Abstract
A simultaneous on-tissue proteolytic digestion and extraction method is described for the in-situ analysis of proteins from spatially distinct areas of a tissue section. The digestion occurs on-tissue within a hydrogel network, and peptides extracted from this gel are identified with liquid chromatography tandem MS (LC-MS/MS). The hydrogels are compatible with solubility agents (e.g. chaotropes and detergents) known to improve enzymatic digestion of proteins. Additionally, digestions and extractions are compatible with Imaging Mass Spectrometry (IMS) experiments. As an example application, an initial IMS experiment was conducted to profile lipid species using a traveling wave ion mobility mass spectrometer. On-tissue MS/MS was also performed on the same tissue section to identify lipid ions that showed spatial differences. Subsequently, the section underwent an on-tissue hydrogel digestion to reveal 96 proteins that co-localized to the rat brain cerebellum. Hematoxylin and Eosin (H & E) staining was then performed to provide additional histological information about the tissue structure. This technology provides a versatile workflow that can be used to correlate multiple complementary analytical approaches in the analysis of a single tissue section.
Keywords: Imaging mass spectrometry, matrix-assisted laser desorption ionization (MALDI), on-tissue digestion, ionotropic hydrogels, traveling wave ion mobility spectrometry, protein identification
INTRODUCTION
Since its initial development,1 matrix-assisted laser desorption ionization (MALDI) Imaging Mass Spectrometry (IMS) has become more accessible through streamlined workflows encompassing all steps in an experiment (tissue sectioning, matrix deposition, MS analysis and data/image processing) for pharmaceutical,2 proteomic,3 lipidomic4 and histological5 applications. Recent advances in the field have focused on improved sensitivity of targeted chemical classes and achieving high spatial resolution IMS.6, 7 Concurrent to these developments, the field has addressed challenging clinical and biological applications.8–11
Identification of imaged small molecules (e.g. pharmaceuticals, peptides and lipids) commonly proceeds with on-tissue tandem MS (MS/MS) and accurate mass measurements. These experiments usually follow an initial IMS experiment and thus are carried out on the section already imaged or on an adjacent serial tissue section. This allows for the best co-localization of the initial imaged precursor ion to the confident MS/MS identification from product ions. However, on-tissue protein identification can be laborious. One approach utilizes traditional proteomic methodologies to identify proteins.12,13 The first step may involve one of several approaches: microextraction from the tissue surface with aqueous/organic solvents, tissue homogenization from multiple neighboring tissue sections, laser capture microdissection (LCM) collection of regions of interest, or bulk tissue homogenization depending on the localization and predicted relative amount of the desired protein. High-performance liquid chromatography (HPLC) fractionation of this bulk tissue homogenization often follows where the fractions are collected throughout the gradient. Aliquots of these fractions are spotted onto a MALDI target for MS analysis to determine what fractions contain the proteins of interest. The aliquot may be subjected to additional offline clean-up and separations such as sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to further isolate the approximate molecular weight fraction in a gel band. A standard in-gel proteolytic digestion procedure is performed, and the product peptides are identified via HPLC-MS/MS and database searching. This process works well for many soluble and more abundant protein species and in applications where there is a relatively large amount of tissue available. However, the process is time consuming, sample (tissue) quantities are often limited and isolating distinct micro-regions of tissue for spatially-directed analysis can be difficult.
A second approach for on-tissue protein identification is the use of in-situ proteolytic digestion.14 Typically, serial sections of tissue are employed. On one tissue section, the IMS experiment is performed targeting proteins, peptides, lipids or other classes of molecules. On an adjacent serial tissue section a proteolytic enzyme (e.g. trypsin) is deposited in a similar array to that of the imaged section. After the enzyme is applied and digestion is allowed to proceed, application of a MALDI matrix follows for peptide imaging. The protein and peptide images are correlated using post-processing tools to direct subsequent on-tissue MS/MS analyses. This approach correlates the original ion image with protein identification through peptide analysis at identical locations on the sections. A drawback of this approach includes the poor on-tissue digestion efficiency caused from rapid drying of the small droplets of enzyme solution that are required to achieve high spatial resolution. Increasing the aqueous content of the enzyme solution and/or spraying/depositing larger droplets can mitigate the problem, but this may lead to delocalization of endogenous biological molecules which will make image correlation and confident identification challenging. Additionally, the total number of tryptic peptides relating to the desired protein species may be limited, reducing the confidence of identification. Previously reported methods14,15 for in-situ digestion on tissue sections do not typically probe as deeply into the proteome when compared to bulk homogenates analyzed by HPLC-MS/MS identification.
