Abstract
MicroRNAs (miRNAs) are environmentally sensitive inhibitors of gene expression that may mediate the effects of metal-rich particulate matter (PM) and toxic metals on human individuals. Previous environmental miRNA studies have investigated a limited number of candidate miRNAs and have not yet evaluated the functional effects on gene expression. In this study, we wanted to identify PM-sensitive miRNAs using microarray profiling on matched baseline and postexposure RNA from foundry workers with well-characterized exposure to metal-rich PM and to characterize miRNA relations with expression of candidate inflammatory genes. We applied microarray analysis of 847 human miRNAs and real-time PCR analysis of 18 candidate inflammatory genes on matched blood samples collected from foundry workers at baseline and after 3 days of work (postexposure). We identified differentially expressed miRNAs (fold change [FC] > 2 and p < 0.05) and correlated their expression with the inflammatory associated genes. We performed in silico network analysis in MetaCore v6.9 to characterize the biological pathways connecting miRNA-mRNA pairs. Microarray analysis identified four miRNAs that were differentially expressed in postexposure compared with baseline samples, including miR-421 (FC = 2.81, p < 0.001), miR-146a (FC = 2.62, p = 0.007), miR-29a (FC = 2.91, p < 0.001), and let-7g (FC = 2.73, p = 0.019). Using false discovery date adjustment for multiple comparisons, we found 11 miRNA-mRNA correlated pairs involving the 4 differentially expressed miRNAs and candidate inflammatory genes. In silico network analysis with MetaCore database identified biological interactions for all the 11 miRNA-mRNA pairs, which ranged from direct mRNA targeting to complex interactions with multiple intermediates. Acute PM exposure may affect gene regulation through PM-responsive miRNAs that directly or indirectly control inflammatory gene expression.
Key Words: miRNA expression, integrative analysis, mRNA expression, inflammation, metal-rich particulate matter, microarray.
Epidemiological studies have consistently shown that exposure to inhalable air particles (particulate matter [PM]) is associated with hospitalization and mortality in exposed individuals, predominantly attributable to cardiovascular and respiratory diseases (Hesterberg et al., 2009). In vivo studies (Chan et al., 2005; Chang et al., 2005; Corey et al., 2006) suggest that the transition metal components of PM may be responsible for a substantial proportion of these effects. Although PM-related risks have been associated with chronic exposures (Dockery et al., 1993), the most consistent effects have been shown in the studies of acute short-term exposures in which health-related effects were observed hours or days after peaks of ambient air pollution (Samet et al., 2000). Systemic inflammation has been suggested as a critical step in PM-induced health effects. Alveolar macrophages and pulmonary epithelial cells constitute the first line of defense against inhaled noxious compounds and have been suggested to initiate a cascade of inflammatory reactions upon PM exposure that can rapidly extend to circulating blood leukocytes. These systemic reactions can be assessed through expression analysis in circulating leukocytes of genes related to inflammatory pathways and oxidative stress, which are exquisitely sensitive to PM exposure (Wang et al., 2005). Modifications of gene expression of inflammatory genes have been demonstrated in peripheral blood of exposed individuals (Wang et al., 2005), potentially reflecting PM effects on the cardiorespiratory system. However, the molecular pathways that determine the systemic changes in gene expression after PM exposures are still largely unknown.
MicroRNAs (miRNAs) are single-stranded RNAs of ~22 nt that operate as post-transcriptional gene regulators by base pairing with target mRNAs and leading to mRNA destruction in the RNA-induced silencing complex through argonaute-catalysed cleavage (Guo et al., 2010; Kim, 2005). Approximately 1000 human miRNAs have been identified in mammalian cells through high-throughput biochemical screens (Kasinski and Slack, 2011). Recent microarray and proteomic approaches have revealed that individual miRNAs can regulate hundreds of target genes (Chen and Chen, 2011). MicroRNAs have been implicated in the regulation of a wide variety of biological processes including cell differentiation and proliferation, as well as immunomodulation and inflammation (Sonkoly and Pivarcsi, 2009a). Recent data have suggested that miRNA signaling may be an active component of the cellular response to environmental risk factors. In particular, Jardim et al. (2009) showed altered miRNA expression profiles in human airway epithelial cells exposed to diesel exhaust particles (DEPs) and found that the 12 miRNAs most sensitive to DEP were involved in the regulation of inflammatory pathways. Also, Bollati et al. (2010) recently showed that individuals exposed to metal-rich PM exhibited altered expression of candidate miRNAs, including miR-222, miR-21, and miR-146a, which might contribute to PM-induced inflammation and oxidative stress.
Previous human miRNA studies of environmental toxicants have only investigated a small number of candidate miRNAs. Because of the wide number of miRNAs potentially involved in inflammatory processes or in other pathways potentially activated by environmental exposures, as well as of the limited direct experimental knowledge on miRNA influences on PM-related pathways, candidate-miRNA studies may severely underrepresent the effects of the exposure. In addition, previous studies of environmental effects have been limited to the investigation of the influences of the exposures on miRNA expression and have not evaluated their ultimate effects on gene expression, thus providing limited information as to whether the exposure-dependent miRNA changes have functional effects on gene expression.
In this study, we used a microarray-based approach to investigate the acute effects of metal-rich PM on miRNA expression in pre- and postexposure matched samples from foundry workers with well-characterized exposure. We designed our study to take advantage of temporal short-term variations in the exposure due to the weekly cycle of days of work and days off. We performed expression analysis of a panel of candidate inflammatory genes expressed in blood in order to identify miRNA-driven alterations in gene expression. We used mRNA-target bioinformatic analyses to obtain in silico confirmation of our experimental findings and characterize the molecular links between PM-sensitive miRNAs and mRNA expression.
MATERIALS AND METHODS
Study participants.
