SUMMARY
Macroautophagy is essential to cell survival during starvation and proceeds by the growth of a double-membraned phagophore, which engulfs cytosol and other substrates. The synthesis and recognition of the lipid phosphatidylinositol 3-phosphate (PI(3)P) is essential for autophagy. The key autophagic PI(3)P sensors, which are conserved from yeast to humans, belong to the PROPPIN family. Here we report the crystal structure of the yeast PROPPIN Hsv2. The structure consists of a seven-bladed β-propeller, and unexpectedly, contains two pseudo-equivalent PI(3)P binding sites on blades 5 and 6. These two sites both contribute to membrane binding in vitro and are collectively required for full autophagic function in yeast. These sites function in concert with membrane binding by a hydrophobic loop in blade 6, explaining the specificity of the PROPPINs for membrane-bound PI(3)P. These observations thus provide a structural and mechanistic framework for one of the conserved central molecular recognition events in autophagy.
INTRODUCTION
Macroautophagy (hereafter, “autophagy”) is central to the clearance of damaged organelles and harmful protein aggregates and to the survival of cells during starvation (Nakatogawa et al., 2009). Dysfunction of components of the autophagy pathway is linked to cancer, neurodegeneration, heart disease, microbial infection and ageing (Levine and Kroemer, 2008; Mizushima et al., 2008). The synthesis of PI(3)P by the class III PI 3-kinase complex is critical for the growth and remodeling of the phagophore (Noda et al., 2010; Obara and Ohsumi, 2008; Simonsen and Tooze, 2009), which is the unique membrane structure characteristic of autophagy.
The lipid PI(3)P has multiple cellular functions, acting as a major marker for endosomal and phagocytic membranes in addition to its autophagic role. In endosomal pathways, PI(3)P is recognized by FYVE and PX domain proteins (Misra et al., 2001). Most of the PI(3)P-binding proteins of autophagy lack PX and FYVE domains, with the sole exception of the mammalian FYVE domain protein DFCP1 (Axe et al., 2008). DFCP1 has no known counterpart in S. cerevisiae, and is currently thought to have a specialized rather than fundamental function in autophagy. Instead, the conserved proteins considered fundamental to the progression of autophagy all contain a predicted seven-bladed β-propeller. These proteins have been dubbed “PROPPINS” (Dove et al., 2004). PROPPINs bind to both PI(3)P and the lysosomal and vacuolar lipid PI(3,5)P2.
The PROPPIN family is defined both by its predicted β-propeller fold and the presence of an FRRG motif required for phosphoinositide binding. Four PROPPINs, WIPI-1, 2, 3, and 4 are known in humans (Polson et al., 2010; Proikas-Cezanne et al., 2004; Vergne et al., 2009), and three in yeast: Atg18 (also known as Svp1), Atg21, and Hsv2 (Barth et al., 2001; Georgakopoulos et al., 2001; Guan et al., 2001; Krick et al., 2008; Meiling-Wesse et al., 2004; Obara et al., 2008; Stromhaug et al., 2004). At least some of these proteins, notably Atg18, normally function as subunits of larger complexes. In vacuolar membrane recycling, Atg18 is part of a complex with the PI(3)P 5-kinase Fab1, the lipid phosphatase Fig4, Vac7, and Vac14 (Botelho et al., 2008; Jin et al., 2008). In autophagy, Atg18 is part of a complex with Atg2 (Obara et al., 2008).
Atg18 acts relatively early in the autophagy pathway, and localizes to the preautophagosomal structure (PAS) independent of another critical early actor, Atg1(Reggiori et al., 2004). The recruitment of Atg18 to the PAS requires the synthesis of PI(3)P (Reggiori et al., 2004) but not that of PI(3,5)P2 (Dove et al., 2004). PAS recruitment of Atg18 is required for the putative membrane shuttling activity of the membrane protein Atg9 (Reggiori et al., 2004). Thus, in the absence of the Atg18-PI(3)P interaction, the delivery of membrane to the growing autophagosome is impeded. Another PROPPIN, Atg21, is not essential for starvation-induced autophagy, but is required for the mechanistically related cytoplasm-to-vacuole (Cvt) transport pathway. Most of the core molecular machinery of yeast autophagy and Cvt is conserved, including a key role for the conjugation of a ubiquitin-like protein, Atg8, to phosphatidylethanolamine (PE). Atg21 functions upstream of the Atg8 conjugation system and is presumably involved in its recruitment to membranes (Meiling-Wesse et al., 2004; Stromhaug et al., 2004).
The term “WD-repeat protein Interacting with PhosphoInosides” (WIPI) is applied to human PROPPINs (Proikas-Cezanne et al., 2004). The founding member of this family, WIPI-1α, is aberrantly expressed in various tumor tissues, and accumulates at autophagic membranes (Proikas-Cezanne et al., 2004). Recruitment of WIPIs to autophagic membranes in mammalian cells requires PI(3)P synthesis by the class III PI 3-kinase complex and is antagonized by PI(3)P-specific phosphatases (Taguchi-Atarashi et al., 2010; Vergne et al., 2009). By analogy to yeast, mammalian PROPPIN family members (i.e. WIPI-2) act upstream of the apparatus that conjugates the mammalian Atg8 ortholog (LC3) to PE (Polson et al., 2010), and so define a critical early step in the progress of autophagy in mammalian cells.
The importance of PROPPINs in autophagy, as well as in other pathways such as recycling of vacuolar proteins, and their unusual specificity for both PI(3)P and PI(3,5)2, has spurred efforts to understand their structure. However, when expressed individually, Atg18 and most other members of this family form large soluble aggregates that are intractable to structural studies. This is not surprising since many PROPPINs are subunits of larger assemblies. We expressed and purified PROPPINs from humans, D. melanogaster, S. cerevisiae, and K. lactis in order to identify non-aggregated PROPPINs. We found that K. lactis Hsv2 was monomeric in solution, and crystallized it. On the basis of the crystal structure of Hsv2, we describe a mechanism for the recognition of PI(3)P by the PROPPINs.