Improvements and complementary methods are needed to address difficulties and challenges of the on-tissue identification process. More user-friendly approaches should be adopted to obviate the need of costly robotic liquid extraction, matrix deposition and tissue isolating instruments. Described here is a spatially-directed simultaneous on-tissue proteolytic digestion and extraction technique to be used in conjunction with existing MALDI IMS workflows. This new approach utilizes on-tissue protein identification within a hydrogel microreactor16 network to simultaneously digest and extract proteins and peptides followed by traditional peptide sequence analysis.
EXPERIMENTAL
Reagents
For hydrogel synthesis, alginic acid sodium salt was purchased from Alfa Aesar (Ward Hill, MA USA) and calcium chloride dihydrate was purchased from J.T. Baker (Center Valley, PA USA). The hydrogel additives Triton X-100, ammonium bicarbonate and proteomics grade trypsin from porcine pancreas (dimethylated), the MALDI matrices 2,5-dihydroxybenzoic acid (DHB, 98 %) and sinapinic acid (98 %) and the acids trifluoroacetic and formic acid and were all purchased from Sigma Aldrich (St. Louis, MO USA). Solvents (ethanol, xylenes, methanol and acetonitrile) were all HPLC grade and the histological dyes (hematoxylin and eosin) were purchased from Fisher Scientific (Fairlawn, NJ USA). 18 MΩ water was provided via a Millipore Milli-Q Synthesis A10 (Billerica, MA USA). All reagent listed were used without additional purification.
Hydrogel fabrication
Hydrogels were fabricated using a previously described method utilizing template chromatography paper (Whatman, Buckinghamshire, UK).17–19 Briefly, designs of various shapes and sizes were created in computer software and color laser printed (Bizhub C360, Konica Minolta, Ramsey, NJ USA) onto the chromatography paper (20 × 20 cm). The color levels were increased to maximize the amount of ink printed. The paper was reprinted with the identical pattern for a total of 3 times to ensure a thick ink coating. The paper template was then cut out and heated for 120 sec on each side with a heat gun (low setting ~300 °C, 1400 W Milwaukee heat gun, Brookfield, WI USA). This allowed for the hydrophobic color ink to melt and penetrate into the paper for defined hydrophobic boundaries. The patterned templates were then soaked in 500 mM CaCl2 for 60 sec as the free Ca2+ ions act as a crosslinking agent to penetrate into the applied polymer creating an ionotropic hydrogel. The hydrogel polymer (alginate) was diluted to 2 % w/v in water. The polymer solution was mixed with 200 mM ammonium bicarbonate with 0.02 % Triton X-100 in a 1:1 ratio such that the final polymer solution was 1 % w/v in 100 mM ammonium bicarbonate and 0.01 % Triton X-100. This solution was first mixed with trypsin prior to being spotted onto the calcium soaked pattern templates. The volume deposited (Vtot) and the amounts of trypsin in each digestion are outlined per experiment in the Results and Discussion section.
Tissue Sectioning and Pre-treatment
Brains from Sprague-Dawley rats were collected and stored at −80°C prior to sectioning (Pel-Freez Biologicals, Rogers, AR USA). Coronal and sagittal sections (12 and 9 µm, respectively) were taken at −19°C on a cryostat (Leica Microsystems Inc., Bannockburn, IL USA). Tissue sections were thaw mounted onto premium microscope slides (Fisher Scientific, Fairlawn, NJ USA) which were cleaned previously in 100 % ethanol (twice) and 100 % methanol rinses (60 sec each).