We selected for this study 10 participants with the highest metal-rich PM exposure among a larger study of 63 workers in a steel production plant in Northern Italy (Tarantini et al., 2009). This selection was designed to conduct a cost-efficient study, also in consideration of the need for specific procedures for RNA preservation that required an extra tube of blood to be drawn. All participants had been working in the current job position for at least 1 year. In order to investigate short-term effects of PM, we obtained blood samples at two different times. The baseline sample was collected on the first day of a working week (after a washout of 2 days off work) before the beginning of any work activity; the postexposure sample was collected at the same hour on the fourth day of the same working week, following 3 consecutive days of work. The 10 participants were nonsmokers, were men with a mean age of 46 years (range, 33–55 years), and had a mean body mass index of 25.53kg/m2 (± 2.6). Ten additional male participants matched for age were recruited in the Milan (Italy) area in order to be representative of a set of “naive” unexposed controls. All the participants were nonsmokers with a mean age of 45.3 years (range, 34–56 years). These participants are usually exposed on average to 50 µg/m3 PM10 (Anselmi and Patelli, 2006). Individual written informed consent and approval from the local Institutional Review Board were obtained before the study. Ten healthy men selected in Milano area as control group matched by age and smoking status.
Blood collection and miRNA isolation.
Buffy-coat samples were separated within 30min of blood draw, immediately snap frozen, and stored at −80°C. Total RNA was extracted from the buffy coats using the Ribopure Kit (Ambion, Inc., Austin, TX), modified for miRNA extraction (Supplementary data). RNA was quantified using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). An Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA) was used to assess RNA integrity based on the RNA integrity number (RIN) factor, and presence of low-molecular weight RNA (5S) was also verified. The RIN values ranged from 6.20 to 9, and 13 samples out of 20 had an RIN value over 7.5. We measured white blood cell (WBC) counts, as well as the proportions of monocytes, lymphocytes, and granulocytes in each sample, and found no differences between baseline and postexposure samples (p ≥ 0.44, data not shown).
Microarray miRNA expression analysis.
We performed comprehensive expression profiling of mature miRNA using the Affymetrix GeneChip miRNA Array (Affymetrix, Santa Clara, CA). The miRNA Array included probe sets for all the 847 human miRNAs (i.e., hsa miRNAs) included in the Sanger miRNA registry (http://www.mirbase.org/). Labeling of total RNA samples was performed using the FlashTag Biotin RNA labeling kit (Genisphere Inc., Hatfield, PA) according to manufacturer’s instruction starting from 1 µg of total RNA. Briefly, the tailing reaction was followed by ligation of the biotinylated signal molecule to the target RNA sample. The labeling reaction is based on Genisphere’s proprietary 3DNA dendrimer signal amplification technology. Biotin-labeled samples were hybridized onto the arrays for 16h at 48°C, washed, and stained using Hybridization Wash and Stain Kit (Affymetrix) in Fluidics Station 450. Chips were scanned with the GeneChip Scanner 3000 7G to acquire fluorescent images of each array and analyzed by use of GeneChip Operating Software. Intensity data from CEL files, obtained from scans and calculated on the pixel values, were imported into the Affymetrix miRNA QC Tool software (Version 1.0.33.0) to quantify the signal value. QC procedures included checks based on plotting the average intensity of the oligo spike-in and background probe sets across all the arrays. As recommended by Genisphere, the support website for the FlashTag Biotin RNA labeling kit, oligo spike-in 2, 23, 29, 31, and 36 probe sets should present a value of more than 1000 intensity units to accept array quality. The experimental design included 10 biological replicates at two time points, i.e., pre-exposure and postexposure. We also included a set of 10 matched “naive” unexposed controls. We ran the microarray analysis on blood RNA samples taken from each participant at the two different time points for a total of 20 arrays. We ran 10 additional arrays for the control participants. Affymetrix indicates a technical reproducibility (inter- and intralot) of more than 95% for the GeneChip miRNA Array (http://www.affymetrix.com/estore/browse/products.jsp?productId=131473#1_1). Therefore, it is a common practice to run this specific array without duplicates. However, we assessed the technical reproducibility of using three technical replicates (RNA samples isolated from reference tissue, Ambion, and tissue Human atlas) and obtained a correlation over 95% among replicates (data not shown).
Real-time PCR analysis of mRNA expression of inflammation-related genes.
Given the study cost efficiency and the high correlation between microarray-based technique and qPCR results (Pradervand et al., 2010), we dedicated the qPCR investigation to study selected inflammatory genes rather than to validate the differentially upmodulated miRNA obtained from the microarray analysis. Using TargetScan, we identified a total number of 2363 genes predicted as targets for miR-421, miR-146a, miR-29a, and let-7g (Supplementary table S1). We decided to restrict the mRNA analysis of potential targets to 18 specific genes involved in inflammatory signaling and mediation that are known to be expressed in WBCs. In order to fulfill these criteria, we combined a PubMed literature search with data queries from Aceview (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html), a database developed at NCBI that provides gene expression information by summarizing all quality-filtered human cDNA data from GenBank, dbEST, and RefSeq. We performed real-time PCR to quantify mRNA expression of the 18 preselected genes (Supplementary table S2). We used a custom RT2 PCR Array (SABioscience, Qiagen, Hilden, Germany), a 96-well plate prespotted array containing gene-specific primer sets (RefSeq accession number listed in Supplementary table S2) plus two housekeeping genes (ACTB and GADPDH), two negative controls, and two internal controls (PPC as Positive PCR Control and RTC as Reverse Transcription Control). An eight-channel liquid handler (Microlab Starlet, Hamilton Robotics) was used to increase throughput for preanalytical sample preparation and to reduce error. The real-time PCR was performed in triplicates, including no-template controls. The array was heat sealed and run on a 7900HT Sequence Detection System (Applied Biosystems, Inc., Foster City, CA). Real-time PCR was performed employing RT2 qPCR MasterMix (Qiagen) using default cycling parameters for 40 cycles (1 cycle of 50°C for 2min, 1 cycle of 95°C for 10min, 40 cycles of 95°C for 15 s, and 60°C for 1min). The relative gene expression was calculated with the ΔCt method.
