Summary
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) guide Argonaute proteins to silence mRNA expression. Argonaute binding alters the properties of an RNA guide, creating functional domains. We show that the domains established by Argonaute— the anchor, seed, central, 3′ supplementary, and tail regions—have distinct biochemical properties that explain the differences between how animal miRNAs and siRNAs bind their targets. Extensive complementarity between an siRNA and its target slows the rate at which fly Argonaute2 (Ago2) binds to and dissociates from the target. Highlighting its role in antiviral defense, fly Ago2 dissociates so slowly from extensively complementary target RNAs that essentially every fully paired target is cleaved. Conversely, mouse AGO2, which mainly mediates miRNA-directed repression, dissociates rapidly and with similar rates for fully paired and seed-matched targets. Our data narrow the range of biochemically reasonable models for how Argonaute-bound siRNAs and miRNAs find, bind, and regulate their targets.
Introduction
Biochemical, computational, and structural studies suggest that Argonaute proteins divide their microRNA (miRNA) or small interfering RNA (siRNA) guides into functionally distinct domains. The most important domain is the seed sequence, which comprises guide nucleotides 2–7 or 2–8 (g2–g8; Lewis et al., 2005; Lewis et al., 2003; Grimson etal., 2007; Doench and Sharp, 2004). Argonaute proteins create the seed by displaying its nucleotides in a prehelical structure that lowers the entropic barrier to target binding (Ma et al., 2005; Parker et al., 2005; Wang et al., 2008a; Parker et al., 2009; Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012). The seed sequence is the primary determinant of binding specificity for both miRNAs and siRNAs (Wightman et al., 1993; Lai and Posakony, 1998; Lai, 2002; Haley and Zamore, 2004; Brennecke et al., 2005; Krek et al., 2005; Lim et al., 2005).
In the RNAi pathway, siRNAs direct Argonaute proteins to cleave complementary target RNAs at the phosphodiester bond linking target nucleotide t10 to t11 (i.e., the nucleotides paired to g10 and g11; Elbashir et al., 2001). In addition to seed pairing, target cleavage requires guide:target base pairing in this central region and the adjacent 3′ nucleotides (Ding et al., 2003; Haley and Zamore, 2004; Martinez and Tuschl, 2004; Schwarz et al., 2006). Unlike siRNAs, animal miRNAs rarely pair extensively with their targets (Bartel, 2009), although for some miRNAs, base pairs 3′ to the center of the miRNA supplement the seed sequence (Wightman etal., 1993; Lai and Posakony, 1998; Brennecke et al., 2005; Friedman et al., 2009). In mammals, ∼5% of evolutionary conserved seed-matching miRNA-binding sites have been estimated to contain such 3′ supplementary pairing (Friedman et al., 2009). How this 3′ supplementary region physically contributes to target recognition remains to be established.
Structures of achaeal, eubacterial, yeast, and human Argonaute proteins suggest that the fundamental properties of Argonautes are conserved (Song et al., 2004; Rivas et al., 2005; Yuan et al., 2005; Wang et al., 2008a, 2009; Elkayam et al., 2012; Nakanishi et al., 2012; Schirle and MacRae, 2012). To define the biochemical properties of this class of small RNA-binding proteins, we used Drosophila melanogaster Ago2 and Mus musculus AGO2 as models. We find that Argonaute divides the small RNA guide into domains—the anchor, seed, central, 3′ supplementary and tail regions—with distinct biochemical properties that explain the differences between how animal miRNAs and siRNAs bind their target mRNAs. Extensive complementarity between an siRNA and its target slows the rate at which fly Ago2 forms a catalytically competent complex and the rate at which it dissociates from an mRNA. In fact, siRNAs tether Argonaute to a highly complementary target so well that nearly all binding events end with cleavage rather than target dissociation. In contrast, Ago2-bound miRNAs paired through the seed sequence bind ≥ 4-fold more rapidly and dissociate 500-fold more quickly than a cleavage-directing siRNA. Both seed-matched and fully paired small RNAs bound to mouse AGO2 associate with and dissociate from a target RNA at similar rates.
Results
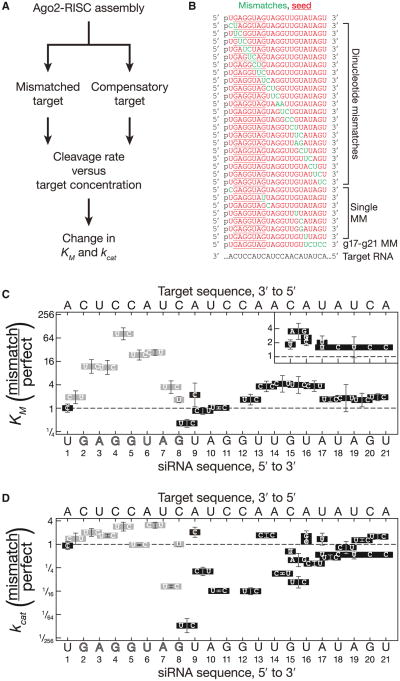
To determine how siRNA:target pairing affects Ago2 function, we systematically altered the sequence of an siRNA whose guide strand corresponds to the let-7 miRNA. We measured the rate of cleavage of a target RNA that was fully complementary to let-7 for 45 variants of the siRNA. (Figure S1 available online). The use of a common target eliminated the influence on Ago2 activity of target site accessibility (Brown et al., 2005; Ameres et al., 2007; Long et al., 2007; Tafer et al., 2008).
Of 26 overlapping dinucleotide mismatches, 22 reduced the rate of target cleavage by Drosophila Ago2 (Figure S1). To understand why some mismatches were tolerated but others were not, we determined the Michaelis-Menten parameters, KM and kcat, for 59 siRNA:target combinations comprising seven single-nucleotide mismatches, 21 dinucleotide mismatches, a contiguous g17–g21 mismatch, and 30 fully complementary siRNA:target pairs (Figures 1A and 1B and Table S1). Each siRNA was assembled into Ago2-RNA-induced silencing complex (RISC) in Drosophila embryo lysate. Half the assembly reaction was used to measure the initial rates of cleavage for a mismatched target and half for a fully complementary target (Figures 1A and S2). Because the RISC concentration was identical for the two targets, the change in kcat attributable to the mismatches corresponded to mismatched Vmax/fully complementary Vmax; similarly, the change in KM equaled mismatched KM/fully complementary KM.
Figure 1. Drosophila Ago2 Divides an siRNA into Functional Domains.
(A) Kinetic analysis scheme.
(B) siRNAs (red) were used with a single-target RNA to examine the effect of mismatches (green) on target cleavage.
(C and D) A fully complementary target analyzed in parallel for each siRNA was used to calculate the change (mismatched target/fully complementary target) in KM (C) and kcat (D). Mismatched bases are indicated in boxes. Gray, mismatches that disrupt seed pairing. Data are mean ± SD for ≥ three independent experiments. No cleavage was detected for g11g12 dinucleotide mutations (AA, UU, UA, and UC) or a UCU trinucleotide mutation at g15–g17.
