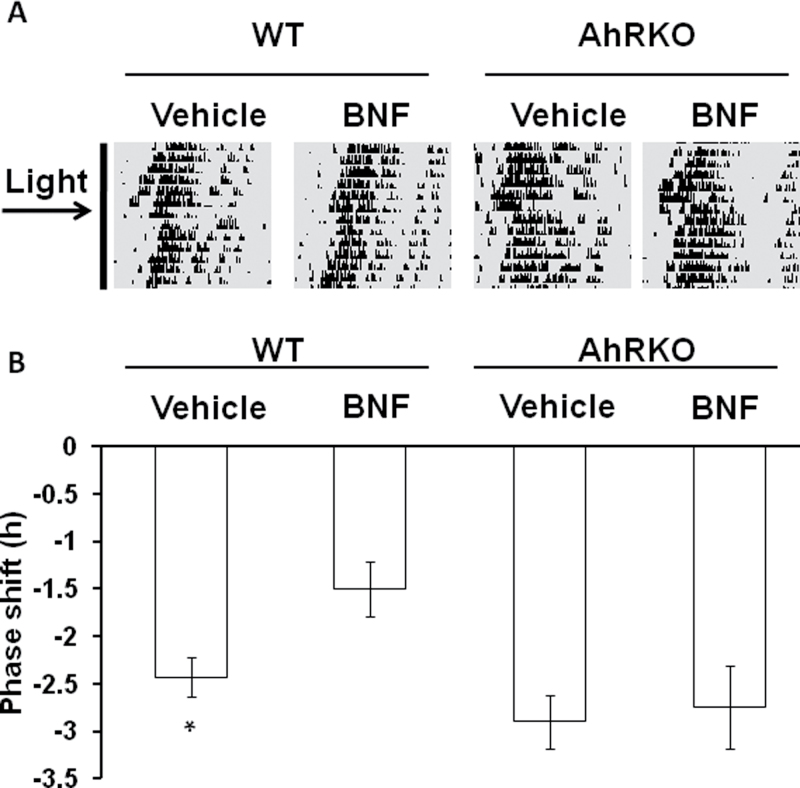
The effects of AhR on light-induced phase resetting of the behavioral circadian rhythm were assessed using wheel running in BNF- and vehicle-treated WT and AhRKO mice (Fig. 1A). In WT mice, the light pulse–induced phase shift was reduced following BNF treatment compared with vehicle-treated controls (Fig. 1B), and WT mice displayed mildly smaller phase shifts than AhRKO mice. In addition, BNF treatment did not change the phase shifts in the AhRKO mice (Fig. 1B), suggesting that the effect of BNF on light-induced phase shift is dependent on AhR. BNF or light treatment did not affect the free-running period of the mice.
FIG. 1.
AhR activation reduces light-induced phase shifts. (A) Representative actogram of wheel-running activity. Mice were entrained to a 12L:12D schedule and then were placed under DD for 7–10 days. Mice were injected with BNF or vehicle at 100 μg/kg of body weight; then after 1 week, mice were given a 30-min light pulse at CT16 as shown by the horizontal arrow. (B) Phase shift after light exposure at CT16 of vehicle- and BNF-treated WT and AhRKO mice. n = 6–8 per group, *p < 0.05, comparing WT with BNF and AhRKO vehicle–treated mice by t-test.
AhR Activation Represses Light-Induced Per1 Gene Expression
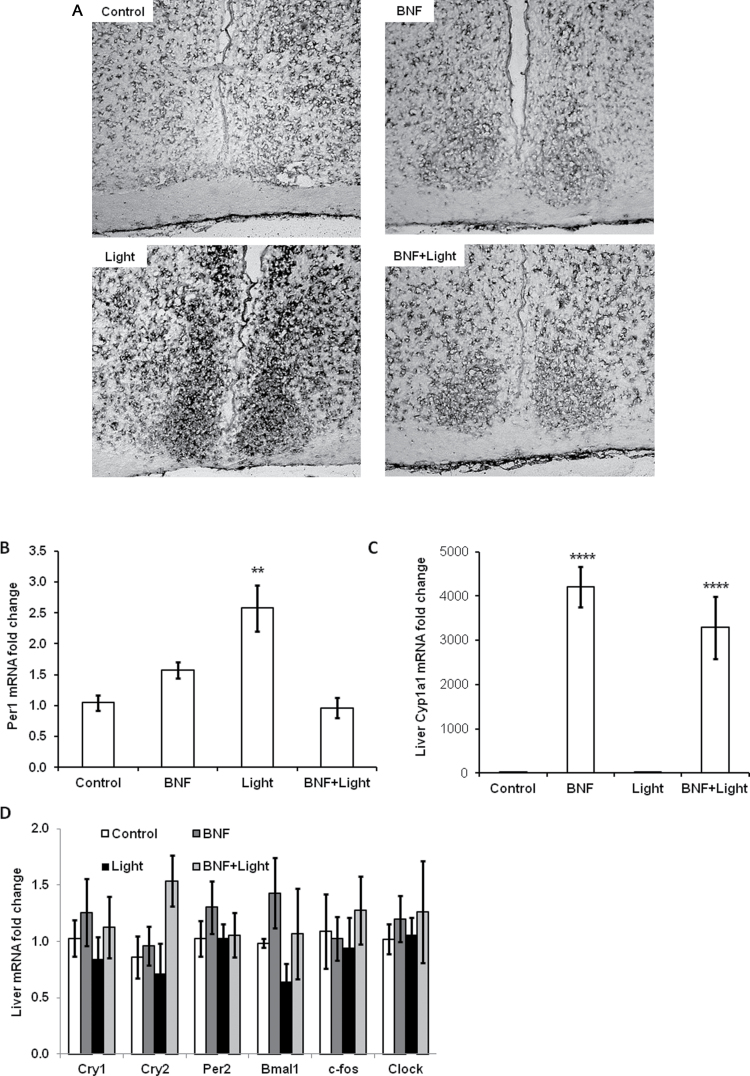
Per1 gene induction is the major molecular mechanism that mediates light-induced phase resetting during the subject night (Akiyama et al., 1999; Albrecht et al., 2001). To determine whether the effect of AhR activation on light-induced phase resetting is associated with reduction of Per1 gene induction, Per1 transcripts were measured in SCN using in situ hybridization after light pulse and/or BNF treatment. Per1 transcripts were induced in the SCN by the light pulse during the early night, and BNF treatment substantially reduced the light-induced Per1 transcripts in SCN (Fig. 2A).