Protein digestion, extraction and identification
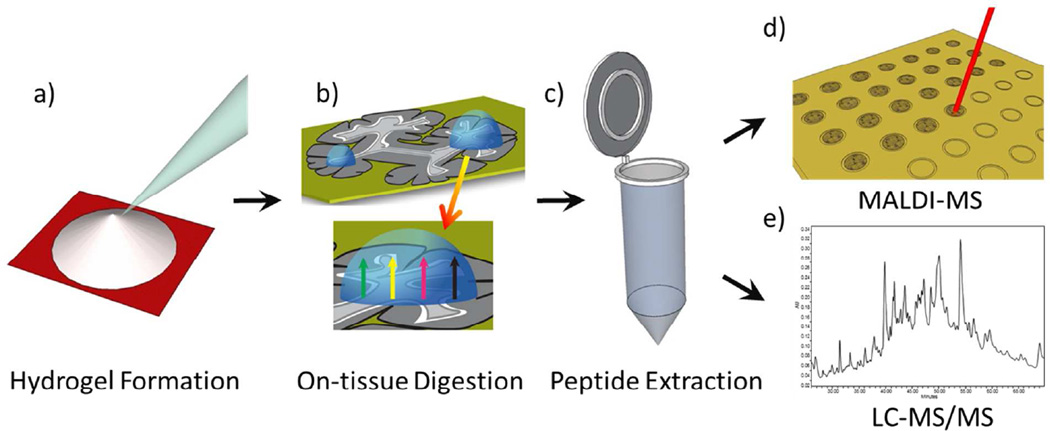
The ionotropic hydrogel mixture was spotted onto the paper target where it was polymerized (< 5 minutes) (Figure 1a). The gel was then removed from the paper pattern via a spatula, thin nosed tweezers or suction via a pipette tip and placed onto a mounted piece of tissue (Figure 1b). To retain the enzyme activity and improve digest efficiency, the slide was placed in a glass petri dish with a small (4 cm × 4 cm) wetted (250–1000 µL depending on gel and tissue sizes to minimize condensation on tissue) piece of paper towel underneath the slide. The petri dish was closed with the glass lid and wrapped with parafilm to make an air-tight seal. The petri dish was placed in an oven and heated to a desired temperature (e.g. 50 °C) for 6–12 hours depending on the tissue thickness, size/shape of the gel, enzyme, etc. This process ensured that the hydrogel did not become dry during digestion. After the digestion, the gel was removed and placed in a micro-centrifuge tube to undergo organic (50 % acetonitrile/5 % formic acid) and aqueous (50 mM NH4HCO3, 25 mM CaCl2) solvent extractions (process repeated twice) to shrink and swell the hydrogel to extract digested peptides (Figure 1c). The supernatants of each extraction are combined and dried in a centrifugal vacuum concentrator (SPD Speedvac, Thermo Scientific, Waltham, MA USA). The reconstituted extract (50–75 µL in 0.1 % formic acid) was then spotted for MALDI MS (Figure 1d) and LC-MS/MS (Figure 1e) and could be stored at −80 °C. All microscopy was performed with an Olympus BX 50 (Center Valley, PA USA) with the following histology procedure: 95 % ethanol 30 sec, purified water 30 sec, hematoxylin 120 sec, water 15 sec, 70 % ethanol 15 sec, 95 % ethanol 15 sec, eosin 60 sec, 95 % ethanol 15 sec, 100 % ethanol 15 sec, xylenes 120 sec.
Figure 1.
Hydrogel-mediated proteomic digestion and extraction workflow beginning with a) hydrogel synthesis on a laser printed piece of chromatography paper, b) on-tissue placement of gel for proteolysis and incubation, c) solvent extraction in aqueous and organic solvents, and analysis of reconstituted extracts with d) MALDI MS and/or e) LC-MS/MS.
Mass spectrometry analysis
MALDI MS profiling was conducted by spotting 2 µL of a 1:1 mix of reconstituted digest mix with 20 mg mL−1 sinapinic acid in 90 % acetonitrile and 0.25 % trifluoroacetic acid. A Bruker Ultraflextreme mass spectrometer (Bremen, Germany) was used to acquire spectra with the spot size set to medium, laser energy at 78 % and laser frequency set to 500 Hz. Resulting spectra were an average of multiple laser shots. This data was processed using FlexAnalysis v3.3.
MALDI IMS experiments were performed with a Waters Synapt G2 instrument (Manchester, UK) at laser frequency 1000 Hz, 1 scan sec−1 and 125 µm × 125 µm laser step. MS acquisition was in Resolution mode with trap and transfer collision energies set to 4 V and 2 V, respectively. Traveling wave ion mobility settings were as follows: nitrogen gas flow at 65 mL min−1 (2.5 mbar), helium cell gas flow at 165 mL min−1, 450 µs pulse delay, ion mobility wave height 40 V, variable ion mobility wave velocity starting at 1100 m s−1 and ending 400 m s−1 with the velocity ramped over 100 % of the cycle. DHB was applied with an automated sprayer at a concentration of 5 mg mL−1 in 90 % acetonitrile and 0.25 % trifluoroacetic acid for a total of 4 passes (HTX Technologies TM Sprayer, Carrboro, NC USA). On-tissue MS/MS was performed at designated collision energies (32.5 V for PC 36:1, 35 V for PC 34:1 and 37.5 V for PC 36:4) applied in the trap triwave after precursor ion selection in the quadrupole. Data was processed in Waters HD Imaging software (images normalized to total ion current), Driftscope v2.2 and Masslynx.