A g1 Mismatch Does Not Alter KM or kcat
In early studies of fly Ago2, a mismatch between siRNA nucleotide g1 and the corresponding t1 position of its target did not impair target cleavage (Haley and Zamore, 2004). Subsequent studies of archaeal (Ma et al., 2005; Parker et al., 2005) and eubacterial (Wang et al., 2008a, 2008b, 2009) Argonautes revealed that binding of the siRNA 5′ phosphate to Argonaute forces the first nucleotide to be unpaired (Ma et al., 2005; Parker et al., 2005; 2008b). Consistent with these findings, a g1C:t1A mismatch had no detectable effect on the KM or kcat of fly Ago2 (Figures 1C and 1D).
The Seed Sequence Behaves Like a Small Helix
Seed sequence mismatches increased KM (Figure 1C). The effect of mismatches on KM was not constant across the seed (Figure 1C); mismatches at the center of the seed (g4g5) increased KM 82-fold, whereas the flanking dinucleotide mismatches (g2g3; g3g4; g5g6; and g6g7) increased KM 11-to 27-fold. These data suggest that base pairs g4:t4 and g5:t5 lie at the center of a 6 or 7 nucleotide RNA helix because central mismatches should disrupt coaxial stacking more than mismatches closer to the ends of the helix. Dinucleotide and single mismatches at the seed periphery (g1g2; g7g8 and g8) had the smallest effect, increasing KM 1.5- to 3.5-fold. The small effect of peripheral seed mismatches helps explain how miRNAs can regulate their targets through some imperfectly seed-matching sites (Ha et al., 1996; Yekta et al., 2004) and through an “offset 6-mer seed,” in which seed pairing begins at g3 and extends to g8 (Friedman et al., 2009).
Dinucleotide mismatches in the seed were generally accompanied by a small increase in kcat; central mismatches caused the greatest effect (e.g., 2.8-fold for a g4g5:t4t5 mismatch). Thus, seed mismatches decreased target binding but enhanced enzyme turnover, perhaps by accelerating release of the 3′ fragment of the cleaved target (Figure 1D).
Central Mismatches Perturb kcat
Target cleavage requires that the center of the siRNA pair with its substrate (Elbashir et al., 2001; Holen et al., 2002; Amarzguioui et al., 2003; Ding et al., 2003; Haley and Zamore, 2004). Central pairing positions the scissile phosphate of the target near the amino acid side chains that catalyze cleavage (Ma et al., 2005; Parker et al., 2005). Structures of eubacterial Argonaute bound to a DNA guide paired to RNA targets of different lengths suggest that base pairing at the center of the guide moves the three catalytic residues—and, presumably, the Mg2+ they bind—closer to the target (Wang et al., 2009). For yeast Argonaute, the rearrangement brings a fourth conserved glutamate into the catalytic site (Nakanishi et al., 2012). For fly Ago2, mismatches spanning g8 to g12 all reduced target cleavage, albeit to widely varying extents (Figures 1D and S1).
Although single-nucleotide mismatches at g8 or g9 had little effect on KM or kcat, a g8g9 dinucleotide mismatch reduced kcat by 93-fold (Figure 1D). Dinucleotide mismatches at g8g9 had a similar effect on kcat for a luciferase-targeting siRNA (Figures S3A-S3C; p value = 1.7 × 10−8; two-tailed, unpaired Student's t test). The effects on kcat of dinucleotide mismatches at g9g10 (5.0-fold reduction) and g10g11 (16-fold reduction) were more modest (Figure 1D). We saw no target cleavage for a g11g12 dinucleotide mismatch. (Our assay can detect ∼500-fold decrease in kcat.) Mismatches at positions g9g10 or g10g11 did not alter KM. Our data support the idea that central pairing enables Ago2 to achieve a catalytically competent conformation but contributes little to target binding.
Only a Subset of 3′ Base Pairs Contribute to KM or kcat
Target pairing 3′ to the center of the small RNA has been proposed to enable Argonaute to achieve a catalytically competent conformation (Haley and Zamore, 2004). Consistent with this view, a dinucleotide mismatch at g12g13 reduced kcat 16-fold, although dinucleotide mismatches at g14g15, g15g16, g16g17, or g17g18, as well as a single mismatch at g15, reduced kcat 1.5- to 9.4-fold (Figure 1D). Similarly, a g15g16 dinucleotide mismatch in a luciferase-targeting siRNA decreased kcat 7.6-fold, compared to a fully matched target RNA (Figure S3). A dinucleotide mismatch at g13g14 however, did not decrease kcat (Figure 1D). We do not know why this dinucleotide mismatch alone among the six had no detectable effect. We note that this atypical dinucleotide mismatch (CC:AA) lies between a GG:CC (g11g12) dinucleotide and G:C pair (g15). These flanking base pairs may mitigate the helical disruption caused by the intervening pyrimidine:purine dinucleotide mismatch.
The effect on KM of dinucleotide mismatches from g12 to g17 was qualitatively similar to mismatches in the seed sequence (Figure 1C). Pairing to miRNA bases g13-g16 (“3′ supplementary base pairing”) is a computational hallmark of a high confidence miRNA-binding site (Brennecke et al., 2005; Grimson et al., 2007; Bartel, 2009; Friedman et al., 2009). We observed a small but significant increased in KM for dinucleotide mismatches at g13g14 (3.6-fold, p value = 0.022), g14g15 (4.2-fold, p value = 0.017) and g15g16 (3- to 4-fold, p value = 6.6 × 10−3) and for a single-nucleotide mismatch at g16:t16 (3.4-fold, p value = 4.7 × 10−3; Figure 1C and Tables S1 and S2). A g15g16 dinucleotide mismatch also increased the KM of the luciferase siRNA by 12-fold (p value = 7.6 × 10−4; Figure S3B). Notably, the 7 nt seed of this siRNA is predicted to pair more weakly with its target (ΔGseed(25°C) = −7.7 kcal mol−1) than the seed of the let-7 siRNA (DGseed(25°C) = −11.2 kcal mol−1). Weaker seed pairing likely makes 3′ supplementary base pairing more important (Brennecke et al., 2005).
Mismatches at the center of the g12–g17 region had the greatest effect on KM, with a g14g15 dinucleotide mismatch increasing KM 4.2-fold. The g14g15 base pairs probably lie at the center of a small RNA helix, much as the g4g5 base pairs do for the seed.