FIG. 2.
AhR activation blocks the Per1 induction in SCN and liver following a light pulse at night. Mice were entrained to a 12L:12D schedule and then were placed under DD for 7–10 days. Mice were injected with BNF or vehicle at 100 μg/kg of body weight; then after 1 week, mice were given a 30-min light pulse at CT16 and then were sacrificed. The whole brain was fixed in DD. (A) In situ hybridization was performed with a digoxygenin-labeled cDNA probe to detect Per1 mRNA in SCN. Representative images are shown; experiment was repeated for total n = 6–8. (B) qPCR for Per1 in liver, n = 7–10 per group. (C) qPCR for cyp1a1 in liver, n = 6–7 per group. (D) qPCR for clock genes in liver, n = 3–4 per group. Results are shown as fold change relative to control, mean ± SEM, **p < 0.01 and ****p < 0.0001 by one-way ANOVA with Tukey’s post hoc comparison.
The master clock in the SCN synchronizes peripheral clock through endocrine and neuronal connections to ensure internal synchrony of mammalian physiological behavior and metabolic rhythms. Light pulses also influence expression of peripheral organ clock genes during subject night (Cailotto et al., 2009). Hepatic Per1 gene expression was significantly increased after the light pulse, and this increase was blocked by BNF treatment (Fig. 2B). As a positive control, AhR target gene cyp1a1 was raised 3000- to 4000-fold with BNF treatment in the liver regardless of whether there was a light pulse (Fig. 2C). In addition, Per1 was the only circadian clock gene whose transcript was altered in the liver following a light pulse; no changes in Cry1, Cry2, Per2, Bmal1, c-fos, or clock were detected (Fig. 2D). These results indicate that BNF represses light-induced Per1 gene expression in the SCN and in the liver (Figs. 2A and 2B), providing a potential mechanism for the reduction of light-induced phase shifts (Fig. 1).
AhR Activation Inhibits FSK-Induced Per1 Gene Expression
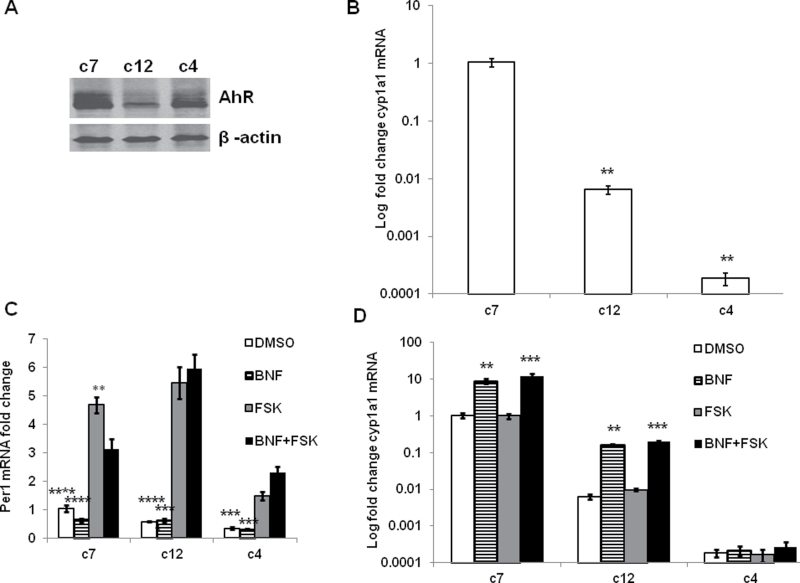
Hepatoma cell lines c7, c12, and c4 were utilized to evaluate the molecular mechanisms by which AhR/ARNT signaling contributes to the inhibition of light-induced Per1 in response to BNF. As indicated in Figure 3A, c7 and c4 cells express high levels of AhR protein, whereas AhR protein levels are dramatically reduced in c12 cells; c4 cells lack functional ARNT (Numayama-Tsuruta et al., 1997). Thus, the AhR/ARNT target gene, cyp1a1, is significantly lower in both c12 and c4 than in c7 cells (Fig. 3B). These results indicate that the AhR/ARNT signaling pathway is intact in c7 cells but not in c12 or c4 cells.
FIG. 3.
AhR activation reduces FSK-induced Per1 levels. (A) Immunoblot was performed to determine basal level of AhR in c7, c12, and c4 cell lines. (B) qPCR was used to test the basal level of AhR target gene cyp1a1 mRNA in c7, c12, and c4 cells. All values were normalized to c7 values, with the fold change of cyp1a1 mRNA expressed on a logarithmic scale because of the large variation in values. n = 3, **p < 0.01, compare c7 control with others by one-way ANOVA with Tukey’s post hoc comparison. (C) c7, c12, and c4 cells were treated with 10μM of BNF for 30min and then 10μM of FSK for 1h. Per1 mRNA was examined by qPCR. n = 5, **p < 0.01 comparing BNF + FSK or BNF group with FSK or DMSO group by Student’s t-test. (D) cyp1a1 was tested in c7, c12, and c4 cells treated as in C. Results are expressed with fold change cyp1a1 mRNA on a logarithmic scale. β-Actin is used as internal control for qPCR. n = 3, **p < 0.01 and ***p < 0.001 comparing DMSO with others within a cell type by one-way ANOVA with Tukey’s post hoc comparison.