Exposure measurement.
Individual exposure levels of the study participants are shown in Table 1. Measures of PM mass that included levels of PM with aerodynamic diameters < 10 μm (PM10) and < 1 μm (PM1) were obtained using a GRIMM 1100 light-scattering dust analyzer (Grimm Technologies, Inc. Douglasville, GA). Measures of PM metal components were performed on the PM10 fraction of PM mass through multielemental analysis by means of inductively coupled plasma mass spectrometer (ELAN DRC II, Perkin Elmer, Waltham). We measured 14 different PM metal components: aluminum, arsenic, barium, cobalt, chromium, copper, iron, manganese, molybdenum, nickel, lead, antimony, tin, and zinc. Inorganic or elemental carbon is the main constituent of PM10 net of metals, so we estimate the elemental carbon levels as the difference between PM10 and the sum of all metals. Levels of organic carbon were listed as polycyclic aromatic hydrocarbon (PAH) levels. We believe that organic PAHs are ideal tracers of organic carbon because of their prominent toxicity. Measures of airborne PM mass and PM metal components obtained in each of the 11 work areas of the steel production facility were used to estimate individual exposures. During the 3 work days between baseline and postexposure, each of the study participants recorded the time that he spent in each of the work areas. Individual exposure was calculated as the average of area-specific PM levels weighted by the time spent in each area.
Table 1.
Individual Exposure of the Study Participants to PM10, PM1, Metal Components (Measured in PM10), Elemental Carbon, and Organic Carbon
| Exposure | Mean | SD | Interquartile range | Min | Max |
|---|---|---|---|---|---|
| PM10 (µg/m3) | 169.29 | 48.37 | 43.80 | 73.72 | 222.86 |
| PM1 (µg/m3) | 6.67 | 3.69 | 5.80 | 1.71 | 11.83 |
| Aluminum (µg/m3) | 2.31 | 1.85 | 1.01 | 1.00 | 7.38 |
| Arsenic (µg/m3) | 0.10 | 0.11 | 0.12 | 0.01 | 0.30 |
| Barium (µg/m3) | 0.23 | 0.12 | 0.22 | 0.07 | 0.42 |
| Cobalt (µg/m3) | 0.01 | 0.01 | 0.01 | 0.00 | 0.02 |
| Chromium (µg/m3) | 0.07 | 0.03 | 0.03 | 0.00 | 0.11 |
| Copper (µg/m3) | 2.05 | 1.81 | 2.24 | 0.11 | 5.36 |
| Iron (µg/m3) | 31.55 | 18.20 | 27.55 | 1.70 | 59.81 |
| Manganese (µg/m3) | 6.16 | 6.06 | 6.89 | 0.16 | 17.05 |
| Molybdenum (µg/m3) | 0.08 | 0.08 | 0.09 | 0.00 | 0.23 |
| Nickel (µg/m3) | 0.28 | 0.16 | 0.16 | 0.02 | 0.52 |
| Lead (µg/m3) | 3.74 | 3.64 | 4.27 | 0.13 | 10.30 |
| Antimony (µg/m3) | 0.03 | 0.02 | 0.03 | 0.00 | 0.07 |
| Tin (µg/m3) | 0.31 | 0.30 | 0.37 | 0.03 | 0.86 |
| Zinc (µg/m3) | 12.14 | 12.03 | 13.36 | 0.29 | 33.59 |
| Elemental carbon (µg/m3) | 110.23 | 53.00 | 67.95 | 31.91 | 194.34 |
| Organic carbon (µg/l) | 6.26 | 10.29 | 8.97 | 0.03 | 31.55 |
Note. Exposure levels were estimated for the 3 days between baseline and postexposure samples.
Statistical analysis.
Data from the miRNA arrays were processed using the Affymetrix detection algorithm based on nonparametric Wilcoxon rank-sum tests, which was applied independently on each array and probe/probe set. Probe sets with p ≥ 0.06 were considered “not-detected above background.” For data normalization, we used robust multiarray, a common method for normalizing and summarizing probe-level intensity measurements from Affymetrix Gene Chips. Starting with the probe-level data from a set of Gene Chips, the perfect-match values were background corrected, normalized, and finally summarized, resulting in a set of expression measures.
We used LIMMA in the R-based Bioconductor package to calculate the level of differential expression simultaneously for all miRNAs by comparing matched postexposure versus baseline samples. Briefly, for each miRNA, we performed a t-test and considered as differentially expressed the miRNAs that showed p < 0.05 and at least a twofold expression change (FC > 2) in baseline versus postexposure samples (McCarthy and Smyth, 2009; Zhao et al., 2010). Empirical Bayes shrinkage was used to compute moderated t-statistics by shrinkage of the standard errors toward a common value. This method has the advantage of providing robust results even when the number of arrays in an experiment is small. For each of the differentially expressed miRNAs, we assessed the correlation with 18 inflammatory associated genes (Table 3) using Spearman’s correlation coefficients, corrected for multiple testing using false discovery rates (FDRs). miRNA-mRNAs pairs with FDR < 0.20 were further considered for in silico network analysis.
Table 3.