The siRNA 3′ End Contributes Little to KM or kcat
The g17:t17 base pair marks the end of the 3′ supplementary binding site: a single-nucleotide mismatch at g17 and dinucleotide mismatches at g18g19 and g19g20 caused no significant change in KM or kcat. A g17g18 dinucleotide mismatch decreased kcat by 2.2-fold (p value = 0.037), whereas a g17–g21 contiguous mismatch decreased kcat by 1.9-fold (p value = 0.024), but neither had an effect on KM. In contrast, a trinucleotide mismatch within the 3′ supplementary region (g15–g17) completely inhibited target cleavage (Figures 1C, 1D, and S1).
Notably, a dinucleotide mismatch at g20g21 caused a modest increase in both KM (1.9-fold, p value = 1.7 × 10−3) and kcat (1.6-fold, p value = 6.9 × 10−3), consistent with earlier suggestions that terminal mismatches facilitate product release from plant and animal RISC (Tang et al., 2003; Haley and Zamore, 2004). We conclude that the final four nucleotides of the small RNA guide—the “tail”—form base pairs only after the target RNA is cleaved.
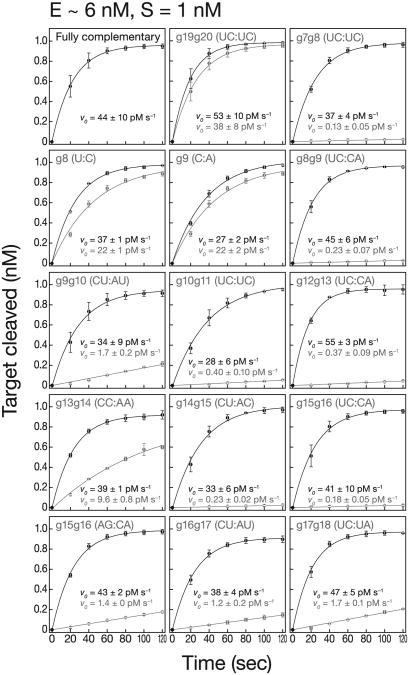
Mismatches that Reduce kcat Reflect a Defect in Catalysis
Mismatches that reduce kcat could reflect a defect in catalysis, product release, or regeneration of RISC to an active state. For these mismatches, we measured the initial rate of target cleavage (v0) under conditions of enzyme excess. When [E] > [S], v0 is largely uninfluenced by product release or enzyme regeneration because most RISCs cleave just a single molecule of target.
All mismatches that reduced the multiple turnover cleavage rate also decreased the rate when [E] > [S] (Figure 2 and Table S1). Thus, a defect in the catalytic step suffices to explain the reduced kcat. In fact, the effects of mismatches were greater when [E] > [S] than when [E] ≪ [S], suggesting that the deleterious effect of mismatches on the inherent rate of target cleavage is partially offset by a favorable effect of mismatches on steps present only when each RISC catalyzes many successive rounds of target cleavage (Table S1, relative kcat/relative v0). In other words, mismatches inhibited catalysis but promoted product release or enzyme regeneration. This was most pronounced for mismatches in the seed and 3′ supplementary region (Table S1, relative kcat/relative v0), favoring the idea that mismatches in these domains promote product release, just as they facilitate the release of miRNA* from pre-Ago1-RISC inflies and humans (Tomari et al., 2007; Kawamata et al., 2009; Yoda et al., 2010).
Figure 2. Mismatches that Impair kcat Disrupt Catalysis but Promote Turnover.
Target cleavage with [S] < [E]. Initial rates, v0, for mismatched (gray) and fully complementary targets (black) were determined by fitting the data to a single exponential. Table S1 lists the change in initial rates (mismatched versus fully complementary). Data are mean ± SD for ≥ three independent experiments.
The Standard Rules for RNA Base Pairing Apply to RISC
Might Ago create a special environment for seed base paring? To test whether the standard rules for RNA base pairing apply, we used the change in KM between mismatched and fully complementary siRNA:target pairs to calculate the free energy cost of mismatches in the seed. We compared this to the cost predicted by nearest neighbor analysis (Xia et al., 1998).
First, we tested whether nearest neighbor values determined in 1 M sodium (pH 7.0; Schroeder and Turner, 2009), changed in our more physiological conditions (100 mM potassium, 4 mM magnesium [pH 7.4]). Values obtained in our conditions agreed well with the published data (Figures S4A and S4B and Table S3). Second, an increase in KM may reflect an increase in kcat because KM = (koff + kcat)/kon. For mismatches in the seed and 3′ supplementary regions, we detected no correlated changes between KM and kcat, justifying our use of the change in KM as a surrogate for relative KD.
The free energy cost, ΔΔG25°C, calculated from the change in KM for both seed (r = 0.93, p value = 4.1 × 10−4) and 3′ supplementary (r = 0.76, p value = 2.0 × 10−3) mismatches correlated well with the values predicted by the nearest neighbor values for RNA base pairing (Figures S4C and S4D). Thus, the relative contributions of each base pair in RISC are similar to those in an RNA:RNA duplex.
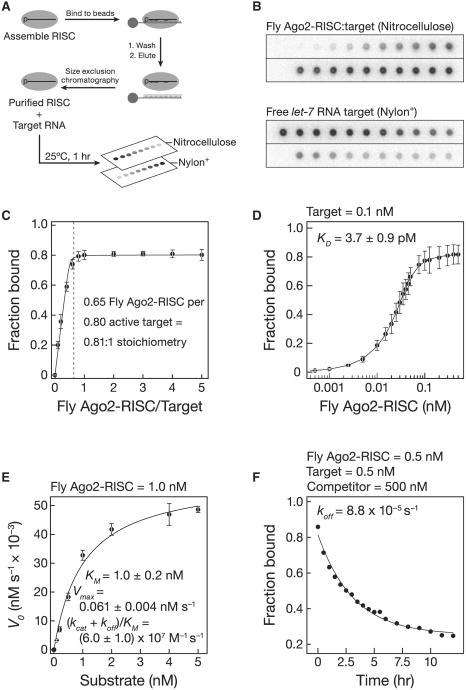
Ago2 Reduces the Affinity of a Guide RNA for Its Target
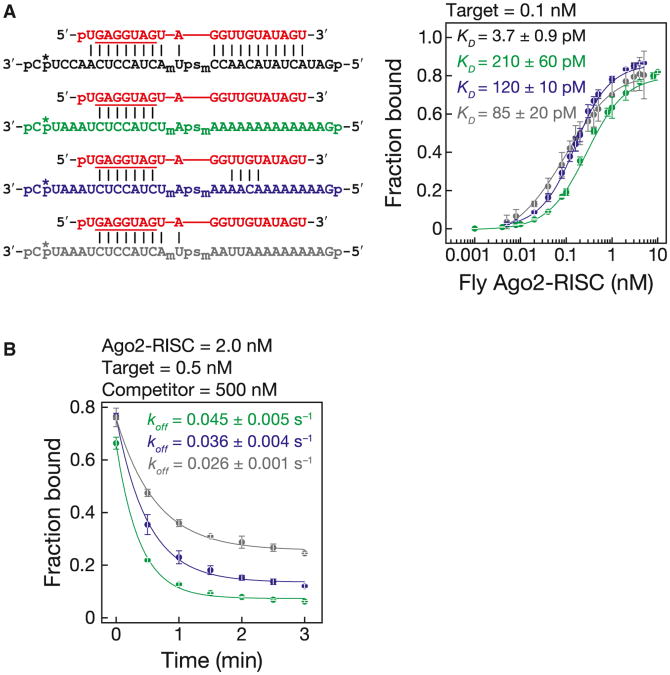
A key obstacle to measuring the binding affinity of Ago2-RISC has been the inability to purify Ago2 bound to a single siRNA guide sequence. We recently developed a simple and efficient method for purifying mature RISC assembled in Drosophila embryo lysate or mouse embryonic fibroblast S100 lysate (C.F.F.-J. and P.D.Z., unpublished data; Figures 3A and 3B). (Mouse and human AGO2 are 99% identical.) We used nitrocellulose filter binding to measure the binding affinity of both fly Ago2-RISC and mouse AGO2-RISC purified by this procedure (fly, Figures 3, 4, and 5; mouse, Figure 6). RISC concentration was determined by quantitative northern hybridization and pre-steady-state analysis (Figures S5A–S5C). To block cleavage, the target RNA contained a phosphorothioate linkage flanked by 2′-O-methyl ribose at positions t10 and t11 (Figures S5D and S5E). Stoichiometric titration showed that 0.81 fly Ago2-RISC and 1.4 mouse AGO2-RISCs bound each molecule of target, consistent with one RISC per target (Figures 3C, 6A, and 6B).