FSK mimics the effect of a light pulse on Per1 by stimulating adenylate cyclase, generating cAMP and increasing Per1 (Travnickova-Bendova et al., 2002). Figure 3C clearly shows that FSK increased the Per1 transcript from DMSO control in c7, c12, and c4 cells. BNF treatment decreased basal levels of the Per1 transcript only in c7 cells, which is consistent with our previous report (Xu et al., 2010). Interestingly, pretreatment with BNF significantly reduced FSK-induced Per1 transcript in c7 cells but not in c12 and c4 cells. These results reaffirm that BNF reduces light-induced Per1 expression through the AhR/ARNT signaling pathway.
The AhR/ARNT target gene cyp1a1 was examined to further evaluate the intact AhR/ARNT signaling pathway. c12 cells displayed a lower basal and BNF-induced expression of cyp1a1 compared with c7 cells, and cyp1a1 was not induced by BNF in c4 cells as shown in Figure 3D. Because of the widely disparate values of cyp1a1 in the three cell lines (likely related to the functionality of the AhR pathway), these data are plotted on a semilog graph and cannot be analyzed by two-way ANOVA. One-way ANOVA was used for each cell type and demonstrated significant increases in cyp1a1 with BNF and BNF + FSK in c7 and c12 cells but not in c4 cells. These data further demonstrate that c12 and c4 cells lack a functional AhR/ARNT signaling pathway.
AhR Activation Reduces FSK-Induced Per1 Through CRE but Not Through E-box Elements
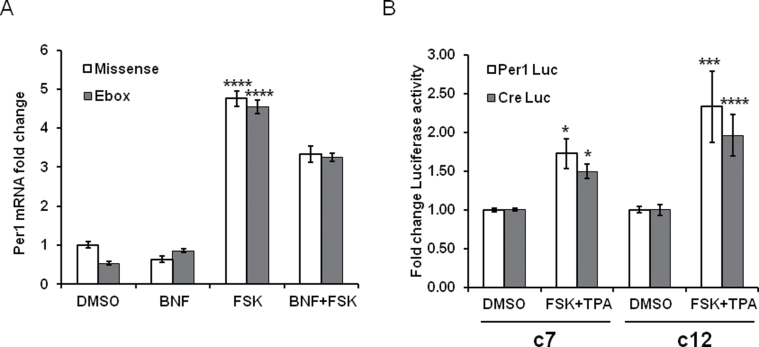
Previously, we demonstrated that AhR disrupts BMAL1/CLOCK activity and attenuates the endogenous Per1 rhythm by forming complex with BMAL1 (Xu et al., 2010). In the next experiment, we used an E-box decoy to exclude that AhR inhibits FSK-induced Per1 through interaction with CLOCK/BMAL1 binding E-boxes. A double-stranded oligonucleotide E-box decoy was employed to block activation of the E-box elements in the Per1 promoter. The increase in Per1 levels following FSK treatment was not changed by the addition of the E-box decoy. BNF reduced the FSK induction of Per1 transcript levels, and addition of the E-box decoy did not alter BNF-induced inhibition (Fig. 4A).
FIG. 4.
AhR activation reduces FSK-induced Per1 through CRE but not through E-box elements. (A) c7 cells were seeded in a 12-well plate. After 24h, E-box decoy or mis-sense ODN was transfected into cells with Lipofectamine. After overnight incubation, cells were pretreated for 30min with 10µM of BNF or vehicle and then exposed to 10µM of FSK or vehicle for 1h. Per1 mRNA was examined by qPCR. n = 3, ****p < 0.0001 comparing FSK with all other groups by two-way ANOVA using Bonferroni’s post hoc comparison. (B) c7 and c12 cells were transfected with Per1-Luc or CRE-Luc using lipofectamine. The Per1-Luc served as a positive control. After overnight incubation, cells were treated with 100ng/ml of TPA + 10μM of FSK for 6h. Promoter activity was evaluated by luciferase assay. Firefly activity was normalized to Renilla luciferase. n = 3, *p < 0.05, ***p < 0.001, and ****p < 0.0001 comparing DMSO with FSK + TPA for both cell types and luciferase constructs with two-way ANOVA and Bonferroni’s post hoc comparison.
Light and/or FSK induction of Per1 is dependent on the cAMP-CREB-CRE pathway. To verify whether the inhibitory effects of BNF on FSK-induced Per1 transcripts occur via the CRE elements, we performed a luciferase reporter assay. The Per1 promoter (Per1-Luc) and only CRE-binding site elements (CRE-Luc) luciferase reporter constructs were transfected into c7 or c12 cells. Both Per1-Luc and CRE-Luc activities were increased after FSK and TPA exposure (1.73±0.19 and 1.50±0.09, respectively, in c7 cells compared with DMSO Per1-Luc and CRE-Luc activities of 1.00±0.02 and 1.00±0.02, respectively, both p < 0.01, Fig. 4B). Per1-Luc and CRE-Luc were increased in both cell lines. Per1-Luc and CRE-Luc were increased significantly more in the c12 cells that lack a functional AhR (2.33±0.45 and 1.96±0.27, respectively, in c12 cells compared with DMSO Per1-Luc and CRE-Luc activities of 1.00±0.04 and 1.00±0.07, respectively, p < 0.001 for Per1-Luc and p < 0.0001 for CRE-Luc, Fig. 4B) suggesting that AhR activation may obstruct CRE activity induced by FSK.