Correlations of Differentially Expressed miRNAs With mRNA Levels of Candidate Inflammatory Genes
| miR-421 | miR-146a | miR-29a | let-7g | |||||
|---|---|---|---|---|---|---|---|---|
| Target-gene mRNAs | ρa | FDR | ρa | FDR | ρa | FDR | ρa | FDR |
| CCL2 | 0.37 | 0.52 | −0.68 | 0.19 | 0.20 | 0.90 | 0.00 | 1.00 |
| CCL5 | −0.67 | 0.22 | 0.83 | 0.09 | −0.28 | 0.90 | 0.35 | 0.69 |
| CDKN1B | −0.57 | 0.40 | 0.40 | 0.429 | −0.35 | 0.90 | 0.40 | 0.64 |
| CDKN1C | −0.50 | 0.44 | 0.73 | 0.15 | 0.05 | 0.93 | 0.45 | 0.58 |
| ICAM1 | −0.17 | 0.71 | 0.17 | 0.80 | 0.03 | 0.93 | 0.12 | 0.92 |
| IFNAR2 | −0.92 | 0.009 | 0.42 | 0.43 | −0.62 | 0.35 | 0.47 | 0.58 |
| IFNG | −0.18 | 0.71 | 0.10 | 0.89 | 0.18 | 0.90 | −0.28 | 0.75 |
| ITGA2 | −0.33 | 0.52 | −0.35 | 0.49 | −0.27 | 0.90 | −0.33 | 0.69 |
| ITGA5 | −0.33 | 0.52 | 0.18 | 0.80 | −0.53 | 0.50 | 0.47 | 0.58 |
| ITGAX | −0.32 | 0.52 | 0.55 | 0.28 | −0.27 | 0.90 | 0.80 | 0.15 |
| ITGB2 | −0.42 | 0.52 | 0.42 | 0.43 | −0.20 | 0.90 | −0.23 | 0.82 |
| NFKB1 | 0.40 | 0.52 | −0.42 | 0.43 | 0.15 | 0.90 | −0.73 | 0.15 |
| NOS2 | 0.75 | 0.18 | −0.58 | 0.28 | 0.63 | 0.34 | −0.17 | 0.86 |
| PDGFRB | 0.72 | 0.18 | −0.58 | 0.28 | 0.63 | 0.35 | −0.53 | 0.58 |
| PTEN | −0.32 | 0.52 | −0.02 | 0.97 | −0.85 | 0.07 | 0.07 | 0.97 |
| TGFB1 | −0.53 | 0.42 | 0.80 | 0.09 | 0.07 | 0.93 | 0.73 | 0.15 |
| TGFB2 | 0.10 | 0.80 | −0.02 | 0.97 | 0.10 | 0.93 | 0.18 | 0.86 |
| TNF | −0.25 | 0.62 | 0.55 | 0.28 | −0.15 | 0.90 | −0.02 | 1.00 |
Note. miRNA-mRNA pairs with FDR < 0.20 (boldface) were considered noteworthy.
aSpearman correlation coefficients.
In silico network analysis.
We performed a network analysis in MetaCore v6.9 (Thomson Reuters, New York), a web-based computational platform for multiple applications in systems biology. MetaCore is primarily designed for analysis of high-throughput molecular data in the context of human and mammalian networks, canonical pathways, diseases, and cellular processes. MetaCore analyses are based on MetaBase, an integrated database of mammalian biology that contains over 6 million experimental findings on protein-protein, protein-DNA, protein-RNA, and protein-compound interactions; metabolic and signaling pathways; proprietary ontologies; and controlled vocabulary.
We built a single-network pathway using the Dijkstra’s algorithm, which efficiently finds the shortest paths that link each miRNA-mRNA pair. Using this algorithm, when, for a given from-to pair, there are more than one paths of the same minimal length, all of them are shown in the network. We set the miRNA as the initial node and the correlated mRNA as the ending node of the path. We fixed the maximum number of steps in the path (i.e., the maximum length for a path) to 3. The analysis was run on homo sapiens data in MetaBase.
RESULTS
Analysis of Differentially Expressed miRNAs
Microarray profiling of the 847 human miRNAs analyzed in matched baseline versus postexposure samples showed that 432 miRNAs had a negative fold change (FC) and 415 miRNAs had a positive FC (Supplementary fig. S1). In the negative FC group, 53 miRNAs had a p-value < 0.05. In the positive FC group, 57 miRNAs had a p-value < 0.05 (Supplementary fig. S1). To limit false positives, we considered as differentially expressed those miRNAs that showed absolute FC > 2 (i.e., absolute log2 FC > 1) and p < 0.05. Using these criteria, we identified four miRNAs that exhibited differential expression between baseline and postexposure samples, including miR-421 (FC = 2.81, p < 0.001), miR-146a (FC = 2.62, p = 0.007), miR-29a (FC = 2.91, p < 0.001), and let-7g (FC = 2.73, p = 0.019) (Table 2). All the four miRNAs were upregulated in postexposure samples compared with baseline. We chose a stringent cutoff (FC > 2 and p < 0.05) because of the limited sample size. Using a smaller FC cutoff on a small sample size may lead to the risk of losing a reliable detection of differentially expressed miRNAs. Supplementary table S3 shows the differentially expressed miRNAs with FC > 1.5 and the miRNA-related diseases annotated in the miR2Disease database. We also measured the miRNA expression levels from 10 naive unexposed controls for the selected miRNAs that were found to exhibit differential expression in post- versus pre-exposure samples among foundry workers. Three out of four of the differentially expressed miRNAs (miR-146a, miR-29a, and let-7g) showed higher expression in postexposure samples from exposed workers versus samples from naive unexposed individuals. However, only hsa-miR-421 was significantly higher (FC = 1.68, p = 0.0181) based on a nominal p-value cutoff of 0.05 (Supplementary table S4).
Table 2.
Microarray Results on Selected miRNAs With Significant Differential Expression Between Matched Baseline and Postexposure Samples (n = 10)
| miRNA | Log FC | FC | p-value |
|---|---|---|---|
| hsa-miR-421 | 1.49 | 2.81 | < 0.001 |
| hsa-miR-146a | 1.39 | 2.62 | 0.007 |
| hsa-miR-29a | 1.54 | 2.91 | < 0.001 |
| hsa-let-7g | 1.45 | 2.73 | 0.019 |
Note. This table reports data for the four miRNAs that showed FC > 2 in the postexposure samples and p < 0.05.