Figure 3. Fly Ago2-RISC Binding.
(A) RISC was assembled and then purified with a partially complementary, tethered 2′-O-methyl oligonucleotide.
(B) Purified Ago2-RISC was then used in filter-binding assays.
(C) Stoichiometric binding titration of target RNA with increasing amounts of purified fly Ago2-RISC. Data are mean ± SD.
(D) Equilibrium binding assays. Data are mean ± SD for 15 independent experiments with three preparations of fly Ago2-RISC.
(E) Kinetics of purified fly Ago2-RISC with a 29 nt fully complementary target RNA. Data are mean ± SD for three independent experiments.
(F) Dissociation rate for a fully complementary target RNA.
See also Figures S4 and S5, and Table S3.
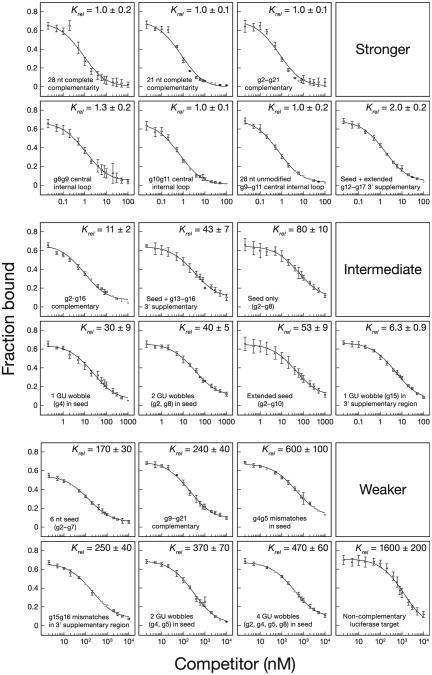
Figure 4. Fly Ago2-RISC Equilibrium Competition.
The equilibrium dissociation constant of fly Ago2-RISC for the competitor, relative to that of a fully complementary target, is reported as the mean Krel ± SD for ≥ three independent experiments. See also Table S4.
Figure 5. Fly Ago2-RISC Binds Seed-Matched Targets at the Rate of Diffusion.
(A) Binding and dissociation was analyzed for target RNAs (left) that were complementary (black) to the entire siRNA (red), the seed (green), the seed plus 3′ supplementary region (blue), or positions g2–g10 (gray). Asterisk, 32P radiolabel; subscript “m”, 2′-O-methyl ribose; “ps”, phosphorothioate linkage.
(B) Dissociation rates for the RNAs in (A). For the dissociation rate curve for the fully complementary RNA, see Figure 3F.
Data are mean ± SD. See also Figure S6.
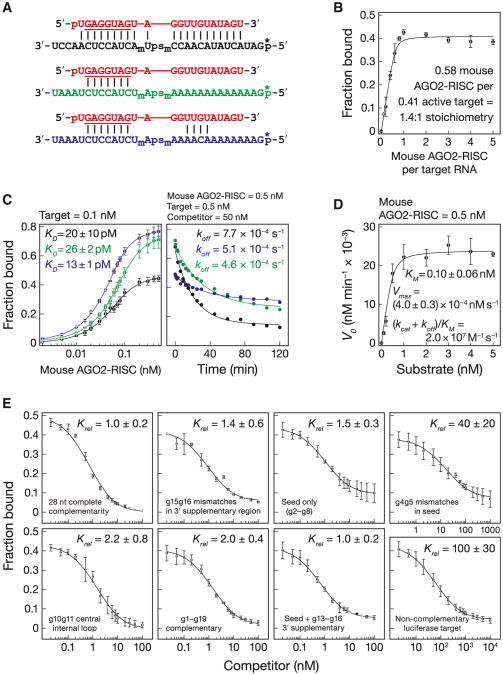
Figure 6. Mouse AGO2-RISC is Specialized for miRNA Regulation.
(A) Binding and dissociation analyses for target RNAs that were complementary (black) to the entire siRNA (red), the seed (green), or the seed plus 3′ supplementary region (blue).
(B) Stoichiometric binding titration with increasing amounts of mouse AGO2-RISC.
(C) Equilibrium binding (left) and dissociation assays (right). Data are mean ± SD for ≥ three independent experiments.
(D) Kinetics of purified mouse AGO2-RISC with a 28 nt fully complementary target. Data are mean ± SD for three independent experiments fitted to the quadratic equation for tight binding.
(E) The equilibrium dissociation constant of mouseAGO2-RISC for the competitor, relative to that of a fully complementary target, is reported as the mean Krel ± SD for ≥ three independent experiments.
Fly Ago2- and mouse AGO2-RISC bound tightly to a fully complementary RNA (Figures 3D and 6C). Our KM data and published Argonaute structures (Wang et al., 2009) suggest that 16–17 base pairs form between the guide and the target RNAs, yet the binding affinity of fly Ago2-RISC (kD = 3.7 ± 0.9 pM, mean ± S.D.; ΔG25°C− 16 kcal mol−1) and mouse AGO2-RISC (kD = 20 ± 10 pM, mean ± S.D.; ΔG25°C− 15 kcal mol−1; see below) for a fully complementary target was comparable to that of a 10 bp RNA:RNA helix. Thus, Argonaute functions to weaken the binding of the 21 nt siRNA to its fully complementary target: without the protein, the siRNA, base paired from positions g2 to g17, is predicted to have a KD∼3.0 × 10−11pM (ΔG25°C = −30.7 kcal mol−1). Argonaute raises the KD of the 16 bp RNA:RNA hybrid by a factor of > 1011.