AhR Reduces FSK-Induced Per1 Transcripts Through the JNK Pathway
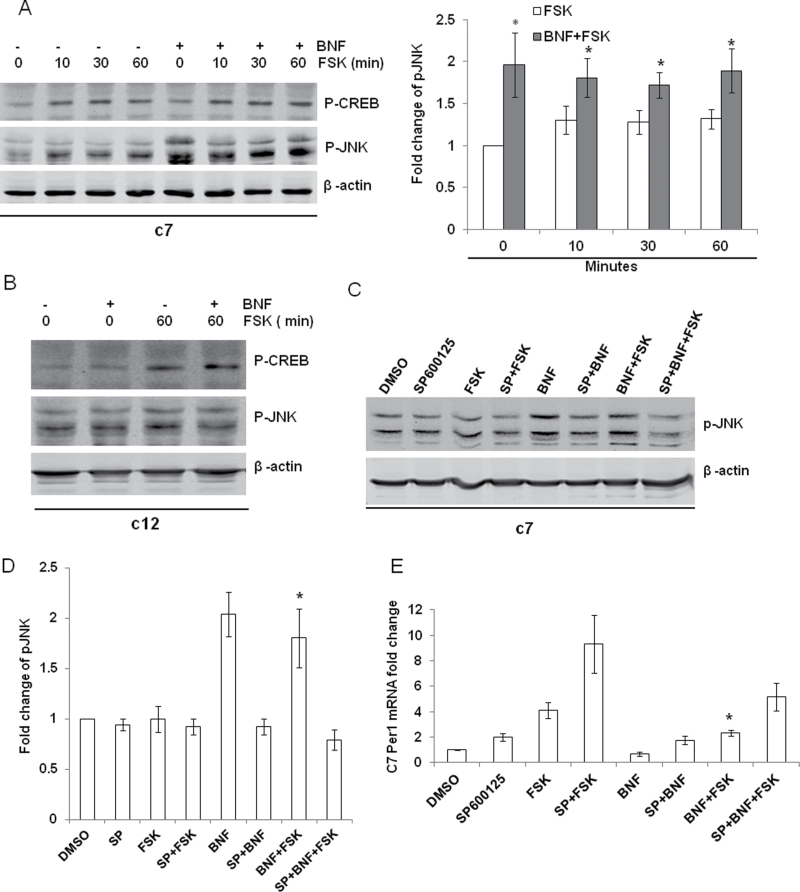
Light exposure acts through the MAPK pathway leading to phosphorylated CREB (p-CREB), increased CRE activity, and induction of Per1 transcripts (Obrietan et al., 1999). To investigate the signaling pathway involved in the inhibitory effect of AhR on FSK-induced Per1, we examined p-CREB after BNF and/or FSK treatment. As shown in Figures 5A and 5B, p-CREB was increased by FSK treatment and the increase was maintained for up to 60min in both c7 and c12 cells. BNF had no effect on the p-CREB induction by FSK in either cell line. These data demonstrate that BNF does not inhibit FSK-induced Per1 through interfering with p-CREB.
FIG. 5.
AhR activation reduces FSK-induced Per1 through the JNK pathway. (A) c7 cells were pretreated for 30min with vehicle or 10µM of BNF and then were treated with 10μM of FSK for 0, 10, 30, and 60min. *p < 0.05 compared with FSK group at individual different time points with Student’s t-test. (B) c12 cells were pretreated for 30min with vehicle or 10µM of BNF and then were treated with 10μM of FSK for 0 or 60min. (C) c7 cells were treated for 30min with vehicle or pJNK inhibitor 10μM of SP600125, then for another 30min with vehicle or 10µM BNF, and finally with vehicle or 10µM FSK for 1h. Protein was collected and immunoblot performed for pCREB and p-JNK (representative blot). (D) Densitometry for immunoblotting experiment described in C. (E) Cells were treated as in C; RNA was collected; and qPCR was performed to examine Per1. Immunoblot was performed to examine p-CREB and p-JNK. β-Actin is used as internal control for immunoblot and qPCR. n = 3–5, *p < 0.05 compared with FSK or SP + BNF + FSK by one-way ANOVA with Tukey’s post hoc comparison for C and D.
It has been reported that c-Jun binds to CRE and inhibits its activity (Zhang et al., 2004) and the AhR agonist TCDD activates JNK in an AhR-dependent manner (Diry et al., 2006; Weiss et al., 2005, 2008), but it is also reported that TCDD activates ERK and JNK independent of AhR (Hoffer et al., 1996; Puga et al., 1992; Tan et al., 2002). We evaluated the effect of BNF on phosphorylation of JNK (p-JNK). In both c7 and c12 cells, FSK treatment slightly increased p-JNK. BNF + FSK treatment further increased p-JNK over the level from FSK treatment alone in c7 cells but not in c12 cells (Figs. 5A and 5B). To determine whether this AhR-dependent increase in p-JNK can explain the inhibitory effect of BNF on FSK-induced Per1, the p-JNK inhibitor SP600125 was used in c7 cells. BNF was unable to increase p-JNK in the presence of SP600125 (Fig. 5C), and SP600125 significantly reversed the ability of BNF to inhibit the FSK-induced Per1 (Fig. 5D).