RESULTS
Phosphoinositde binding and crystal structure of the PROPPIN Hsv2
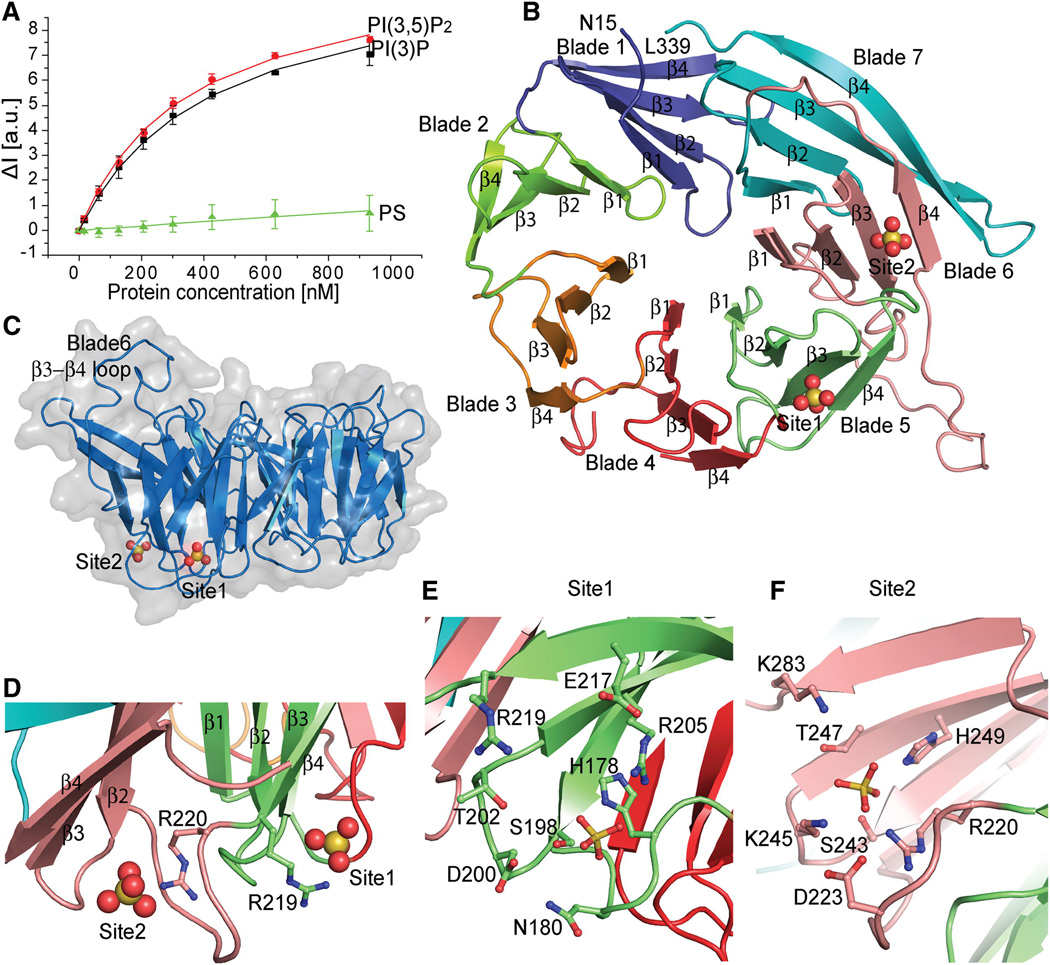
Hsv2 has been until now the least-studied of the three PROPPINs of S. cerevisiae. Hsv2 is involved in micronucleophagy (Krick et al., 2008). Hsv2 was selected as the model system for this study because it does not aggregate in solution, and therefore quantitative analyses of liposome binding can be carried out with confidence. In vitro, Hsv2 binds to both PI(3)P and PI(3,5)P2- containing liposomes with submicromolar affinity (Figure 1A). As a control for the specificity of phosphoinositide recognition, the interaction of Hsv2 with liposomes containing an equivalent negative charge density in the form of another acidic lipid, phosphatidylserine (PS), was measured. Hsv2 manifests virtually no binding to this highly acidic lipid (Figure 1A).
Figure 1. PROPPIN structure.
- FRET assay for Hsv2 binding to PI(3)P, PI(3,5)P2 and control PS liposomes. Error bars represent the standard deviation of triplicate measurements.
- Ribbon model of the Hsv2 structure, colored from blade 1–7 and the sulfates shown as spheres.
- Surface representation of Hsv2 showing the discoidal shape and location of bound sulfate ions.
- The conserved arginines of the FRRG motif coordinating the sulfates that are 17 Å apart in the two sites are shown in ball and stick model.
- Ribbon representation of Hsv2 with the sulfates and binding site 1 (E) and 2 (F) residues in ball and stick model.
The crystal structure of Hsv2 was determined at 3.0 Å (Figure 1B, S1A, B, Table 1). The structure conforms to the predicted seven-bladed β-propeller fold. The overall structure is discoidal, with the only irregularity in the disk a protrusion from the edge of blade 6 where a large loop connects β3 and β4 (Figure 1C). Hsv2 possesses the FRRG motif that is diagnostic of a phosphoinositide-binding PROPPIN (Dove et al., 2004). Typically, phosphoinositide binding domains contain a single prominent Arg-rich binding pocket for the phosphorylated lipid headgroup (Moravcevic et al., 2012). To our surprise, the structure revealed that the side-chains of Arg 219 and Arg 220 of the FRRG motif participate in not one, but two different pockets (Figure 1D–F).
Table 1.
Statistics of Crystallographic Data Collection and Refinement
| SeMet | Native | |
|---|---|---|
| Data Collection | ||
| Space group | P4132 | P4132 |
| Cell dimensions | ||
| a, b, c (Å) | 157.26, 157.26, 157.26 | 157.24, 157.24, 157.24 |
| α, β, γ(°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-3.2 (3.26-3.20) | 50.0-3.0 (3.05-3.0) |
| Rmerge(%) | 8.6 (36.2) | 10.3 (82.4) |
| I /σ(I) | 80.6 (14) | 18.1 (2.8) |
| Completeness (%) | 100 (100) | 99.9 (100) |
| Redundancy | 41.1 (38.8) | 10.3 (9.5) |
| Refinement | ||
| Resolution (Å) | 3.0 | |
| No. reflections | 13757 | |
| Rwork/Rfree | 21.8/27.0 | |
| No of atoms | ||
| Protein | 2550 | |
| Ligand/Ion | 15 | |
| B-factor (Å2) | ||
| Protein | 79.1 | |
| Ligand/Ion | 89.9 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angle (°) | 1.273 |