Extracted peptides were analyzed by a 75 min data dependent LC-MS/MS analysis. Briefly, peptides were loaded via pressure cell onto a 40 mm by 0.1 mm self-packed reversed phase (Jupiter 5 µm, 300 Å C18 – Phenomenex, Torrance, CA USA) trapping column fritted into an M520 filter union (IDEX, Lake Forest, Illinois USA). After loading and equilibration, this trapping column was attached to a 200 mm by 0.1 mm (Jupiter 3 µm, 300 Å C18 - Phenomenex, Torrance, CA USA), self-packed analytical column with a laser pulled tip (i.d. 1 µm) coupled directly to an LTQ linear ion trap (Thermo Scientific, Waltham, MA USA) using a nanoelectrospray source. Reverse phase separation was performed on a Waters nanoAcquity UPLC (Milford, MA USA) using the following gradient run at 500 nL min−1 flow rate: initial flow of 98% A (0.1 % formic acid), 2 % B (acetonitrile, 0.1 % formic acid) ramped to 25% B over 45 min, to 90% B at 60 min where it remained for 5 min, and then a 2 min ramp back to 2% B where it remained for an additional 8 min. A series of a full scan mass spectra followed by 5 data-dependent MS/MS spectra was collected throughout the run and dynamic exclusion was enabled to minimize acquisition of redundant spectra. MS/MS spectra were searched via SEQUEST against a rat database (UniprotKB taxon 10116 - reference proteome set) that also contained reversed versions for each of the entries.20 Identifications were filtered and collated at the protein level using Scaffold (Proteome Software, Portland, OR USA) with a 0.5 % false discovery recovery rate.
RESULTS and DISCUSSION
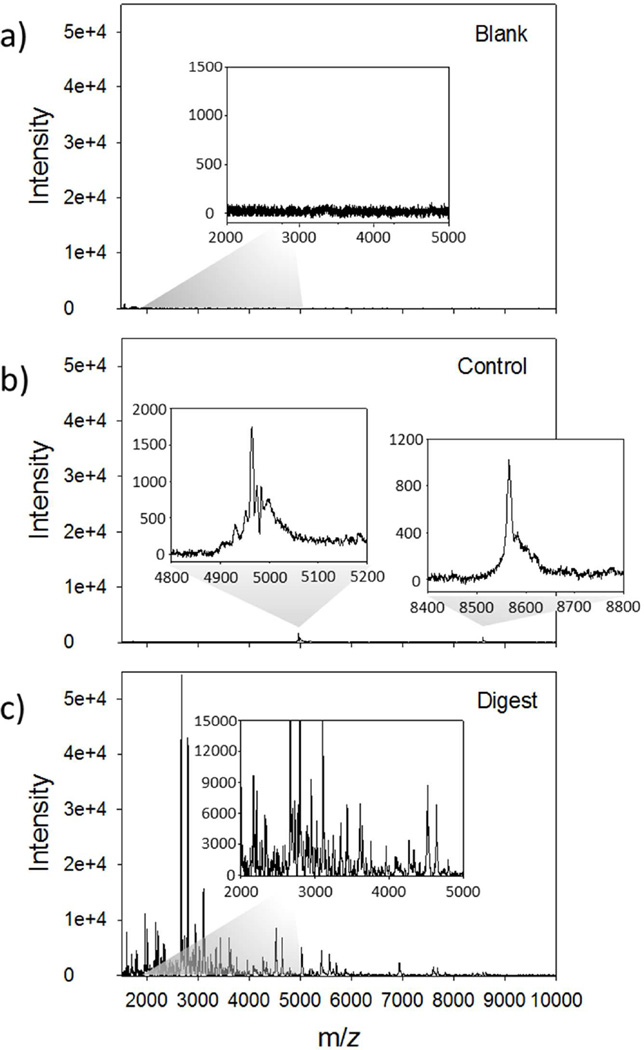
A workflow for a typical hydrogel mediated on-tissue digestion is shown in Figure 1. A circular 4 mm hydrogel containing trypsin (Vtot = 18 µL, 20 µg trypsin in 100 mM NH4HCO3) was placed on a 12 µm thick section of rat cerebrum and allowed to react for 6 hours in a humidity chamber at 50 °C. Following digestion, the gel underwent solvent extraction for peptide recovery, and the resulting peptides were dried down and reconstituted to a volume of 75 µL. MALDI MS spectra (2000 shots summed, linear mode) of a blank (trypsin loaded off-tissue, Figure 2a), control (no trypsin on-tissue, Figure 2b) and digested (trypsin loaded on-tissue, Figure 2c) hydrogels obtained from 2 % (by volume) of the total extract. Abundant signals were present throughout the mass range demonstrating extensive on-tissue digestion and efficient extraction from the hydrogel network of the trypsin loaded on-tissue hydrogel (Figure 2c). The absence of signals from the hydrogel polymer is the result of the presence of CaCl2 in the extraction buffer which maintains the integrity of the ionotropic hydrogel, and prevents contamination of the supernatant. For a blank a hydrogel with trypsin was placed on an empty microscope slide (Figure 2a) and as a control, a gel without trypsin was placed on a serial section of tissue (Figure 2b). As shown in the blank spectrum (Figure 2a), no signal is present indicating autolytic trypsin peptides are below the detection limit for the amount of trypsin used in this study. The control spectrum shows a lack of peptide signals in the lower m/z 1500–4000 range, but some low intensity signals in the higher mass range (m/z 4500–9000) were observed, e.g. m/z 4964 and 8566 (Figure 2b