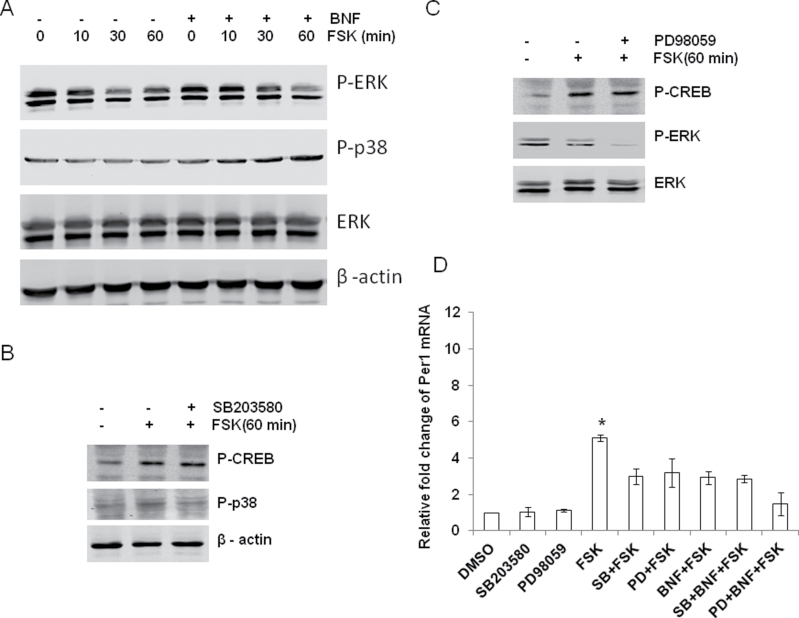
AhR activation also affects p38 and ERK (Park et al., 2005; Tan et al., 2002); thus, we examined their response to FSK and BNF + FSK stimulation. The phosphorylation of p38 (p-p38) was not changed after FSK exposure, but addition of BNF increased p-p38 in a time-dependent manner (Fig. 6A). The addition of BNF had no significant effect on phosphorylation of ERK (Fig. 6A). As seen in Figure 6D, the inhibition of BNF on FSK-induced Per1 was not reversed by specific inhibitors of either the p38 MAPK cascade (SB203580) or the ERK MAPK cascade (PD98059). Neither inhibitor affected the increase in p-CREB caused by FSK (Figs. 6B and 6C). BNF inhibited FSK-induced Per1 transcripts more than either of these inhibitors alone (3.20±0.29 and 2.98±0.34, respectively, for SB203580 and PD98059, p < 0.05 compared with FSK alone 5.11±0.41) suggesting that these inhibitors may act through AhR to inhibit FSK induction (Fig. 6D). Together, these data suggest that BNF blocks the FSK-induced Per1 through an increase in p-JNK.
FIG. 6.
MAPK had no significant role in the inhibition of BNF on FSK-induced Per1. (A) c7 cells were pretreated for 30min with vehicle, or 10µM of BNF for 30min, and then cells were treated with either vehicle or 10μM of FSK 1h. (B and C) c7 cells were pretreated for 30min with vehicle, or 10μM of SB203580 (a p38 inhibitor, p38 I), or 10μM of PD98059 (an ERK inhibitor, ERK I), and then cells were treated with 10μM of FSK for 60min. (D) c7 cells were pretreated with SB203580 or PD98059, then cells were treated for 30min with vehicle or 10µM BNF, and finally cells were treated with FSK for 60min. Immunoblot was performed to examine the phosphorylation of ERK and p38. Real-time PCR was performed to test Per1 mRNA. β-Actin is used as internal control for immunoblot and qPCR. n = 3–5, *p < 0.05 compared with other groups with Student’s t-test.
DISCUSSION
Previously, we demonstrated that the response of the circadian clock to light was altered in mice that had been exposed to TCDD 8 days prior to the light exposure (Mukai et al., 2008). Recently, we have also shown that TCDD treatment can alter the clock’s endogenous rhythm (Xu et al., 2010). Similar to our previous findings, another AhR agonist, BNF, also reduced the phase shift after a light pulse in early night in WT mice. To test whether this effect was AhR dependent, we treated AhRKO mice with BNF and light pulses. The phase shift in AhRKO mice was unaffected by BNF treatment. These results demonstrate that BNF acts through AhR in its effects on light-induced phase shifts. Furthermore, these data suggest that AhR activation affects the signaling pathway through which light resets the circadian clock.
Induction of the Per1 transcript is required for light-induced phase resetting of the circadian clock both in vivo and in vitro (Akiyama et al., 1999; Shigeyoshi et al., 1997; Tischkau et al., 2003). Our current results support an induction of the Per1 transcript along with a phase delay in response to early night light exposure. Furthermore, we have demonstrated that AhR activation by BNF inhibits the increase in Per1 levels caused by light and significantly reduces the light-induced phase delay in vivo, suggesting a modulatory effect of AhR activation on light-induced phase shifting through inhibition of the Per1 transcript.
It can be argued that the most important function of the SCN is to decode environmental light signals and communicate these changes to the rest of the organism. In this manner, the SCN acts as the master clock for the body, communicating changes in environmental lighting to the peripheral organs through autonomic or endocrine signals (Buijs et al., 1998; de la Iglesia et al., 1995; Kennaway et al., 2002; Nagai et al., 1996). Nocturnal light exposure immediately affects corticosterone and melatonin secretion, locomotor activity, body temperature, heart rate, and glucose metabolism (Buijs et al., 1999; Ishida et al., 2005; Klein and Weller, 1972; Loh et al., 2008; Scheer et al., 2001).
The Per genes play an essential role in allowing the SCN clock to adapt to changes in environmental lighting; increased Per expression after light stimuli synchronizes peripheral rhythms to the central clock (Bae et al., 2001; Shearman et al., 2000; Zheng et al., 1999, 2001). In the liver, Per1 and Per2 increase after light exposure in early night, but other clock components including Per3, Cry1, Cry2, and the clock controlled gene, Dpb, do not change (Cailotto et al., 2009). Similarly, in this study, we saw that Per1 increased in the liver after a light pulse at night, but all other clock genes tested, including Per2, Bmal1, Clock, Cry1, and Cry2, as well as c-fos, had no significant change. Although Cailotto et al. (2009) reported that Per1 and Per2 but not Per3 transcript were increased after 1h light exposure at CT14, it is possible that we did not detect changes in Per2 due to 30min light exposure at CT16.