Correlation of miRNAs With Inflammatory Genes: Experimental Data
We calculated the correlation of postexposure expression levels of the four upregulated miRNAs with postexposure mRNA expression of the 18 candidate inflammatory genes. Using a Benjamini-Hochberg FDR adjustment for multiple comparisons (FDR < 0.2), we found correlations among 11 miRNA-mRNA pairs (Table 3). miR-421 was negatively correlated with mRNA levels of interferon alpha receptor 2 (IFNAR2) (ρ = −0.92, FDR = 0.009) and positively correlated with nitric oxide synthase 2 (NOS2) (ρ = 0.75, FDR = 0.18) and platelet-derived growth factor receptor, beta polypeptide (PDGFRB) (ρ = 0.72, FDR = 0.18) mRNAs. Expression of miR-146a was negatively correlated with mRNA levels of the chemokine C-C-motif ligand 2 (CCL2) (ρ = −0.68, FDR = 0.19) and positively correlated with chemokine C-C-motif ligand 5 (CCL5) (ρ = 0.83, FDR = 0.09), cyclin-dependent kinase inhibitor 1C (CDKN1C) (ρ = 0.73, FDR = 0.15), and transforming growth factor beta 1 (TGFB1) (ρ = 0.8, FDR = 0.09) mRNAs. Expression of miR-29a was negatively correlated with mRNA levels of phosphatase and tensin homolog (PTEN) (ρ = −0.85, FDR = 0.07). Expression of let-7g was positively correlated with mRNA levels of the integrin alpha X (ITGAX) (ρ = 0.80, FDR = 0.15) and TGFB1 (ρ = 0.73, FDR = 0.15) and negatively correlated with the NF-kB1 (ρ = −0.73, FDR = 0.15) mRNA.
Correlation of miRNAs With Inflammatory Genes: In Silico Network Analysis
We performed MetaCore pathway analysis on the 11 noteworthy correlations observed between miRNA and mRNA expression levels to generate visual representation schemes of biological molecules linking the miRNA-mRNA pairs. The in silico network analysis identified biological functional interactions for all the 11 miRNA-mRNA pairs. Representative examples of miRNA-mRNA interactions selected from the complete in silico network analyses are shown in Figures 1–3. The results for the remaining miRNA-mRNA pairs are reported in Supplementary figures S2 and S3.
Fig. 1.
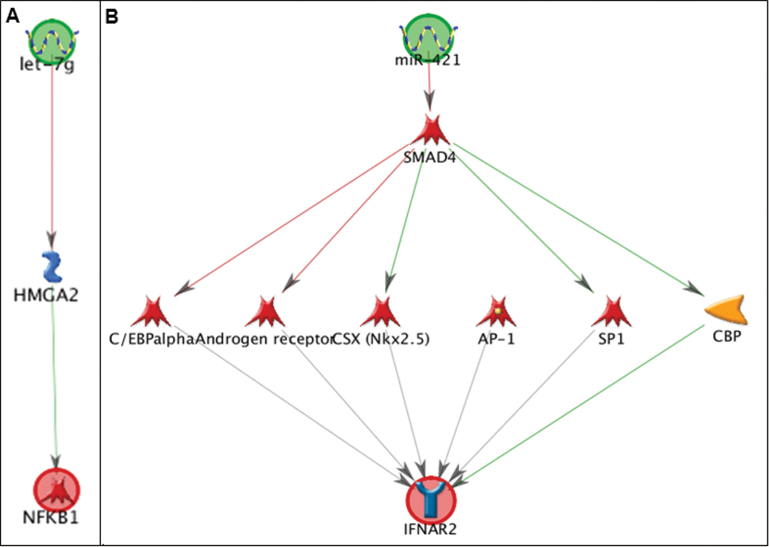
Network analysis showing the pathways linking miR-29a and PTEN. The annotations of symbols and abbreviations in the regulatory network are shown. Individual miRNAs and genes are represented as nodes; the different shapes represent the functional class of genes. The arrow heads indicate the direction of the predicted interaction. Green lines represent upregulation and red lines represent downregulation. Gray lines represent an unspecified effect. Known targets are highlighted in blue.
Fig. 3.
Network analysis showing the pathways linking miR-146a and TGFB1.
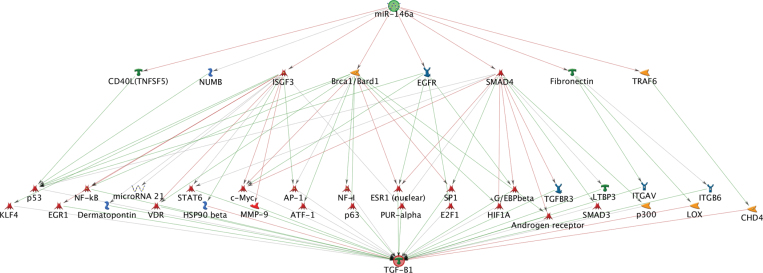
The in silico network analysis showed interactions between miRNA and mRNA pairs with different degrees of complexity. For the negative correlation between miR-29a and PTEN, the in silico network analysis showed a direct interaction (Fig. 1), consistent with direct targeting of the PTEN mRNA by miR-29a. We found three pathways—linking miR-146a and CCL2, miR-146a and CCL5 (Supplementary fig. S2, panels A and B), and let-7g and NF-kB1 (Fig. 2, panel A)—that were connected by a one-step interaction, with one or more intermediate molecules that directly interacted with both the mRNA and the miRNA. For example, let-7g was shown to target the mRNA of the HMGA2-binding protein, which in turn influences the expression of the NF-kB1 mRNA (Fig. 2, panel A). An additional six pathways—linking miR-421 and IFNAR2 (Fig. 2, panel B); miR-421 and NOS2; miR-421 and PDGFRB; let-7g and ITGAX; let-7g and TGFB1; and miR-146a and CDKN1C (p57) (Supplementary fig. S3)—connected the miRNA to the corresponding mRNA through two-step interactions. For instance, miR-421 was shown to target the SMAD4 transcription factor (Fig. 2, panel B), which in turn controls the expression of five other genes including four transcription factors (C/EBP-alpha, AR, CSX, and SP1) and one kinase (CBP) that ultimately affect IFNAR2 mRNA expression. The remaining pathway—linking miR-146a and TGFB1 (Fig. 3)—showed a higher degree of complexity, with multiple intermediates and hubs of interactions between the miRNAs and the mRNAs. miR-146a was shown to target as many as eight mRNAs, from which a complex network of interactions is originated that converge to ultimately influence TGFB1 mRNA expression.