Hsv2 has two phosphoinositide binding sites
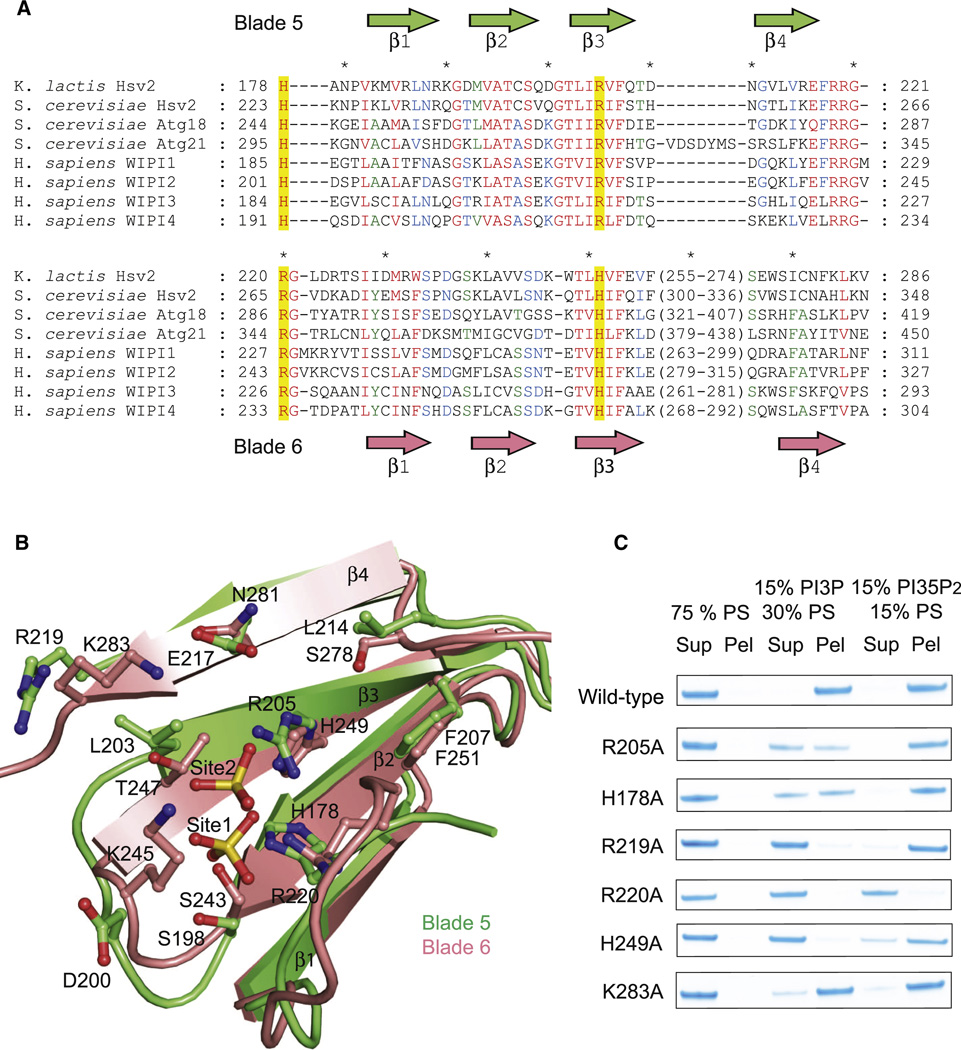
Each of the two sites is marked by a sulfate ion from the crystallization medium. The two sites, as defined by the bound sulfate ions, are separated by 17 Å. Each of the sites contains a small constellation of conserved charged and polar residues. Site 1 is built from His 178, Asn 180, Ser 198, Thr 202, Arg 205, Glu 217, and Arg 219 of propeller blade 5 (Figure 1E). Site 2 is formed by blade 6 of the propeller, and includes Arg 220, Ser 243, Lys 245, Thr 247, and His 249 (Figure 1F). The portions of blades 5 and 6 forming the two sites correspond to one another (Figure 2A, B) such that when they are overlaid, the two sulfate ions are within 2 Å of one another (Figure 2B). The high conservation of these residues (Figure 2A, Figure S2) and their physical chemical properties led us to infer that these were the sites for PI(3)P headgroup binding. It proved impossible to obtain co-crystals of Hsv2 with monomeric short-chain 3-phosphoinositides or the soluble inositol phosphates Ins(1,3)P2 and Ins(1,3,5)P3 corresponding to the lipid ligands. In order to validate the putative PI(3)P binding sites, key residues were mutated and the ability of the mutant proteins to bind to liposomes was tested (Figure 2C). Site 1 mutations R205A and H178A reduced binding to PI(3)P-containing liposomes, and R219A nearly eliminated binding (Figure 2C). Site 2 mutant R220A had no detectable PI(3)P binding, while site 2 mutants H249A and K283A had reduced binding (Figure 2C). These data show that both sites are necessary to bind efficiently to PI(3)P-containing membranes, since mutation of either site impacts binding.
Figure 2. The PROPPIN family has two conserved lipid binding sites.
- Structurally annotated alignment of PROPPIN sequences for lipid-binding blades 5 and 6. Sequences were aligned with T-Coffee within each blade based on multiple alignment of primary sequences. Residues showing 100, 80 and 60% conservation across the different proteins are highlighted in red, blue and green respectively.
- Ribbon representation of the structural superposition of blades 5 (green) and 6 (pink). The sulfates and binding sites residues are shown in ball and stick model.
- Mutations in the indicated site 1 and site 2 residues reduce or eliminate binding to liposomes.
In order to probe whether PI(3)P and PI(3,5)P2 bind to the same sites, the panel of mutants was tested against PI(3,5)P2-containing liposomes. The net charge on the liposomes was kept constant by adjusting the concentration of PS. Site 1 mutants have no effect on PI(3,5)P2 binding (Figure 2C), even though they reduce or eliminate binding to PI(3)P. This suggests that site 1 is not involved in PI(3,5)P2 binding. PI(3,5)P2 binding is, however, eliminated by the site 2 mutations R220A, and perturbed to a small extent by other site 2 mutations. We interpret these results to mean that the PI(3)P and PI(3,5)P2 overlap, and that site 2 is the more important one for PI(3,5)P2 binding.