KM is not KD
The KD measured in our binding assay (3.7 ± 0.9 pM) was ∼270-fold smaller than the KM (1.0 ± 0.2 nM) determined with purified fly Ago2 (Figure 3E). By definition, KM = (koff + kcat)/kon. When kcat≪koff, KM∼KD. To understand why KM so dramatically underestimates the affinity of fly Ago2 for a fully complementary target, we measured koff directly (Figure 3F). For fly Ago2-RISC, the dissociation rate constant, koff = 8.8 × 10−5 s−1, was much slower than the turnover rate, kcat = 6.1 × 10−2 s−1. Consequently, KM ∼kcat/kon. Hence, for fly Ago2-RISC, KM is not KD.
In contrast, the KD for mouse AGO2 (20 ± 10 pM) was only ∼5-fold smaller than the KM(0.10 ± 0.06 nM), because for mouse the dissociation rate (koff = 7.7 × 10−4 s−1) is comparable to kcat(8.1 × 10−4 s−1; Figures 6C and 6D). For mouse AGO2-RISC, KM∼ KD.
We used a competition assay to determine the contributions to binding of the anchor, seed, central, 3′ supplementary, and tail regions of the siRNA. For the fully complementary let-7 target, this assay gave values similar to those measured in the direct binding assay: 10 ± 1 pM (Figure 4 and Table S4) versus 3.7 ± 0.9 pM (Figure 3D) for fly and 36 ± 5 pM (Figure 6E and Table S4) versus 20 ± 10 pM (Figure 6C) for mouse. Binding was specific: a noncomplementary luciferase RNA target competed ∼1,600-fold less tightl y for fly Ago2 (Figure 4) and ∼100-fold less efficiently for mouse AGO2 (Figure 6E). Single-stranded sequences flanking the RISC-binding site in a target RNA have been reported to have no effect on the KM of human AGO2-RISC (Ameres et al., 2007), and we detected no difference in binding between a 28 nt (kD = 3.9 ± 0.9 pM) and a 21 nt (KD = 3.6 ± 0.7 pM) competitor for fly Ago2 (Figure 4 and Table S4).
The Fly Ago2 Seed Does Not Tolerate GU Wobble Pairs
GU wobble pairs between miRNAs and their targets have been reported to be tolerated, and some miRNA target prediction algorithms permit GU wobbles even in the seed (John et al., 2004; Miranda et al., 2006; Kertesz et al., 2007). We measured the effect of seed GU wobble pairs on target binding by fly Ago2-RISC (Figure 4). A GU wobble at g4 decreased KD by 30-fold; two GU wobbles (g2, g8) decrease KD 40-fold (Figure 4). Two GU wobbles at the center of the seed (g4, g5) reduced binding 370-fold, and four GU wobbles (g2, g4, g5, g8) decreased binding 470-fold. We conclude that GU wobbles behave like mismatches and are not tolerated in the seed. Our data explain earlier reports that GU wobbles interfere with Argonaute function (Doench and Sharp, 2004; Brennecke et al., 2005; Grimson et al., 2007) and suggest that GU pairs in the seed should not be allowed by miRNA target prediction algorithms.
Just Two-Thirds of siRNA Nucleotides Contribute to Binding for Fly Ago2
Mismatches at g1, g8g9, or g10g11 had little or no effect on binding. Likewise, a target lacking phosphorothioate and 2′-O-methyl modifications but mismatched with the siRNA from positions g9–g11 bound with an affinity similar to that of the fully complementary, modified RNA (Krel = 1.0–1.3; Figure 4). A target complementary to only siRNA nucleotides g2–g16 bound just 11-fold less tightly than a target with complete, 21 nt complementarity. In contrast, a g4g5 dinucleotide mismatch in the seed weakened binding 600-fold; a g15g16 mismatch in the 3′ supplementary region reduced binding 250-fold (Figure 4).
Thus, more than a third of the nucleotides in an siRNA guide make little or no contribution to target binding. Supporting this view, a target RNA complementary to only g2–g8 (the seed) and g12–g17 (extended 3′ supplementary pairing) bound nearly as tightly as the fully complementary RNA (Krel = 2.0 ± 0.2; Figure 4). Yet, a target complementary only to the seed and the 3′ supplementary region (g2–g8; g13–g16) bound 43-fold less tightly than the fully complementary target; a target complementary only to the seed bound 80 times less tightly. Direct binding measurements yielded essentially the same results as the competition assay (Figure 5A). Although the seed and 3′ supplementary regions supply much of the energy used by RISC to bind targets, nucleotides adjacent to the 3′ supplementary region also contribute to binding for fly Ago2-RISC.
For Fly Ago2-RISC, a 7 nt Seed Binds Better Than a 6-mer
Computational analysis in flies suggested that in the absence of 3′ supplementary pairing, 7 nt (g2–g8) but not 6 nt (g2–g7) seed complementarity can distinguish authentic miRNA-binding sites from chance complementarity (Brennecke et al., 2005), unlike in mammals, where both types of seed-matching sites have predictive power (Lewis et al., 2005). Intriguingly, fly Ago2-RISC bound a 6-mer seed-matching target 2-fold less tightly than the 7-mer seed (Figure 4). Because most miRNAs function through Ago1 in flies, it remains to be tested whether Ago1 behaves similarly.
Mouse AGO2 Is Optimized for miRNA Regulation, Not RNAi
Like fly Ago2, competition assays performed with mouse AGO2-RISC showed that central (g10g11) and terminal mismatches (g20g21) had no detectable effect on binding, whereas g4g5 seed mismatches reduced binding 40-fold (Figure 6E). Surprisingly, g15g16 mismatches did not impair binding for mouse AGO2-RISC (Krel = 1.4 ± 0.6; Figure 6E). Moreover, direct binding assays found no substantive difference in affinity between a seed-matching (KD = 26 ± 2 pM) and a fully complementary target (20 ± 10 pM; Figure 6C). We did observe a small but significant (p value = 3.2 × 10−4) increase in affinity for a target with seed and 3′ supplementary pairing (kD = 13 ± 1 pM), compared to the affinity of a target with seed pairing alone. The modest contribution of the 3′ supplementary region to target binding helps explain why in mammals less than 5% of evolutionary conserved, predicted miRNA-binding sites include conserved 3′ pairing (Friedman et al., 2009). We conclude that seed complementarity and, to a far lesser extent, 3′ supplementary base pairing, provide all the binding energy tethering mouse AGO2-RISC to its targets. Our data suggest that evolution has optimized mammalian AGO2 for miRNA-based regulation. In contrast, fly Ago2 binds far more tightly to fully complementary targets than to those matching only the seed, as might be expected for an enzyme responsible for binding and destroying viral and transposon transcripts.