Fig. 2.
Network analysis showing the pathways linking (A) let-7g and NF-KB1 and (B) miR-421 and IFNAR2.
DISCUSSION
In this study, we identified four differentially expressed miRNAs after short-term exposure to metal-rich PM. We evaluated the correlations of the four differentially expressed miRNAs with a panel of 18 candidate inflammatory mRNAs and found 11 miRNA-mRNAs pairs that were correlated in our experimental data. In silico network identified biological functional interactions for all the 11 miRNA-mRNA pairs.
To the best of our knowledge, this is the first human study applying a high-throughput array to measure metal-rich PM-induced changes in miRNA expression in humans. Based on the hypothesis that miRNA analysis might generate disease biomarkers, there have been a growing number of studies that have shown that miRNAs are expressed and can be measured in circulating blood (Bianchi et al., 2011; Fichtlscherer et al., 2011; Mitchell et al., 2008). Based on effect-size and statistical significance criteria, we found four differentially expressed miRNAs, i.e., miR-421, miR-29a, miR-146a, and let-7g, which all showed upregulation in postexposure samples. Based on previous literature, all these four miRNAs have roles in inflammation or responses to environmental toxicants (Chen et al., 2011; Izzotti et al., 2011; Kumar et al., 2011; Polikepahad et al., 2010; Sonkoly and Pivarcsi, 2009b; Takahashi et al., 2012).
The roles of miRNAs as part of the cellular response to air toxicants are yet largely underinvestigated. A recent in vitro microarray study by Jardim et al. (2009) identified 197 miRNAs that were either upregulated or downregulated at least 1.5-fold after DEP exposure. Also, network analyses presented by Jardim et al. showed that the molecular networks mapped to the DEP-sensitive miRNAs were highly enriched for inflammatory response pathways. In previous work on the same study population from which the 10 individuals for this study were selected, Bollati et al. (2010) measured three candidate miRNAs, miR-146a, miR-222, and miR-21 involved in oxidative stress and inflammatory pathways, and showed that miR-146a expression was associated with the content of individual metals, such as lead and cadmium, in the PM10 fraction. In our transcriptome-wide study, we screened a wide number of miRNAs using a microarray approach. This type of study, particularly when considering the limited sample size, is expected to identify those miRNAs with the largest effects. Consistent with Bollati’s results, we observed a significant upregulation of miR-146a between baseline and postexposure samples. However, this study did not reveal any significant effects on miR-222, and miR-21 expression, which were found to be upregulated in postexposure samples by Bollati et al. The discrepancies between the previous and present reports may be due to differences such as sample size, the approach used for miRNA analysis, miRNA responses in bronchial cells versus blood leukocytes, and the different types of investigated exposures. Both diesel exhausts (Edling et al., 1987) and metal-rich PM particles (Cavallari et al., 2008) have been related to cardiovascular and respiratory effects in humans but are different in chemical compositions and physical properties. Our results suggest that miRNAs may show exposure-specific responses to different environmental triggers.
We characterized the functionality of differentially expressed miRNAs by measuring mRNA expression of a panel of inflammatory genes. The changes in miRNA expression might be the result of systemic inflammation or direct effects of PM. The in silico network analysis identified biological interactions for the 11 miRNA-mRNA pairs, ranging from direct miRNA targeting of mRNA to complex interactions. We found direct targeting as the determinant of the negative association between miR-29a and PTEN expression. PTEN is a phosphoinositide phosphatase that was originally identified as a tumor suppressor frequently mutated or deleted in various human cancers to promote tumorigenesis (Li et al., 1997). PTEN overexpression decreases inflammatory cytokine levels, whereas a reduced PTEN activity promotes a proinflammatory response (Furgeson et al., 2010; Koide et al., 2007). A previous study in Hep G2 cells showed that miR-29a directly inhibits the expression of PTEN by binding the 3′UTR of PTEN mRNA (Kong et al., 2011). Assuming a total context score (TCS) < −0.2 (Sualp and Can, 2011), TargetScan also predicts PTEN as a target for miR-29a (TCS = −0.55).
The in silico network analysis on the remaining miRNA-mRNA correlated pairs revealed no direct targeting but indirect interactions with different degrees of complexity. Three miRNA-mRNA pairs, i.e., miR-146a and CCL2; miR-146a and CCL5; and let-7g and NF-kB1, were connected through a one-step interaction. For instance, miR-146a can directly suppress the expression of signal transducer and activator of transcription 1 (STAT1), a transcription factor that enhances CCL2 proinflammatory gene transcription, thus resulting in reduced CCL2 mRNA levels (Supplementary fig. S2, panel A). Yang et al. (2011) have shown that in oxLDL-stimulated macrophages the secretion of CCL2 is dramatically inhibited by overexpression of miR-146a. A recent functional study confirmed that STAT1 is an miR-146a target in human peripheral blood mononuclear cells (Tang et al., 2009). An additional six pathways—linking miR-421 and IFNAR2; miR-421 and NOS2; miR-421 and PDGFRB; let-7g and ITGAX; let-7g and TGFB1; and miR-146a and CDKN1C—connected miRNAs to the corresponding mRNAs through two-step interactions. Also, those correlations are consistent with previous evidence that support our in silico network analysis. For instance, targeting of SMAD4 (SMAD family member 4) by miR-421, which was found as the initial step in the pathways linking miR-421 with IFNAR2 and miR-421 with PDGFRB, has been previously demonstrated by induction of ectopic expression of miR-421 in pancreatic cancer cell lines and consequent reduction of SMAD4 expression (Hao et al., 2011). The remaining pathway linking miR-146a and TGFB1 showed a high degree of complexity, with multiple intermediates and hubs of interactions between the miRNAs and the mRNAs.