Membrane binding mechanism
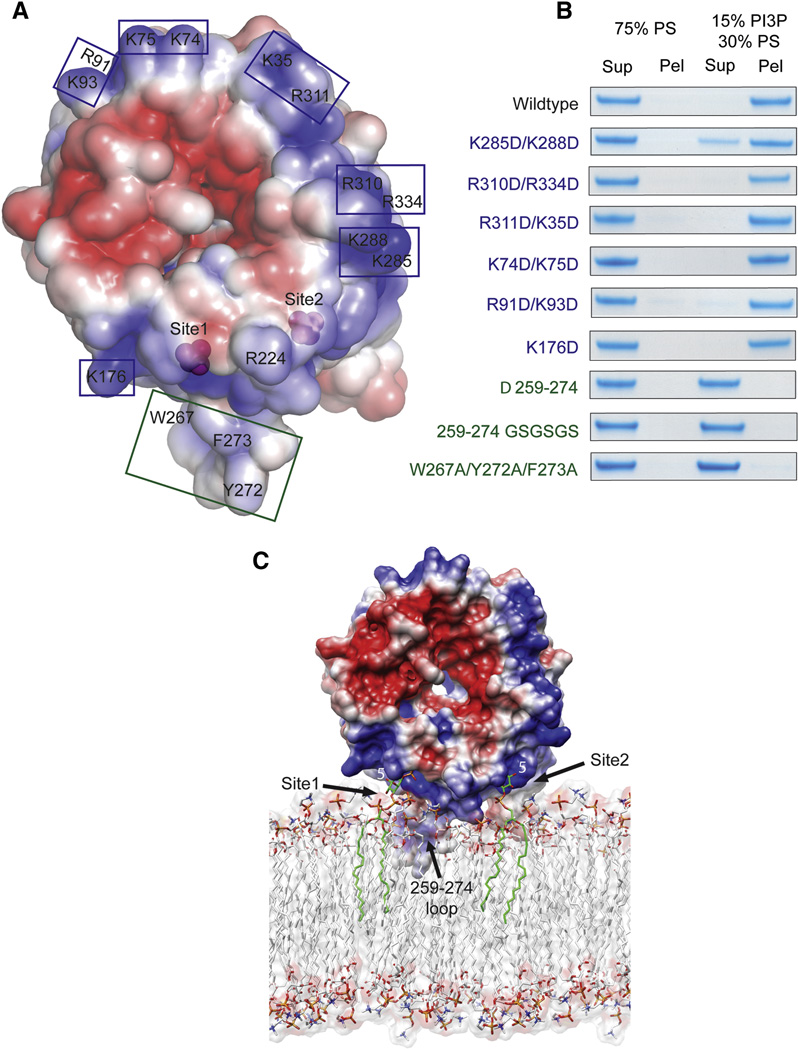
We sought to model the binding of two molecules of PI(3)P to sites 1 and 2 in the context of the membrane. The region of this face adjoining sites 1 and 2, and extending around much of the rim of the disk, is electropositive (Figure 3A, blue region). Such basic surfaces are characteristic of membrane binding domains. However, other regions of this surface, notably at the center of the disk, are electronegative (Figure 3A, red region). In one potential model, the flat face of the Hsv2 disk might dock onto the membrane surface. To test this possibility, six charge-reversal mutants were engineered at points covering a wide swath of the basic face (Figure 3A, B). Of the six charge reversal mutants, five showed no effect on binding. Only the K285D/K288D mutant, which abuts site 2, showed a small reduction in binding. The face-first membrane docking model was therefore discarded. In a second potential model, the disk could interact edge-on to the membrane such that hydrophobic residues of the blade 6 β3-β4 loop might penetrate the membrane (Figure 3C). To test this model, the blade 6 β3-β4 loop was either deleted or replaced by the sequence (Gly-Ser)3. Both of these mutants completely failed to bind liposomes (Figure 3B). Moreover, mutation of the three prominent hydrophobic residues in the blade 6 β3-β4 loop, W267A/Y272A/F273A, also completely eliminated binding.
Figure 3. How PROPPINs bind to membranes.
- Surface of Hsv2 colored according to electrostatic potential, with saturating blue and red at ±3 kT/e.
- Charge reversal mutations (blue boxes and type) in the flat basic face of the disk do not perturb binding to liposomes, but hydrophobic loop mutations (green boxes and type) eliminate binding.
- Docking of Hsv2 to a model PI(3)P-containing membrane.
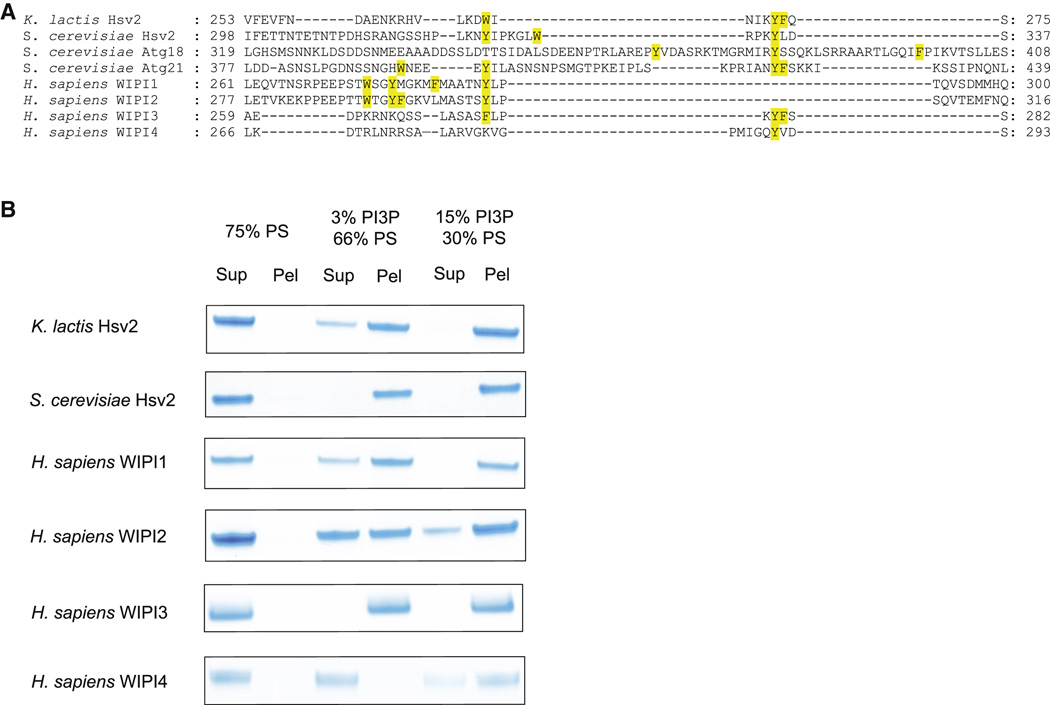
We conclude that Hsv2, and most likely other PROPPINs, bind to membranes in an edge-on geometry involving the insertion of loop residues into the membrane (Figure 3C). This binding mode leaves a putative ubiquitin binding site, which coincides with the flat face of Atg18 (Pashkova et al., 2010) fully accessible even when bound to membranes. In other PROPPINs, this loop has variable length and composition, but in all cases except for WIPI-4, the loop contains at least three aromatic residues (Figure 4A). Indeed, WIPI-4 is the only PROPPIN tested that did not bind to liposomes containing 3% PI(3)P (Figure 4B). However, even WIPI-4 binds to liposomes containing 15 % PI(3P) (Figure 4B). These observations led us to conclude that multiple membrane-penetrating aromatic residues within the β3-β4 loop play a role in the membrane targeting of the PROPPINs.
Figure 4. Hydrophobic in blade 6 augments lipid binding of PROPPINS.
- Structurally annotated alignment of PROPPIN sequences for β3-β4 region in blade 6. The hydrophobic residues in the β3-β4 loop are highlighted in yellow.
- Human WIPI4 which has one hydrophobic residue in the loop region binds more weakly to PI(3)P-containing liposomes than other PROPPINs.