Essentially Every Target that is Fully Paired to Fly Ago2-RISC Is Cleaved
To understand the molecular basis for the difference between mouse and fly Ago2-RISC, we measured the rate of dissociation of let-7-programmed fly Ago2-RISC for several prototypical RNA targets (Figure 5A).
Fly Ago2-RISC dissociated slowly from a fully complementary target: koff = 8.8 × 10−5 s−1, corresponding to a half-life (t1/2) ∼2.2 hr (Figure 3F). Given that kcat for let-7-programmed fly Ago2-RISC was 6.1 × 10−2 s−1(t1/2 ∼11 s), essentially every fly Ago2-RISC that binds a target will slice it rather than dissociate from the un-cleaved RNA (Figure 3E).
RISC dissociated far more rapidly from targets paired to the seed sequence (g2–g8) or the seed plus the 3′ supplementary region (g13-g16): koff = 4.5 × 10−2 s−1 (t1/2 ∼15 s) for seed-matched and koff = 3.6 × 10−2 s−1 (t1/2 ∼19 s) for seed plus 3′ supplementary pairing (Figure 5B). Such rapid dissociation from partially paired targets may minimize titration of RISC by seed-matching off-targets. Intriguingly, fly Ago2-RISC dissociated more slowly from a target that paired with an extended seed-match (g2–g10; koff = 2.6 × 10–2s–1, t1/2 ∼27 s) than from a target complementary to both the seed and 3′ supplementary region (Figure 5B).
Mouse AGO2-RISC Often Dissociates before It Cleaves
Mouse AGO2-RISC dissociated ∼90-fold more slowly from a seed-matched target (koff = 5.1 × 10−4 s−1; t1/2 ∼23 min) than did fly Ago2. Moreover, the mouse AGO2 dissociation rate constants for targets matching the seed, seed plus 3′ supplementary region (koff = 4.6 × 10−4 s−1; t1/2∼25 min), and the entire RNA guide (koff = 7.7 × 10−4 s−1; t1/2∼15 min) were quite similar (Figure 6C), consistent with their similar KD values. Given that the kcat for purified mouse AGO2-RISC was 8.1 × 10−4 s−1 (t1/2 ∼14 min; Figure 6D), a fully complementary target is as likely to dissociate as to be cleaved.
Our data also suggest that in both flies and mammals, the typical miRNA:Argonaute complex is in rapid equilibrium between the target-bound and unbound states, explaining why RNA-binding proteins can compete with miRNAs for overlapping binding sites (Bhattacharyya et al., 2006; Huang et al., 2007; Kedde et al., 2007; Elcheva et al., 2009; Takeda et al., 2009; Goswami et al., 2010; Jafarifar et al., 2011; Toledano et al., 2012).
miRNAs in RISC Find Their Targets at Rates that Approach that of Diffusion
We used our experimentally determined KD and koff to calculate kon(=koff /KD), the bimolecular association rate constant for RISC binding its target. For both fly and mouse AGO2-RISC, kon for targets matching only the seed and the seed plus the 3′ supplementary region were similar: kon (seed) = 2.1 × 108 M−1 s−1 and kon (seed plus 3′ supplementary) = 3.1 × 108 M−1 s−1 for fly Ago2; kon (seed) = 2.0 × 107 M−1 s−1 and kon (seed plus 3′ supplementary) = 3.6 × 107 M−1 s−1 for mouse AGO2. These rates suggest that miRNA-programmed Argonautes find their target RNAs near the limits of macromolecular diffusion (Hammes and Schimmel, 1970; Berg and von Hippel, 1985).
For fly Ago2-RISC, a dinucleotide mismatch that disrupts seed pairing (g4g5) reduced kon( = kcat + koff/KM) by ∼30-fold (Table S1) and increased koff by ∼40-fold (kon = 7.0 × 105 M−1 s−1; koff = 3.6 ± 0.9 × 10−3 s−1; Figures S6A and S6B). The KD value (5.2 nM) calculated from these kon and koff values agrees well with the KD (2.3 ± 0.6 nM) measured by equilibrium competition experiments (Figure 4 and Table S4). Our data provide strong support for the idea that in flies seed pairing must precede the formation of base pairs between the target and the 3′ half of the siRNA.
Base Pairing Beyond the Seed Proceeds at a Slower Rate for Fly Ago2-RISC
In contrast, the calculated kon (kon = 2.4 × 107 M−1 s−1) for fly Ago2-RISC binding a fully complementary target is ∼10 times slower than for a seed-matching target. For fully complementary targets in flies, (kcat + koff)/KM approximates kon and should reflect the rate at which RISC attains a catalytically active conformation, i.e., pairing from g2 to g17. Calculating kon from enzyme kinetics yields a similar value: 5.9 × 107 M−1 s−1 (Figure 3E). Taken together, our data suggest that seed pairing occurs more rapidly than the subsequent propagation of base pairs across the center of the siRNA and through the 3′ supplementary region.
We imagine that complete base pairing to fully complementary targets requires conformational rearrangement of the siRNA within fly Ago2-RISC. Structural studies of eubacterial and eukaryotic Argonautes support this idea. They reveal a conformational rearrangement of the protein near the center of the guide when it is extensively paired to its target and release of its 3′ end from the PAZ domain of Argonaute (Wang et al., 2009; Boland et al., 2011). In this view, cleavage of a target by fly Ago2-RISC is not limited by the search for a complementary sequence among the RNAs in a cell but rather by the rate at which the siRNA, bound to Argonaute, can form an additional ∼8 base pairs beyond the seed.
In contrast, mouse AGO2-RISC associates with a fully paired target at a rate (kon = 3.6 × 107 M−1 s−1) indistinguishable from seed (kon = 2.0 × 107 M−1 s−1) or seed plus 3′ supplementary pairing (kon = 3.6 × 107 M−1 s−1). The association rate derived from enzyme kinetics corroborates these measurements: kon = (koff + kcat)/KM = 2.0 × 107 M−1 s−1 (Figure 6D). Thus, fly Ago2 binds rapidly through its seed, then completes pairing of its 3′ bases more slowly, whereas mouse AGO2 binds seed-matching targets more slowly, so that the rate of propagating the helix to the 3′ half of the guide does not limit the rate of target cleavage.
Centrally Bulged Sites
Centrally bulged siRNAs are often used to model miRNA function in cultured mammalian cells (Zeng et al., 2002; Doench and Sharp, 2004; Broderick et al., 2011). This approach typically uses an asymmetric 3 × 2 internal loop at g9–g11. Although we have not measured the binding of 3 × 2 asymmetric internal loops, our results with 3 × 3 symmetric internal loops are likely to be similar. Compared to naturally occurring, seed-match sites, centrally bulged sites bind RISC 80-fold more tightly for fly Ago2 (Figure 4), suggesting an explanation why centrally bulged sites require a lower concentration of RISC to mediate reporter repression (Broderick et al., 2011).