Light exposure at night activates the MAPK signaling pathway, accelerates the phosphorylation of CREB, activates CRE element activity, and consequently induces Per1 in the SCN (Ginty et al., 1993; Obrietan et al., 1998). FSK treatment also dramatically increases p-CREB and induces Per1 transcripts in cell culture (Obrietan et al., 1999; Travnickova-Bendova et al., 2002). To further investigate the molecular mechanism by which AhR activation inhibits the response of Per1 in this pathway, we used hepatoma cell lines, which have been used extensively to investigate molecular mechanisms associated with AhR activation (Boverhof et al., 2005; Hoffman et al., 1991; Probst et al., 1993; Reyes et al., 1992), but we did not use SCN2.2 cell lines here due to their undetectable AhR expression. Per1 was increased after FSK exposure in all cell lines. BNF inhibited this FSK-induced increase in Per1 only in the c7 cell line, which contains an intact and fully functional AhR/ARNT signaling pathway. FSK-induced Per1 is enhanced in c12 cells as a result of the absence of AhR, which may be related to the fact that AhR has a constitutive repressive effect on endogenous Per1 expression, as we demonstrated previously (Xu et al., 2010). However, BNF did not alter the response of Per1 to FSK in c12 cells. In the c4 line, which expresses AhR, but lacks ARNT, BNF also had no effect on FSK-induced increase in Per1 levels. Collectively, these results suggest that both AhR and ARNT are important for the inhibitory effects of BNF.
E-box and CRE cis-elements are key components involved in the regulation of transcription from the Per1 promoter. CLOCK/BMAL1 controls E-box activity and the Per1 endogenous rhythm, whereas the CRE element responds to light signals to mediate the acute increase in Per1 after light exposure (Zylka et al., 1998). In our previous study, we demonstrated that AhR activation inhibits endogenous Per1 levels, thereby disrupting the rhythm of Per1 through actions at the CLOCK/BMAL1-controlled E-box in the Per1 promoter. In this study, BNF blocked the effects of FSK on Per1 in the presence of an E-box decoy, thereby excluding the E-box as the region of the Per1 promoter through which AhR acts. In contrast, activation of CRE-Luc by FSK was significantly reduced when AhR was present, showing that the inhibition induced by AHR activation occurs through its action on the CRE element. Our results suggest that BNF inhibits the FSK increase in Per1 by reducing the CRE element activity rather than the E-box element.
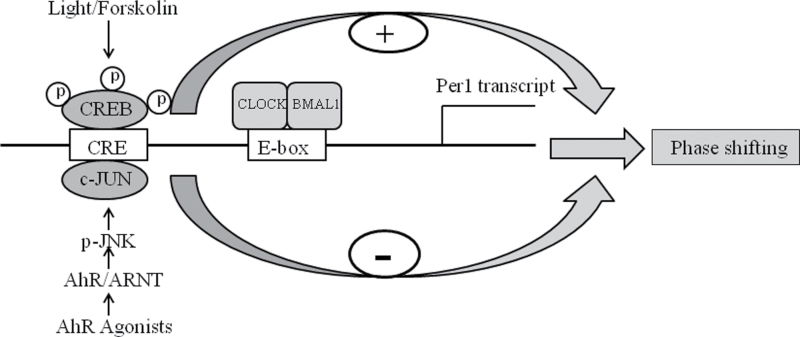
MAPK pathways are reported to acutely regulate the clock gene Per1 (Obrietan et al., 1999). Light causes elevated intracellular calcium to activate ERK in the SCN, leading to enhanced p-CREB and increased Per1 levels (Ginty et al., 1993; Nomura et al., 2006; Obrietan et al., 1998). Similarly, AhR activation has extensive effects on the MAPK signal pathway (Puga et al., 2009). AhR agonists such as TCDD activate JNK through AhR (Diry et al., 2006; Weiss et al., 2005, 2008), but TCDD can also activate p38, ERK, and JNK independent of AhR (Hoffer et al., 1996; Puga et al., 1992; Tan et al., 2002). Interestingly, we found that BNF did not inhibit the increase in p-CREB caused by FSK treatment, but BNF did block the increase in Per1 caused by FSK. Furthermore, our results showed that FSK increased the phosphorylation of JNK, and BNF dramatically augmented the phosphorylation of JNK compared with FSK treatment alone in c7 cells that express AhR, but not in c12 cells whose AhR expression is greatly reduced. The p-JNK inhibitor Sp600125 significantly reversed the ability of BNF to block the increase in Per1 caused by FSK (Fig. 7). Zhang et al. (2004) have reported that c-Jun has a negative effect on CRE activity. Therefore, we conclude that the inhibitory effect of BNF on FSK-induced Per1 resulted from the increase in p-JNK (Fig. 7).
FIG. 7.
Schematic diagram indicating that AhR activation represses the light-induced phase shifting. On one hand, light promotes the phosphorylation of CREB and Per1 transcript causing phase shifting; on the other hand, AhR activation enhances the phosphorylation of JNK and decreases light-induced Per1 transcript and phase shifting.
The light-dark cycle is the major environmental synchronizer of the SCN clock. A daily resetting or phase shifting of the master circadian clock is required to assure it remains synchronized with the external environment (Challet et al., 2003). This phase shifting depends on environmental light acting indirectly on the SCN through activation of specific retinal ganglion cells in the retinohypothalamic tract. Light signals are carried into the SCN through activation of specific signal transduction pathways involving the phosphorylation of CREB and activation of MAPKs (Ginty et al., 1993; Nomura et al., 2006; Obrietan et al., 1998). Rapid phase resetting allows an organism to adapt quickly to changes in external lighting and thus minimizes “jet-lag.” Our results suggest that the activation by the presence of environmental contaminants blunts the responsiveness of the clock to light. Consequently, the diminished ability to reset in response to light may create desynchrony between the organism and that environment and may ultimately precipitate health problems that manifest, such as fatigue, sleep disorders, and endocrine dysfunction (Crowley et al., 2007). This study demonstrates that AhR agonists alter the clock’s response to light. It provides new insights to prevent health problems resulting from ubiquitous environmental contaminants and some chemotherapy agents.
FUNDING
National Institutes of Health (ES017774 to S.A.T.).