PIPn specificity of other PROPPINs
In order to assess how broadly the dual specificity for PI(3)P and PI(3,5)P2 was conserved in the PROPPIN family, several other PROPPINs were tested. PI(3)P and PI(5)P form a pair of structures related by rotation about the axis through the 1 and 4 positions of the inositol ring. Rotation about this axis changes the position of the free hydroxyl groups of inositol, but interconverts the position of the 3- and 5-phosphoryl groups of PI(3)P and PI(5)P, respectively, such the their charges can occupy equivalent positions. We screened those PROPPIN family members that were stable and that did not form soluble aggregates. Specifically, S. cerevisiae Atg18 and Atg21 form large soluble oligomers or aggregates, and human WIPI-2 is marginally stable. Since this experiment was designed to assess the intrinsic lipid specificity of PROPPIN monomers, Atg18, Atg21, and WIPI-2 were omitted. K. lactis and S. cerevisiae Hsv2, and human WIPI-1, -3, and -4 all were found to bind efficiently to PI(3)P, PI(3,5)P2, and PI(5)P (Figure 5). In no case did they bind to control PS liposomes of equal net charge (Figure 5). These data suggest that the ability to bind both PI(3)P and PI(3,5)P2 with similar affinity is an intrinsic property of the PROPPIN fold. The ability to bind PI(5)P efficiently is striking and surprising, but consistent with a previous observation that WIPI-1 binds to PI(5)P (Jeffries et al., 2004). This suggests that the detailed positioning of the inositol hydroxyl groups is not of primary importance, and that it is the position of the phosphoryl group relative to the membrane matters the most.
Figure 5. Different PROPPINs bind similarly to PI(3)P, PI(3,5)P2, and PI(5)P.
Pulldown assays with 15% phosphatidylinositol phosphate liposomes show PROPPIN binding to phosphatidylinositol 3-phosphate, 5-phosphate and 3,5-bisphosphate. The final concentration of the proteins in the pulldown experiment was 1.5 µM, identical to that used for assays shown in Figures 2 and 3.
Subcellular localization of Atg18 and phosphoinositide binding
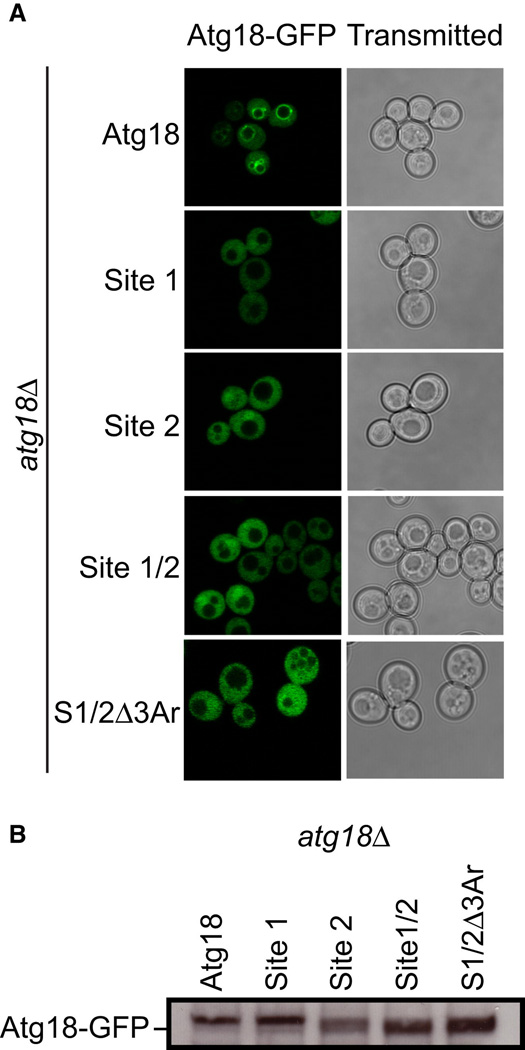
Of the three yeast PROPPINs, Atg18 has the clearest role in starvation-induced autophagy (Reggiori et al., 2004). Therefore, we selected S. cerevisiae Atg18 as the model system to assess the function of the PI(3)P binding sites in vivo. Three mutant ATG18 alleles were designed to disrupt phosphoinositide binding: ATG18Site1 (H244A/R271A), ATG18Site2 (R286A/H315A), and ATG18Site/1Site2 (H244A/R271A/R286A/H315A). In unstarved yeast, Atg18 localizes to the cytosol and the vacuolar membrane (Dove et al., 2004). Vacuolar membrane localization depends on the synthesis of PI(3,5)P2 by Fab1 and on the presence of an intact FRRG motif (Dove et al., 2004; Nair et al., 2010). Disruption of either phosphoinositide binding site results in a complete loss of Atg18 localization to the vacuolar membrane (Figure 6A). In order to investigate whether the blade 6 loop of Atg18 has a role in function, the three aromatic residues highlighted in Figure 4A, Tyr367, Tyr381, and Phe398, were mutated to Ala. This triple aromatic mutation, termed “Δ3Ar” was combined with the ATG18Site1/Site2 construct to generate the allele ATG18Site1/Site2/Δ3Ar (H244A/R271A/R286A/H315A/Y367A/Y381A/F398A). Again, a complete loss of vacuolar localization was observed. The loss of localization that was observed is not due to a decrease in the stability of Atg18 as western blotting analysis against GFP showed similar expression levels between all of the Atg18 mutants tested (Figure 6B). These data confirm that the phosphoinositide binding sites required for in vitro phosphoinositide binding are functional for in vivo membrane localization.
Figure 6. Both phosphoinositide binding sites are required for the proper localization of Atg18 in vivo.
- atg18Δ S. cerevisiae cells transformed with ATG18-GFP, ATG18Site1-GFP, ATG18Site2-GFP, ATG18Site1/Site2-GFP and ATG18Site1/Site2/Δ3Ar-GFP were grown to mid log phase and visualized by confocal microscopy. Representative cells are shown with Atg18-GFP fluorescence images on the left and transmitted light images on the right.
- Western blot analysis was performed using anti-GFP antibodies to determine the overall expression of each Atg18-GFP variant.