Although a target with g9–g11 mismatch bound fly Ago2-RISC as tightly as a fully complementary RNA (kD = 3.0 ± 1.0 pM; Figure 4 and Table S4), the mechanism of binding is clearly different from the fully paired target: its measured koff value of 1.1 ±0.1 × 10−3 s−1 and calculated kon value of 3.1 × 108 M−1 s−1 are 5- to 13-fold faster than the fully complementary target (Figures S6A and S6B). We propose that the g9–g11 mismatch bypasses an energetically unfavorable rearrangement that occurs for a fully complementary target RNA. Interestingly, the crystal structure of eubacterial Argonaute shows that both ends of the guide remain anchored in the presence of a g10g11 mismatch (Wang et al., 2008a).
Discussion
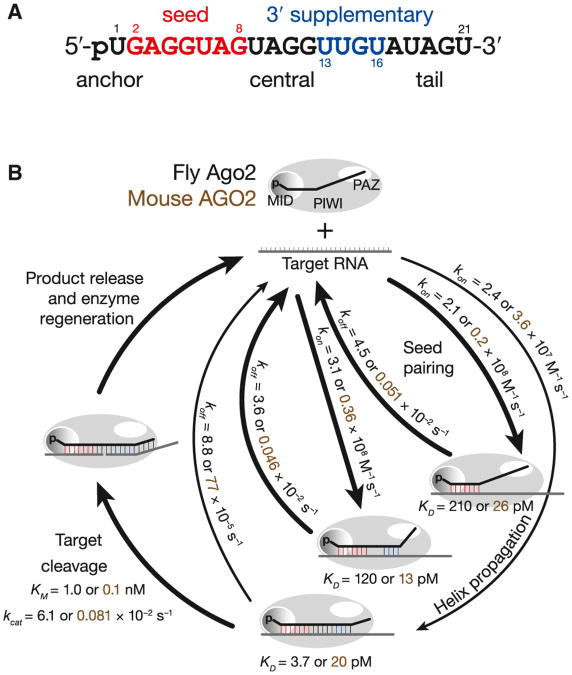
Argonaute divides a small RNA guide into anchor, seed, central, 3′ supplementary, and tail functional domains (Figure 7). Nucleotides in the anchor (g1) and tail (g18–g21) facilitate Argonaute loading and help secure the siRNA or miRNA guide to Argonaute after the passenger or miRNA* strand has been removed. But these terminal domains are unlikely to base pair with a target RNA, even when pairing is predicted by their sequences. In contrast, central base pairing (g9–g12) between the guide and target is required for efficient target cleavage. Mismatches in this central region prevent RISC from attaining a catalytically competent conformation. For fly Ago2-RISC, achieving this conformation takes more time than seed pairing alone. Our data show that nearly every fly Ago2-RISC that reaches this conformation cleaves its RNA target rather than releasing it. For mouse AGO2-RISC, a slow catalytic rate often allows the target to escape before being sliced.
Figure 7. Model for RISC Function.
(A) Loading of an siRNA or miRNA into Argonaute creates distinct functional domains in the RNA guide.
(B) A model for RISC binding and cleavage of target RNA. See also Figure S7.
In contrast, most miRNA:Argonaute complexes rapidly bind to and dissociate from their RNA targets via their seed. Even when RISC binds a target through both its seed and 3′ supplementary regions, it dissociates nearly as rapidly as for seed-only binding. Thus, the properties of RISC are essentially the same for both the typical seed-only and the less common seed plus 3′ supplementary pairing targets. That the rates of association and dissociation are so similar for these two binding modes suggests that pairing between a target and the 3′ supplementary region of a miRNA does not require winding the target RNA around the guide, side-stepping the topological problem that must be solved for siRNAs to direct RISC to cleave a target.
The finding that miRNAs use so little of their sequence to identify their regulatory targets surprised the biological community (Wightman et al., 1993; Lai and Posakony, 1998; Reinhart et al., 2000; Lai, 2002). Our data show that miRNA-programmed RISC binds with a strength and binding site size similar to those of high affinity RNA-binding proteins (e.g., Hall and Stump, 1992; Zamore et al., 1999; Zearfoss et al., 2011; Wright et al., 2011). It is siRNA-programmed RISC whose behavior should surprise us: it binds highly complementary targets far less tightly than a comparable antisense RNA because Argonaute reduces the contribution of most of its nucleotides to target binding.
What do the physical properties of RISC teach us about its cellular function? miRNAs and siRNAs are typically present in cells at dramatically different concentrations. For example, in flies in which the white gene is silenced by RNAi, the abundance of all antisense white siRNAs combined is less than that of any of 29 most abundant miRNAs (Ghildiyal et al., 2008). Previously, the ability of siRNAs to function at low abundance has been ascribed to the catalytic nature of RNAi (Fire et al., 1998; Montgomery et al., 1998; Hutvágner and Zamore, 2002). To achieve a concentration 10-fold greater than the KD for siRNA-like binding (3.7 pM for fly Ago2-RISC) would require only ∼5 molecules of RISC in ovarian terminal filament cells (∼200 μm3; Schneider, 1972) and ∼11 molecules in a cultured S2 cell (∼500 μm3; Sarikaya et al., 2012). Thus, even for Argonaute proteins with no endonuclease activity, small numbers of molecules of RISC can repress highly complementary targets; endonuclease activity is only needed when a small amount of RISC must repress a larger amount of target. The combination of high affinity and catalytic turnover helps explain why the siRNA-directed RNAi pathway provides an effective defense against viral infection in plants and invertebrate animals (Hamilton and Baulcombe, 1999; Wilkins et al., 2005; Galiana-Arnoux et al., 2006; Wang et al., 2006).
Animal miRNAs nearly always repress their targets by binding rather than endonucleolytic cleavage. This explains why animal cells express miRNAs at such high levels. Recent data suggest that only the most abundant cellular miRNAs mediate target repression (Mullokandov et al., 2012). Our data provide a biochemical explanation for this observation.
Consider two abundant miRNAs in a cultured HeLa cell (∼5,000 μm3; Cohen and Studzinski, 1967; Milo et al., 2010): miR-21 (4 nM; Lim et al., 2003) and the let-7 miRNA family, nine highly related miRNAs sharing a common seed sequence (∼3nM; Cole etal., 2009). Both miRNAs are present at a concentration greater than the KD we measured for seed matched targets for fly (∼210 pM) or mouse (∼26 pM) Ago2-RISC. Assuming a mean target mRNA abundance of ten molecules per cell and 50 different mRNA targets per miRNA, miR-21 and let-7 each regulate ∼500 (170 pM) total target mRNA molecules per HeLa cell (Friedman et al., 2009). Under these conditions, nearly every miR-21 or let-7 target mRNA (∼95%–99%) with an accessible seed match will be bound by the complementary miRNA-programmed RISC (Figure S7).