REFERENCES
- Akiyama M., Kouzu Y., Takahashi S., Wakamatsu H., Moriya T., Maetani M., Watanabe S., Tei H., Sakaki Y., Shibata S. (1999). Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J. Neurosci. 19, 1115–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U., Zheng B., Larkin D., Sun Z. S., Lee C. C. (2001). MPer1 and mper2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms 16, 100–104 [DOI] [PubMed] [Google Scholar]
- Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536 [DOI] [PubMed] [Google Scholar]
- Bendova Z., Sumova A. (2006). Photoperiodic regulation of PER1 and PER2 protein expression in rat peripheral tissues. Physiol. Res. 55, 623–632 [DOI] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Chittim B., Harkema J. R., Jump D. B., Zacharewski T. R. (2005). Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 85, 1048–1063 [DOI] [PubMed] [Google Scholar]
- Buijs R. M., Hermes M. H., Kalsbeek A. (1998). The suprachiasmatic nucleus-paraventricular nucleus interactions: A bridge to the neuroendocrine and autonomic nervous system. Prog. Brain Res. 119, 365–382 [DOI] [PubMed] [Google Scholar]
- Buijs R. M., Wortel J., Van Heerikhuize J. J., Feenstra M. G., Ter Horst G. J., Romijn H. J., Kalsbeek A. (1999). Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur. J. Neurosci. 11, 1535–1544 [DOI] [PubMed] [Google Scholar]
- Cailotto C., Lei J., van der Vliet J., van Heijningen C., van Eden C. G., Kalsbeek A., Pévet P., Buijs R. M. (2009). Effects of nocturnal light on (clock) gene expression in peripheral organs: A role for the autonomic innervation of the liver. PLoS ONE 4, e5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E., Caldelas I., Graff C., Pévet P. (2003). Synchronization of the molecular clockwork by light- and food-related cues in mammals. Biol. Chem. 384, 711–719 [DOI] [PubMed] [Google Scholar]
- Crowley S. J., Acebo C., Carskadon M. A. (2007). Sleep, circadian rhythms, and delayed phase in adolescence. Sleep Med. 8, 602–612 [DOI] [PubMed] [Google Scholar]
- de la Iglesia H. O., Blaustein J. D., Bittman E. L. (1995). The suprachiasmatic area in the female hamster projects to neurons containing estrogen receptors and GnRH. Neuroreport 6, 1715–1722 [DOI] [PubMed] [Google Scholar]
- Ding J. M., Faiman L. E., Hurst W. J., Kuriashkina L. R., Gillette M. U. (1997). Resetting the biological clock: Mediation of nocturnal CREB phosphorylation via light, glutamate, and nitric oxide. J. Neurosci. 17, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diry M., Tomkiewicz C., Koehle C., Coumoul X., Bock K. W., Barouki R., Transy C. (2006). Activation of the dioxin/aryl hydrocarbon receptor (AhR) modulates cell plasticity through a JNK-dependent mechanism. Oncogene 25, 5570–5574 [DOI] [PubMed] [Google Scholar]
- Ginty D. D., Kornhauser J. M., Thompson M. A., Bading H., Mayo K. E., Takahashi J. S., Greenberg M. E. (1993). Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241 [DOI] [PubMed] [Google Scholar]
- Hoffer A., Chang C. Y., Puga A. (1996). Dioxin induces transcription of fos and jun genes by Ah receptor-dependent and -independent pathways. Toxicol. Appl. Pharmacol. 141, 238–247 [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. (1991). Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252, 954–958 [DOI] [PubMed] [Google Scholar]
- Ishida A., Mutoh T., Ueyama T., Bando H., Masubuchi S., Nakahara D., Tsujimoto G., Okamura H. (2005). Light activates the adrenal gland: Timing of gene expression and glucocorticoid release. Cell Metab. 2, 297–307 [DOI] [PubMed] [Google Scholar]
- Karman B. N., Tischkau S. A. (2006). Circadian clock gene expression in the ovary: Effects of luteinizing hormone. Biol. Reprod. 75, 624–632 [DOI] [PubMed] [Google Scholar]
- Kennaway D. J., Voultsios A., Varcoe T. J., Moyer R. W. (2002). Melatonin in mice: Rhythms, response to light, adrenergic stimulation, and metabolism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, R358–R365 [DOI] [PubMed] [Google Scholar]
- Klein D. C., Weller J. L. (1972). Rapid light-induced decrease in pineal serotonin N-acetyltransferase activity. Science 177, 532–533 [DOI] [PubMed] [Google Scholar]
- Ko C. H., Takahashi J. S. (2006). Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277 [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- Loh D. H., Abad C., Colwell C. S., Waschek J. A. (2008). Vasoactive intestinal peptide is critical for circadian regulation of glucocorticoids. Neuroendocrinology 88, 246–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey P. L., Takahashi J. S. (2004). Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5, 407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer J. H., Watanabe K., Schaap J., Albus H., Détári L. (1998). Light responsiveness of the suprachiasmatic nucleus: Long-term multiunit and single-unit recordings in freely moving rats. J. Neurosci. 18, 9078–9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M., Lin T. M., Peterson R. E., Cooke P. S., Tischkau S. A. (2008). Behavioral rhythmicity of mice lacking AhR and attenuation of light-induced phase shift by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Rhythms 23, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M., Tischkau S. A. (2007). Effects of tryptophan photoproducts in the circadian timing system: Searching for a physiological role for aryl hydrocarbon receptor. Toxicol. Sci. 95, 172–181 [DOI] [PubMed] [Google Scholar]
- Nagai K., Nagai N., Shimizu K., Chun S., Nakagawa H., Niijima A. (1996). SCN output drives the autonomic nervous system: With special reference to the autonomic function related to the regulation of glucose metabolism. Prog. Brain Res. 111, 253–272 [DOI] [PubMed] [Google Scholar]