Autophagic function of the two sites
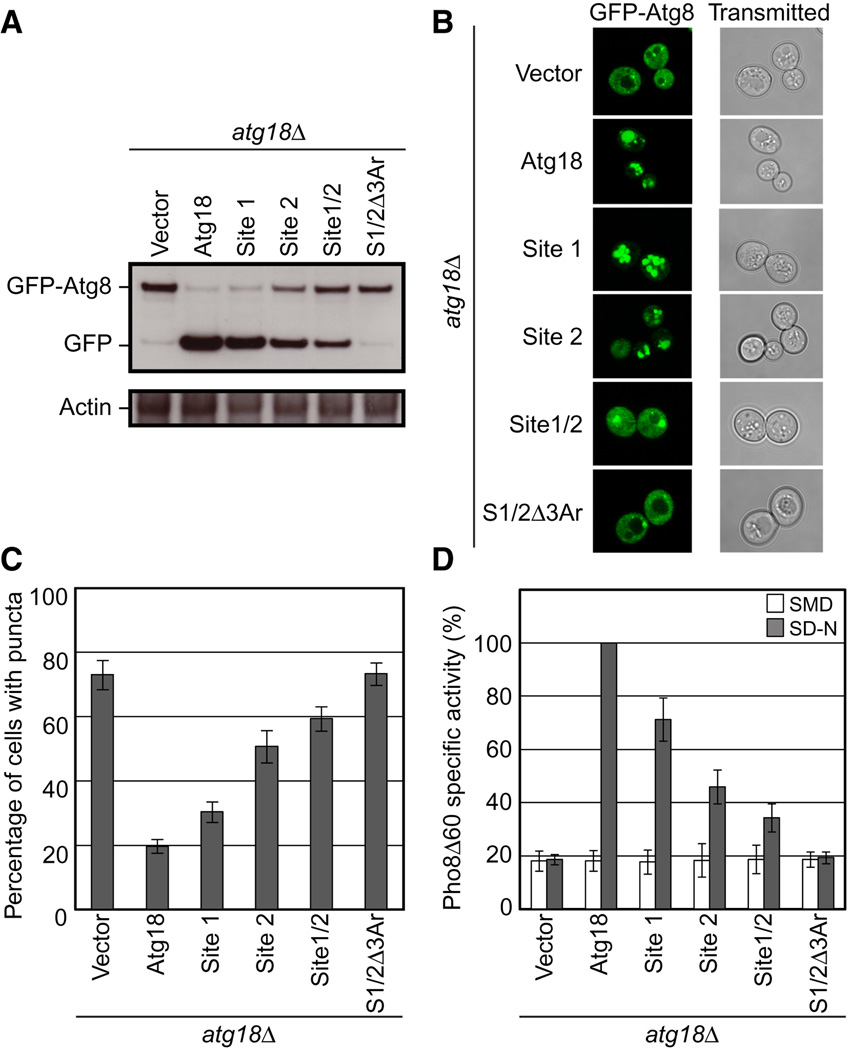
All four mutant ATG18 alleles described above showed defects in autophagy as judged by three different commonly used functional assays (Klionsky et al., 2007): GFP-Atg8 processing, Pho8Δ60 activity, and GFP-Atg8 localization (Figure 7). Atg8, an essential autophagy protein, becomes conjugated to phosphatidylethanolamine within the autophagosome and is degraded once the autophagosome fuses with the vacuole. As GFP degrades slowly in the vacuole, a fusion protein of GFP-Atg8 can be used to monitor autophagy where the production of free GFP is indicative of proper delivery of autophagic cargo to the vacuole (Figure 7A). The mutant alleles were defective in the order ATG18Site1/Site2/Δ3Ar>ATG18Site1/Site2 > ATG18Site2 > ATG18Site1. While ATG18Site1/Site2 had some residual activity, ATG18Site1/Site2/Δ3Ar has no detectable activity above background.
Figure 7. Sites 1 and 2 and the blade 6 loop aromatic residues are important for autophagy.
- GFP-Atg8 processing monitored by western blot against GFP. The full length GFP-Atg8 and GFP bands are labeled. The actin loading control is shown below.
- Representative microscopy images for GFP-Atg8 shown on the left and transmitted light images shown on the right.
- The percentage of cells displaying GFP-Atg8 puncta with a total of 100 cells counted.
- Pho8Δ60 alkaline phosphatase specific activity normalized to starved cells expressing Atg18-myc. Data for non-starved cells are shown in white and starved cells are shown in gray. Error bars in (C) and (D) represent the standard deviation of triplicate experiments.
The localization of GFP-Atg8 can be used to monitor autophagy in live cells using fluorescence microscopy (Figure 7B,C). A high percentage of cells grown in rich media contain GFP-Atg8 puncta. Once autophagy is initiated, the percentage of cells displaying GFP-Atg8 puncta decreases as Atg8 becomes targeted to autophagosomes and ultimately the vacuole. However, strains defective in autophagy show little to no decrease in the number of cells displaying Atg18 puncta, as was observed in the ATG18Site1/Site2/Δ3Ar mutant. Lastly, the engineered alkaline phosphatase, pho8Δ60, is activated only upon its arrival at the vacuole during autophagy, and the total phosphatase activity in a PHO8Δ60 pho13Δ strain provides a secondary measure for the delivery of autophagic cargo to the vacuole (Figure 7D). All mutant alleles showed the same pattern of defective autophagy as in the GFP-Atg8 processing and localization assays.
All three assays strongly confirm that both phosphoinositide binding sites are required for full Atg18 function in autophagy. In each case, ATG18Site2 had a stronger phenotype than ATG18Site1, consistent with the greater effect seen for mutants at these sites in liposome binding assays. It is important that ATG18Site1/Site2 has a stronger phenotype than either single site mutant allele, indicating that the two sites are non-redundant and functionally cooperative. In addition, the ATG18Site1/Site2/Δ3Ar resulted in a complete loss of Atg18 autophagic function, which demonstrates that aromatic residues of the blade 6 loop are important for autophagic function, as expected on the basis of the membrane anchoring role of their putative counterparts in Hsv2. This outcome also shows that a complete loss of all membrane binding determinants translates into a complete loss of autophagic function.
DISCUSSION
The crystal structure of Hsv2 and its bound sulfate ions provides the basis for an atomistic model of one of the most ancient and fundamental events in autophagy, recognition of the PI(3)P signal. While PI(3)P is a key marker of autophagosomal, endosomal and phagosomal membranes, the read-out of this signal in autophagy is unique. The FYVE domain is a key mediator of PI(3)P recognition on endosomes and is the classical example of a PI(3)P-directed conditional membrane binding domain (Moravcevic et al., 2012). The FYVE domains of Vps27 and EEA1, for example, contain stereospecific binding pockets for the PI(3)P headgroup which are insufficient for membrane targeting on their own, but are bolstered by the insertion of a hydrophobic tip into the bilayer. FYVE (Dumas et al., 2001) and PX (Bravo et al., 2001; Pylypenko et al., 2007; Zhou et al., 2003) domains have conserved interactions with both the phosphoryl and hydroxyl moieties of the inositol phosphates headgroups of their partner lipids. These hydroxyl interactions confer a high specificity for PI(3)P over its closest structural cognate, PI(5)P. For example, the PX domain of p40phox has negligible affinity for PI(5)P (Ellson et al., 2001). In contrast, we and others (Jeffries et al., 2004) find that PROPPINs bind to PI(5)P with similar to modestly lower affinity than PI(3)P. This observation, taken together with the structural model for PIP recognition (Figure 3) leads us to propose a measuring stick model for PIP recognition by the PROPPIN core. In this model, the most important binding determinant is the distance between the singly-bonded, membrane-distal phosphoryl group and the membrane core. While there are likely to be some interactions with inositol hydroxyl groups, this model posits that these are of secondary importance.