Our study design allows for capturing variations in miRNA expression that are rapidly induced during the 4 days of exposure between the two time points at which blood samples were drawn and disappear or wane during the 2 days off work. Because of the limited number of study participants, it is possible that some of the findings in our study were due to chance. To limit false positives in the detection of differentially expressed miRNAs, we used a widely accepted strategy based on both effect-size and statistical significance cutoffs to reduce type II error (Patterson et al., 2006). The combination of a level of statistical significance of p < 0.05 and an FC with a cutoff of 2 has been previously shown to improve agreement among microarray platforms in the discovery of differentially expressed transcripts than the p-value alone (McCarthy and Smyth, 2009).
In this study, the high levels of exposures, coupled with the matched study design, provided us with a unique combination to efficiently study the PM acute effects in humans. The study design is suited to reflect the conditions of most workers, as well as of individuals exposed to outdoor PM, who are customarily exposed to variable levels of PM exposure over time. The levels of PM exposure in the steel production facility were substantially higher than the expected outdoor exposures to ambient PM. The mean level of PM10 exposure observed in our study, which was equal to 169.29 μg/m3, was approximately three times higher than the ambient PM10 levels measured in the geographic area where the plant is located (average annual ambient PM10 levels between 41 and 57 μg/m3) (Anselmi and Patelli, 2006). Although the study participants were in a modern facility with state-of-the-art systems for exposure reduction, we cannot exclude that exposures other than PM might have contributed to the observed effects. In addition to PM, workers may have additional exposures, including heat, PAHs, carbon monoxide, and nonionizing radiations. In order to avoid the possible confounding such as smoke, we selected only nonsmoker participants. We address that our study was design to mimic the real life exposure of a human population to outdoor pollution. It is worth noting that limiting our investigation to individuals who have all been working in the same facility avoided potential concerns related to the selection of external referents who might have differed from the exposed population in terms of socioeconomic factors and other characteristics determining hiring into the plant (Pearce et al., 2007).
In this integrative analysis, the data potentially provide novel insights into molecular processes that regulate cellular responses, such as proinflammatory signaling, to a representative particulate material abundant in certain forms of metal. Our results provide further evidence on the impact of acute PM exposure on gene regulation mechanisms and identify novel PM-responsive miRNAs that may control inflammatory gene expression. We acknowledge that this study is generating hypothesis in terms of mechanistic knowledge. However, future studies, using other approaches, are warranted to validate either the functional regulatory significance of the specific miRNA-gene target associations or the effects of the individual PM components on the observed responses. In vitro studies may test miRNA-gene interference, which would be expected to result in loss of miRNA-mediated gene expression (Vasudevan, 2012) and the specific mechanisms activated by single metal component in order to identify the metal with the most dramatic health effects.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences (R21ES020010, R01ES020268, R21ES019773, and R01ES00 0002); TOSCA (2010-1607); ESSIA Regione Lombardia (DGR VIII/10462); Ministero dell’Istruzione, dell’Università e della Ricerca (PRIN 2007Y84HTJ).
Supplementary Material
REFERENCES
- Anselmi U., Patelli R. (2006). Rapporto sulla qualità dell’aria di Brescia e provincia. ARPA Lombardia, Milan (in Italian) [Google Scholar]
- Bianchi F., Nicassio F., Marzi M., Belloni E., Dall’olio V., Bernard L., Pelosi G., Maisonneuve P., Veronesi G., Di Fiore P. P. (2011). A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol. Med. 3, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V., Marinelli B., Apostoli P., Bonzini M., Nordio F., Hoxha M., Pegoraro V., Motta V., Tarantini L., Cantone L., et al. (2010). Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health Perspect. 118, 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari J. M., Fang S. C., Eisen E. A., Schwartz J., Hauser R., Herrick R. F., Christiani D. C. (2008). Time course of heart rate variability decline following particulate matter exposures in an occupational cohort. Inhal. Toxicol. 20, 415–422 [DOI] [PubMed] [Google Scholar]
- Chan G. C., Fish J. E., Mawji I. A., Leung D. D., Rachlis A. C., Marsden P. A. (2005). Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J. Immunol. 175, 3846–3861 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Hwang J. S., Chan C. C., Wang P. Y., Hu T. H., Cheng T. J. (2005). Effects of concentrated ambient particles on heart rate variability in spontaneously hypertensive rats. J. Occup. Health. 47, 471–480 [DOI] [PubMed] [Google Scholar]
- Chen S. J., Chen H. C. (2011). Analysis of targets and functions coregulated by microRNAs. Methods Mol. Biol. 676, 225–241 [DOI] [PubMed] [Google Scholar]
- Chen T., Li Z., Tu J., Zhu W., Ge J., Zheng X., Yang L., Pan X., Yan H., Zhu J. (2011). MicroRNA-29a regulates pro-inflammatory cytokine secretion and scavenger receptor expression by targeting LPL in oxLDL-stimulated dendritic cells. FEBS Lett. 585, 657–663 [DOI] [PubMed] [Google Scholar]
- Corey L. M., Baker C., Luchtel D. L. (2006). Heart-rate variability in the apolipoprotein E knockout transgenic mouse following exposure to Seattle particulate matter. J. Toxicol. Environ. Health Part A. 69, 953–965 [DOI] [PubMed] [Google Scholar]