- Nomura K., Takeuchi Y., Fukunaga K. (2006). MAP kinase additively activates the mouse Per1 gene promoter with CaM kinase II. Brain Res. 1118, 25–33 [DOI] [PubMed] [Google Scholar]
- Numayama-Tsuruta K., Kobayashi A., Sogawa K., Fujii-Kuriyama Y. (1997). A point mutation responsible for defective function of the aryl-hydrocarbon-receptor nuclear translocator in mutant Hepa-1c1c7 cells. Eur. J. Biochem. 246, 486–495 [DOI] [PubMed] [Google Scholar]
- Obrietan K., Impey S., Smith D., Athos J., Storm D. R. (1999). Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J. Biol. Chem. 274, 17748–17756 [DOI] [PubMed] [Google Scholar]
- Obrietan K., Impey S., Storm D. R. (1998). Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat. Neurosci. 1, 693–700 [DOI] [PubMed] [Google Scholar]
- Park S. J., Yoon W. K., Kim H. J., Son H. Y., Cho S. W., Jeong K. S., Kim T. H., Kim S. H., Kim S. R., Ryu S. Y. (2005). 2,3,7,8-Tetrachlorodibenzo-p-dioxin activates ERK and p38 mitogen-activated protein kinases in RAW 264.7 cells. Anticancer Res. 25, 2831–2836 [PubMed] [Google Scholar]
- Pittendrigh C. S., Daan S. (1974). Circadian oscillations in rodents: A systematic increase of their frequency with age. Science 186, 548–550 [DOI] [PubMed] [Google Scholar]
- Probst M. R., Reisz-Porszasz S., Agbunag R. V., Ong M. S., Hankinson O. (1993). Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol. Pharmacol. 44, 511–518 [PubMed] [Google Scholar]
- Puga A., Ma C., Marlowe J. L. (2009). The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem. Pharmacol. 77, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga A., Nebert D. W., Carrier F. (1992). Dioxin induces expression of c-fos and c-jun proto-oncogenes and a large increase in transcription factor AP-1. DNA Cell Biol. 11, 269–281 [DOI] [PubMed] [Google Scholar]
- Ralph M. R., Foster R. G., Davis F. C., Menaker M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 [DOI] [PubMed] [Google Scholar]
- Reppert S. M., Weaver D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- Reyes H., Reisz-Porszasz S., Hankinson O. (1992). Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science 256, 1193–1195 [DOI] [PubMed] [Google Scholar]
- Scheer F. A., Ter Horst G. J., van Der Vliet J., Buijs R. M. (2001). Physiological and anatomic evidence for regulation of the heart by suprachiasmatic nucleus in rats. Am. J. Physiol. Heart Circ. Physiol. 280, H1391–H1399 [DOI] [PubMed] [Google Scholar]
- Schmidt J. V., Su G. H., Reddy J. K., Simon M. C., Bradfield C. A. (1996). Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. U.S.A. 93, 6731–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman L. P., Sriram S., Weaver D. R., Maywood E. S., Chaves I., Zheng B., Kume K., Lee C. C., van der Horst G. T., Hastings M. H., et al. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y., Taguchi K., Yamamoto S., Takekida S., Yan L., Tei H., Moriya T., Shibata S., Loros J. J., Dunlap J. C., et al. (1997). Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053 [DOI] [PubMed] [Google Scholar]
- Tan Z., Chang X., Puga A., Xia Y. (2002). Activation of mitogen-activated protein kinases (MAPKs) by aromatic hydrocarbons: Role in the regulation of aryl hydrocarbon receptor (AHR) function. Biochem. Pharmacol. 64, 771–780 [DOI] [PubMed] [Google Scholar]
- Tischkau S. A., Gallman E. A., Buchanan G. F., Gillette M. U. (2000). Differential cAMP gating of glutamatergic signaling regulates long-term state changes in the suprachiasmatic circadian clock. J. Neurosci. 20, 7830–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkau S. A., Mitchell J. W., Tyan S. H., Buchanan G. F., Gillette M. U. (2003). Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J. Biol. Chem. 278, 718–723 [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. U.S.A. 99, 7728–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C., Faust D., Dürk H., Kolluri S. K., Pelzer A., Schneider S., Dietrich C., Oesch F., Göttlicher M. (2005). TCDD induces c-jun expression via a novel Ah (dioxin) receptor-mediated p38-MAPK-dependent pathway. Oncogene 24, 4975–4983 [DOI] [PubMed] [Google Scholar]
- Weiss C., Faust D., Schreck I., Ruff A., Farwerck T., Melenberg A., Schneider S., Oesch-Bartlomowicz B., Zatloukalová J., Vondrácek J., et al. (2008). TCDD deregulates contact inhibition in rat liver oval cells via Ah receptor, JunD and cyclin A. Oncogene 27, 2198–2207 [DOI] [PubMed] [Google Scholar]
- Xu C. X., Krager S. L., Liao D. F., Tischkau S. A. (2010). Disruption of CLOCK-BMAL1 transcriptional activity is responsible for aryl hydrocarbon receptor-mediated regulation of Period1 gene. Toxicol. Sci. 115, 98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu S., Perpetua M. D., Walker W. H., Harbrecht B. G. (2004). Cytokines increase CRE binding but decrease CRE-mediated reporter activity in rat hepatocytes by increasing c-Jun. Hepatology 39, 1343–1352 [DOI] [PubMed] [Google Scholar]
- Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z. S., Eichele G., Bradley A., et al. (2001). Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 [DOI] [PubMed] [Google Scholar]
- Zheng B., Larkin D. W., Albrecht U., Sun Z. S., Sage M., Eichele G., Lee C. C., Bradley A. (1999). The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173 [DOI] [PubMed] [Google Scholar]
- Zylka M. J., Shearman L. P., Weaver D. R., Reppert S. M. (1998). Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110 [DOI] [PubMed] [Google Scholar]