The PROPPINs are much larger than FYVE and PX domains, and contain two lipid binding sites rather than one. The presence of two binding sites confers on the PROPPINs the ability to bind avidly and with high (100s of nM) affinity to PI(3)P-containing membranes. Instead of the ordered hydrophobic membrane-binding tip found on the FYVE domain, and recently reported on the autophagy factor Beclin 1 (Huang et al., 2012), Hsv2 augments binding with flexible and exposed loop containing aromatic residues, and it seems probable that the other PROPPINs do so as well. This principle is also used, for example, by the MABP domain involved in endosomal and plasma membrane targeting (Boura and Hurley, 2012). Sequence variability in this loop may serve as a powerful vehicle for additional levels of regulation and specificity.
One puzzle that remains to be resolved is why Atg18 has such a strong preference for PI(3,5)P2 in vitro (Dove et al., 2004 and data not shown), yet functionally interacts with PI(3)P in vivo in autophagy. We speculate that because of the PIP binding sites are on the very edge of the propeller structure, they are unusually susceptible to modulation by other protein partners that could bind along the same edge. These could be heterotypic interactors, such as Atg2, Fab1, Fig4, Vac7, Vac14, or ubiquitin in the case of Atg18, or homotypic interactions within a larger homo-oligomer. These effects might be mediated by direct edge-to-edge protein:protein interactions, or indirectly via modulation of the blade 6 loop conformation.
Genomes from yeast to humans encode multiple PROPPIN genes. The PROPPIN fold, which is several-fold larger than the otherwise comparable PX and FYVE domains, has the advantage of having many more potential loci for protein-protein interactions. Indeed, such a scaffolding role is what the β-propeller fold is best known for. In the course of evolution, the ability of the PROPPIN core to assemble with other proteins, the sequence of the blade 6 loop, and the details of sites 1 and 2 themselves, have become adapted for related but distinct roles in autophagy and membrane remodeling pathways. Eukaryotic proteomes thus contain a repertoire of PROPPIN proteins with finely tuned membrane affinities and abilities to form distinct protein complexes.
EXPERIMENTAL PROCEDURES
Crystallization and Structure Determination
Native and selenomethionyl crystals were grown at 21°C using the hanging-drop vapor diffusion method by combining equal volumes of 300 µM protein and well solution containing 50 mM MES pH 6.2 and 1.8 M magnesium sulfate. The crystals were cryo-protected with 20% glycerol and flash frozen in liquid nitrogen. The Selenium SAD dataset and the native dataset were collected at Selenium edge (0.9793 Å) at the APS SER-CAT 22-ID beamline and processed with HKL2000. The crystals belonged to the P4132 space group and had a single copy of the protein in the asymmetric unit. SHELX (Schneider and Sheldrick, 2002)located all the six ordered Met sites and a partial model was generated using the program Buccaneer (Cowtan, 2006). The partial model was subjected to multiple cycles of manual model building with COOT (Emsley et al., 2010) and refinement with Refmac5 (Murshudov et al., 1999). The native structure was solved by molecular replacement method using the program Phaser (McCoy et al., 2007). The refinements were done with PHENIX (Adams et al., 2010) and the final model was refined to Rwork/ Rfree of 0.22/0.27.
Liposome binding FRET assay
FRET (Förster resonance energy transfer) was used to quantitate K.lacits Hsv2 liposome binding and fluorescence intensity was measured using a Fluorolog-3 (HORIBA Jobin Yvon) at room temperature. Tryptophan was used as the fluorescence donor and a covalently dansylated lipid (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl)) (dansyl-PE) as the acceptor. 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (PS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), and dansyl-PE were dissolved in chloroform. Dipalmitoyl phosphatidylinositol 3-phosphate (PI(3)P) and dipalmitoyl phosphatidylinositol 3,5-bisphosphate PI(3,5)P2 were dissolved in a mixture of methanol, chloroform and water at a ratio of 1:2:0.8. Lipids were mixed at the appropriate ratios to preserve the same net charge on liposomes by varying the PS mole fraction. The charges were assumed to be as follows: PS -1; PI(3)P -3 and PI(3,5)P2 -4. The final compositions in mole percent were as follows: PI(3)P-containing liposomes: 5% PI(3)P, 60% PS, 27.5% PC, 7.5% dansyl-PE; PI(3,5)P2-containing liposomes: 5% PI(3,5)P2, 55% PS, 32.5% PC, 7.5% dansyl-PE; control liposomes: 75% PS, 17.5% PC, 7.5% dansyl-PE. The lipid mixture was pipetted into glass tubes and the organic solvent was evaporated with a stream of nitrogen. Residual solvent was removed by overnight incubation in vacuum. Lipids were hydrated for 5 hr in binding buffer (20 mM Tris pH 7.4, 100 mM NaCl, 3 mM βME, 1 mM EDTA) at 4 °C and multilamellar vesicles were prepared by vigorous vortexing. Unilamellar vesicles were prepared using by extrusion through a 100 nm filter. The final lipid concentration was 0.1 mg/ml and the intensity of fluorescence of liposomes alone (I0) was recorded. Increasing amounts of protein were titrated into the lipid in a 1 ml cuvette. At each concentration, the intensity of fluorescence (I) was recorded (excitation at 280 nm and emission at 524 nm) and the change in intensity (ΔI) was calculated as the different between the recorded intensity and the intensity of fluorescence of liposomes alone (ΔI = I – I0). We observed a low level of nonspecific quenching of liposome fluorescence by protein, presumably due to absorption of excitation energy by the tryptophan residues. The baseline for nonspecific binding was established using the site 2 mutant R220A in combination with control liposomes. The nonspecific binding can be expressed by equation I = − 1.32*c / (388 + c) where c is protein concentration. Protein concentration was plotted against ΔI and fitted to a Langmuir isotherm using Origin (OriginLab Corporation). Each data point is an average of three measurements and the error reported is the error of the best fit.
Liposome sedimentation assay
The liposome sedimentation experiments were performed with the multilamellar vesicles with the specified lipid composition. 25 µl of the liposome was mixed with 25 µl protein and incubated for 30 min at 4°C. The final lipid and protein concentrations were 1 mg/mL and 1.5 µM respectively. Protein bound to the liposomes was sedimented by centrifugation at 16,000 × g for 20 min. Protein present in the pellet and supernatant fractions were analyzed by SDS-PAGE.
Yeast ATG18 Assays
The Saccharomyces cerevisiae strains used in this study are listed in Table S1. HAY1135 (GFP-ATG8) was a generous gift from R. Youle and was modified by replacing ATG18 with the nourseothricin N-acetyl-transferase gene from pAG25 (Goldstein and McCusker, 1999) to create MJR7 (atg18ΔGFP-ATG8). The S. cerevisiae strain YCY45 (atg18Δ PHO8Δ60) was a generous gift from D. Klionsky. Atg18 under the control of its endogenous promoter was cloned from Saccharmocyes cerevisiae genomic DNA (Novagen 69240) and ligated into pJK59 (Prinz et al., 2000) using NotI and XhoI which replaced Sec63 producing Atg18 with a C-terminal GFP tag. ATG18 together with its endogenous promoter was cloned from Saccharmocyes cerevisiae genomic DNA (Novagen 69240) and ligated into the centromeric plasmids yCPLAC111 and yCPLAC33 (Gietz and Sugino, 1988). A c-terminal myc tag was subsequently added using Quickchange mutagenesis. Site directed mutagenesis of the two phosphoinositide binding sites was performed using Quickchange mutagenesis and all mutants were verified by sequencing of the complete gene.