- Dockery D. W., Pope C. A., 3rd, Xu X., Spengler J. D., Ware J. H., Fay M. E., Ferris B. G., Jr., Speizer F. E. (1993). An association between air pollution and mortality in six U.S. cities. N. Engl. J. Med. 329, 1753–1759 [DOI] [PubMed] [Google Scholar]
- Edling C., Anjou C. G., Axelson O., Kling H. (1987). Mortality among personnel exposed to diesel exhaust. Int. Arch. Occup. Environ. Health. 59, 559–565 [DOI] [PubMed] [Google Scholar]
- Fichtlscherer S., Zeiher A. M., Dimmeler S. (2011). Circulating microRNAs: Biomarkers or mediators of cardiovascular diseases?. Arterioscler. Thromb. Vasc. Biol. 31, 2383–2390 [DOI] [PubMed] [Google Scholar]
- Furgeson S. B., Simpson P. A., Park I., Vanputten V., Horita H., Kontos C. D., Nemenoff R. A., Weiser-Evans M. C. (2010). Inactivation of the tumour suppressor, PTEN, in smooth muscle promotes a pro-inflammatory phenotype and enhances neointima formation. Cardiovasc. Res. 86, 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Zhang S., Zhou Y., Liu C., Hu X., Shao C. (2011). MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem. Biophys. Res. Commun. 406, 552–557 [DOI] [PubMed] [Google Scholar]
- Hesterberg T. W., Long C. M., Bunn W. B., Sax S. N., Lapin C. A., Valberg P. A. (2009). Non-cancer health effects of diesel exhaust: A critical assessment of recent human and animal toxicological literature. Crit. Rev. Toxicol. 39, 195–227 [DOI] [PubMed] [Google Scholar]
- Izzotti A., Larghero P., Longobardi M., Cartiglia C., Camoirano A., Steele V. E., De Flora S. (2011). Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat. Res. 717, 9–16 [DOI] [PubMed] [Google Scholar]
- Jardim M. J., Fry R. C., Jaspers I., Dailey L., Diaz-Sanchez D. (2009). Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 117, 1745–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski A. L., Slack F. J. (2011). Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 11, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N. (2005). MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- Koide S., Okazaki M., Tamura M., Ozumi K., Takatsu H., Kamezaki F., Tanimoto A., Tasaki H., Sasaguri Y., Nakashima Y., et al. (2007). PTEN reduces cuff-induced neointima formation and proinflammatory cytokines. Am. J. Physiol. Heart Circ. Physiol. 292, H2824–H2831 [DOI] [PubMed] [Google Scholar]
- Kong G., Zhang J., Zhang S., Shan C., Ye L., Zhang X. (2011). Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE. 6, e19518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. (2011). Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 128, 1077–1085 [DOI] [PubMed] [Google Scholar]
- Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 275, 1943–1947 [DOI] [PubMed] [Google Scholar]
- McCarthy D. J., Smyth G. K. (2009). Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 25, 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O’Briant K. C., Allen A., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson T. A., Lobenhofer E. K., Fulmer-Smentek S. B., Collins P. J., Chu T. M., Bao W., Fang H., Kawasaki E. S., Hager J., Tikhonova I. R., et al. (2006). Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24, 1140–1150 [DOI] [PubMed] [Google Scholar]
- Pearce N., Checkoway H., Kriebel D. (2007). Bias in occupational epidemiology studies. Occup. Environ. Med. 64, 562–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S., Knight J. M., Naghavi A. O., Oplt T., Creighton C. J., Shaw C., Benham A. L., Kim J., Soibam B., Harris R. A., et al. (2010). Proinflammatory role for let-7 microRNAS in experimental asthma. J. Biol. Chem. 285, 30139–30149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand S., Weber J., Lemoine F., Consales F., Paillusson A., Dupasquier M., Thomas J., Richter H., Kaessmann H., Beaudoing E., et al. (2010). Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. BioTechniques. 48, 219–222 [DOI] [PubMed] [Google Scholar]
- Samet J. M., Dominici F., Curriero F. C., Coursac I., Zeger S. L. (2000). Fine particulate air pollution and mortality in 20 U.S. cities, 1987-1994. N. Engl. J. Med. 343, 1742–1749 [DOI] [PubMed] [Google Scholar]
- Sonkoly E., Pivarcsi A.(2009a). Advances in microRNAs: Implications for immunity and inflammatory diseases. J. Cell. Mol. Med. 13, 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E., Pivarcsi A.(2009b). microRNAs in inflammation. Int. Rev. Immunol. 28, 535–561 [DOI] [PubMed] [Google Scholar]
- Sualp M., Can T. (2011). Using network context as a filter for miRNA target prediction. BioSystems. 105, 201–209 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Nakaoka T., Yamashita N. (2012). Profiling of immune-related microRNA expression in human cord blood and adult peripheral blood cells upon proinflammatory stimulation. Eur. J. Haematol. 88, 31–38 [DOI] [PubMed] [Google Scholar]
- Tang Y., Luo X., Cui H., Ni X., Yuan M., Guo Y., Huang X., Zhou H., de Vries N., Tak P. P., et al. (2009). MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 60, 1065–1075 [DOI] [PubMed] [Google Scholar]
- Tarantini L., Bonzini M., Apostoli P., Pegoraro V., Bollati V., Marinelli B., Cantone L., Rizzo G., Hou L., Schwartz J., et al. (2009). Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ. Health Perspect. 117, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S. (2012). Functional validation of microRNA-target RNA interactions. Methods. 58, 126–134 [DOI] [PubMed] [Google Scholar]
- Wang Z., Neuburg D., Li C., Su L., Kim J. Y., Chen J. C., Christiani D. C. (2005). Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ. Health Perspect. 113, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., He Y. S., Wang X. Q., Lu L., Chen Q. J., Liu J., Sun Z., Shen W. F. (2011). MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 585, 854–860 [DOI] [PubMed] [Google Scholar]
- Zhao H, Shen J, Medico L, Wang D., Ambrosone CB., Liu S. (2010). A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One 5(10): e13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.