Target repression by miRNAs can be reduced by the presence of competitor RNAs containing miRNA binding sites that titrate miRNA-RISC away from the mRNAs it regulates (Arvey et al., 2010; Garcia etal., 2011; Mukherji etal., 2011). The fundamental properties of RISC make specific predictions about how the activity of specific miRNAs can be inhibited by the expression of these competitor transcripts. The effect of such competitor RNAs reflects the concentration of both the miRNA and miRNA-binding sites (Ebert and Sharp, 2012), as well as the affinity of miRNA-RISC for those sites. For abundant miRNAs such as miR-21 or the let-7 family, the expression of competitor RNAs containing miRNA binding sites–even highly complementary binding sites–will have little impact on the regulation of their target genes in flies or mammals. Doubling the expression of mRNAs repressed by miR-21, for example, would require ∼7.8 nM seed only competitor and ∼4.0 nM fully paired competitor for fly Ago2-RISC. For mouse AGO2-RISC, it would still require ∼7.7 nM seed only competitor and ∼7.2 nM of the fully paired competitor. Taken together, this translates to ∼22,400 copies of seed only competitor and ∼12,000–21,700 copies of fully paired competitor (Figure S7). If the competitor contained one miRNA-binding site, it would comprise 12%–50% of all the mRNA in the cell (Islam et al., 2011).
In contrast, doubling the expression of the mRNA targets for an intermediate (mir-93; ∼140 pM) or a low abundance miRNA (mir-24; 7.3 pM) would require just 600–800 additional seed-matching sites (Figure S7). For mir-93 whose abundance confers the ability to bind to ∼60% of all potential targets, the competitor must be as abundant as the sum of all the target mRNAs (∼500 copies). Low abundance miRNAs like mir-24 are unlikely to contribute much biologically meaningful regulation because they are present at a concentration less than their KD for seed-matching targets in both flies and mammals: <4% of miR-24 targets are expected to be bound by the miRNA at any given time. Using the conservative assumption that every bound miRNA-RISC completely represses an mRNA target, miR-24 is predicted to reduce the expression of the average seed-matched target by <4% (Figure S7).
Thus, the proposal that “competing endogenous RNAs” (“ceRNAs”) sequester miRNAs, derepressing the authentic targets of that miRNA (Salmena et al., 2011), applies only to a small subset of miRNAs whose cellular concentration and target abundance meet a narrow range of values. The miRNAs with the largest impact on gene expression–the most abundant miRNAs–are not predicted to be regulatable by endogenous, transcribed seed-matched competitor transcripts. Consistent with this view, viral and experimental inhibition of specific miRNA function by transcribed RNA requires the use of extensively complementary miRNA-binding sites that recruit a cellular pathway that actively degrades the targeted miRNA (Ebert et al., 2007; Loya et al., 2009; Ameres et al., 2010; Xie et al., 2012). Absent this target directed, catalytic destruction of miRNAs, RNAs of ordinary abundance are unlikely to compete with mRNAs for binding abundant, biologically functional miRNAs.
Experimental Procedures
General Methods
Target cleavage reactions were performed as described (Haley and Zamore, 2004; Haley et al., 2003) except with 4 mM Mg2+. Cleavage targets (Table S2) were prepared by in vitro transcription and capping (Haley et al., 2003). For binding, synthetic RNAs were 5′ or 3′ 32P radiolabeled.
Binding, Competition, and Dissociation Assays
Ago2-RISC was assembled with let-7 siRNA in 0–2 hr embryo lysate or S100 from immortalized Ago2−/− MEFs expressing mouse AGO2 (O'Carroll et al., 2007). Binding reactions were at 25°C for 1 hr; protein-RNA complexes were captured on nitrocellulose and unbound RNA on Nylon membranes under vacuum and washed with ice-cold buffer. Competition reactions were at 25°C for 1 hr (mouse) or 6 hr (fly). For koff, Ago2-RISC was incubated with 32P-radiolabeled RNA target for 1 hr then competitor RNA was added and dissociation measured.
Supplementary Material
Acknowledgments
We thank D. Turner, J. Chen, A. Carruthers, R. Gilmore, S. Ryder, B. Farley, O. Bilsel, S. Kathuria, and P. Gandhi for help and discussions; the Turner lab for use of equipment; A. Boucher, T. Covello, and G. Farley for technical support; and members of the Zamore lab for critical comments on the manuscript. This work was supported in part by National Institutes of Health grants GM62862 and GM65236. Phillip D. Zamore is a cofounder and scientific advisor to Alnylam Pharmaceuticals.
Footnotes
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.10.036.
References
- Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci USA. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- Cohen LS, Studzinski GP. Correlation between cell enlargement and nucleic acid and protein content of HeLa cells in unbalanced growth produced by inhibitors of DNA synthesis. J Cell Physiol. 1967;69:331–339. doi: 10.1002/jcp.1040690309. [DOI] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schwarz DS, Keene A, Affar B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J Biol Chem. 2010;285:20532–20540. doi: 10.1074/jbc.M110.109298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Hall KB, Stump WT. Interaction of N-terminal domain of U1A protein with an RNA stem/loop. Nucleic Acids Res. 1992;20:4283–4290. doi: 10.1093/nar/20.16.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammes GG, Schimmel PR. In: 2 Rapid Reactions and Transient States. Enzymes D, Paul B, editors. Vol. 2. San Diego: Academic Press; 1970. pp. 67–114. [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, Lönnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. Regulation of Drosophila neurogenesis by RNA: RNA duplexes? Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic micro-RNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′ -end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38(Database issue):D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using micro-RNA sensor and decoy libraries. Nat Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya DP, Belay AA, Ahuja A, Dorta A, Green DA, 2nd, Extavour CG. The roles of cell size and cell number in determining ovariole number in Drosophila. Dev Biol. 2012;363:279–289. doi: 10.1016/j.ydbio.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Schroeder SJ, Turner DH. Optical melting measurements of nucleic acid thermodynamics. Methods Enzymol. 2009;468:371–387. doi: 10.1016/S0076-6879(09)68017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS ONE. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano H, D'Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008a;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008b;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 2011;30:533–545. doi: 10.1038/emboj.2010.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, SantaLucia JJ, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Bartel DP, Lehmann R, Williamson JR. The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry. 1999;38:596–604. doi: 10.1021/bi982264s. [DOI] [PubMed] [Google Scholar]
- Zearfoss NR, Clingman CC, Farley BM, McCoig LM, Ryder SP. Quaking regulates Hnrnpa1 expression through its 3′ UTR in oligodendrocyte precursor cells. PLoS Genet. 2011;7:e1001269. doi: 10.1371/journal.pgen.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.