For analysis of Atg18 localization, YSC1021-554091 (Open Biosystems) transformed with ATG18-GFP, ATG18Site1-GFP, ATG18Site2-GFP, ATG18Site1Site2-GFP or ATG18Site1/Site2/Δ3Ar-GFP subcloned into pJK59were grown to mid log phase in SMD (0.67% yeast nitrogen base with ammonium sulfate, 2% glucose and supplemented with the appropriate amino acids). Wild-type and mutant ATG18-GFP cells were visualized with a 100x oil immersion objective on a LSM780 scanning confocal microscope (Carl Zeiss Microscopy). For quantification of wild-type and mutant Atg18 expression levels, cells were harvested, resuspended in 50 mM Tris pH 8.0, 1% SDS, 6M urea, 1mM EDTA and lysed by the addition of a half volume of 425–600 µm glass beads (Sigma G8772) and vortexed vigorously at 4oC. Cell lystates were subjected to western blot analysis using a GFP antibody (SantaCruz sc9996).
For autophagy assays, YCY45 cells containing yCPLAC33, ATG18-myc yCPLAC33, ATG18Site1-myc yCPLAC33, ATG18Site2-myc yCPLAC33, ATG18Site1/Site2-myc yCPLAC33, or ATG18Site1/Site2/Δ3Ar-myc yCPLAC33 were grown to mid log phase in SMD (0.67% yeast nitrogen base with ammonium sulfate, 2% glucose and supplemented with the appropriate amino acids). Cells were starved for 4 hrs in SD-N (0.17% yeast nitrogen base without ammonium sulfate and amino acids, 2% glucose) and the Pho8Δ60 alkaline phosphatase assay was performed as previously described (Klionsky, 2007). MJR7 cells containing yCPLAC111, ATG18-myc yCPLAC111, ATG18Site1-myc yCPLAC111, ATG18Site2-myc yCPLAC111, ATG18Site1/Site2-myc yCPLAC111 or ATG18Site1/Site2/Δ3Ar-myc yCPLAC33 were grown to mid log phase in SMD. Cells were starved for 4 hrs in SD-N and autophagy was subsequently monitored by following GFP-Atg8 processing, or GFP-Atg8 fluorescence in live cells. For GFP-Atg8 processing, cells were harvested, resuspended in 50 mM Tris pH 8.0, 1% SDS, 6M urea, 1mM EDTA and lysed by the addition of a half volume of 425–600 µm glass beads (Sigma G8772) and vigorous vortexing at 4oC. Cell lysates were subjected to western blot analysis using a GFP antibody (Santa Cruz sc9996). Western blot loading controls were performed against actin (Abcam 8224). Microscopy of GFP-Atg8 live yeast was performed with a 100x oil immersion objective on a LSM780 scanning confocal microscope (Carl Zeiss Microscopy). For each experiment, 100 cells were counted and the final experiment is the result of three independent trials.
Supplementary Material
HIGHLIGHTS.
Crystal structure of the yeast PROPPIN Hsv2
Two sites in propeller blades 5 and 6 bind to phosphatidylinositol 3-phosphate
Aromatic residues of a loop in propeller blade 6 help bind the membrane
Both PI(3)P binding sites in the aromatic loop residues are important in autophagy
ACKNOWLEDGMENTS
We thank D. Klionsky, R. Youle, W. Prinz, M. Lemmon, and L. Weisman for strains and DNA constructs, S. Harrison for suggesting K. lactis, B. Canagarajah for assistance with figures, L. Saidi and B. Beach for technical assistance, and R. Stanley for discussions. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This research was supported by the Intramural Program of the NIH, NIDDK (J.H.H.), an Intramural AIDS Research Fellowship (E.B.), and Ruth Kirschtein NRSA fellowship GM099319 (M. J. R.).
Footnotes
ACCESSION NUMBERS
Crystallographic coordinates have been deposited in the RCSB data bank with accession code 4EXV.
SUPPLEMENTAL INFORMATION
Supplemental information includes extended experimental procedures, two supplemental figures and one supplemental table, which can be found online at:
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Meiling-Wesse K, Epple UD, Thumm M. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 2001;508:23–28. doi: 10.1016/s0014-5793(01)03016-2. [DOI] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 Phosphoinositide Kinase Signaling Complex Requires the Fig4 Phosphoinositide Phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Hurley JH. Structural basis for membrane targeting by the MVB12-associated β-prism domain of the human ESCRT-I MVB12 subunit. Proc Natl Acad Sci U S A. 2012;109:1901–1906. doi: 10.1073/pnas.1117597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJG, Lavenir I, Perisic O, Hawkins PT, et al. The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. Acta Crystallogr Sect D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, et al. Phosphatidylinositol 3-phosphate regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T, Koutroubas G, Vakonakis I, Tzermia M, Prokova V, Voutsina A, Alexandraki D. Functional analysis of the Saccharomyces cerevisiae YFR021w/YGR223c/YPL100w ORF family suggests relations to mitochondrial/peroxisomal functions and amino acid signalling pathways. Yeast. 2001;18:1155–1171. doi: 10.1002/yea.764. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Choi W, Hu W, Min N, Guo Q, Ma M, Liu M, Tian Y, Lu PJ, Wang FL, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell. 2004;15:2652–2663. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang YL, Park S, Duex JE, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Monitoring autophagy in yeast: The Pho8Δ60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy. 2008;4:896–910. doi: 10.4161/auto.6801. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K, Barth H, Voss C, Eskelinen EL, Epple UD, Thumm M. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J Biol Chem. 2004;279:37741–37750. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- Misra S, Miller GJ, Hurley JH. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: Facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr Sect D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Nair U, Cao Y, Xie ZP, Klionsky DJ. Roles of the Lipid-binding Motifs of Atg18 and Atg21 in the Cytoplasm to Vacuole Targeting Pathway and Autophagy. J Biol Chem. 2010;285:11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21:671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Obara K, Ohsumi Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy. 2008;4:952–954. doi: 10.4161/auto.6790. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu LP, Ramaswamy S, Piper RC. WD40 Repeat Propellers Define a Ubiquitin-Binding Domain that Regulates Turnover of F Box Proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de LJ, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr Sect D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of Local PtdIns3P Levels by the PI Phosphatase MTMR3 Regulates Constitutive Autophagy. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CZ, de La Sierra-Gallay IL, Quevillon-Cheruel S, Collinet B, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, Graille M, et al. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J Biol Chem. 2003;278:50371–50376. doi: 10.1074/jbc.M304392200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.