inserts). It is likely that these signals are from the abundant and commonly observed intact proteins thymosin β4 and ubiquitin, respectively.21 Interestingly, the presence of these ions indicates that abundant smaller proteins can passively diffuse into these gels as well. Potential modifications of the procedure to improve the extraction of intact proteins into the gels include increasing the porosity22 or altering the alginate (or other hydrogel) properties.23 This could be of particular usefulness in off-line solution digestions of hydrogel extracted proteins (discussed below) and top-down protein identification workflows.
Figure 2.
MALDI MS profile analysis of 2 % (by volume) of the reconstituted hydrogel mediated digestion extracts from rat brain cerebrum (12 µm thickness). Linear mode full scan spectra (m/z 1500–10000, 2000 shots summed) from a) trypsin loaded hydrogel placed off-tissue (blank), b) on-tissue hydrogel without trypsin (control) and c) an on-tissue trypsin hydrogel (digest).
The LC-MS/MS analysis of this sample digestion identified 211 proteins. This amount is in range with similarly sized punched sections of mouse liver fresh frozen and formalin-fixed and paraffin-embedded tissues analyzed by LC MALDI FTICR and TOF/TOF analysis,24 and comparable to simple protein extractions without detergent additives from bulk homogenizations of mouse brains analyzed by LC-MS/MS.25 There were 85 proteins with extensive sequence coverage, having more than 5 unique peptides identified. Also of interest were high molecular weight proteins that are not readily observed in typical MALDI IMS experiments (Table 1). For example, 26 large proteins were confidently identified with predicted molecular weights greater than 100 kDa. Many of these higher molecular weight proteins were identified as membrane proteins (e.g. several Na+/K+ transporting ATPases, neurofascin, clathrin heavy chain and ankyrin-2), cytoskeletal proteins (e.g. α actinin, microtubule-associated proteins and spectrin), extracellular matrix proteins (restrictin) and cytoplasmic proteins (e.g. puromycin-sensitive aminopeptidase, protein bassoon and dynein heavy chain). Another 77 of the 211 proteins were detected in the 50–99 kDa range, also a challenging intermediate high mass range for MALDI IMS.
Table 1.
High molecular weight (> 100 kDa) proteins identified with on-tissue hydrogel-mediated protein digestion and extraction on rat brain cerebrum.
| Protein Name | UniProt Accession Number |
Molecular Weight (kDa) |
# Unique Peptides |
# Assigned Spectra |
|---|---|---|---|---|
| Hexokinase type 1 | P05708 | 102 | 4 | 6 |
| α actinin 1 | Q6GMN8 | 103 | 5 | 10 |
| Puromycin-sensitive aminopeptidase | P55786* | 103 | 4 | 6 |
| Type II brain 4.1 minor isoform | Q9JMB2 | 107 | 3 | 5 |
| Na+/K+ ATPase α3 subunit | P06687 | 112 | 20 | 67 |
| Na+/K+ transporting ATPase α2 chain precursor | D3ZSA3 | 112 | 3 | 6 |
| Na+/K+ transporting ATPase α1 chain precursor | P06685 | 113 | 10 | 17 |
| Contactin-1 precursor | Q63198 | 113 | 6 | 9 |
| Neurofilament heavy polypeptide | F1LRZ7 | 114 | 8 | 11 |
| Ubiquitin-activating enzyme E1 | Q5U300 | 118 | 3 | 4 |
| Electroneutral K+/Cl− cotransporter 2 | Q63633 | 124 | 2 | 2 |
| Plasma membrane Ca2+ ATPase isoform 2 | P11506 | 137 | 6 | 9 |
| Neurofascin | P97685 | 138 | 2 | 2 |
| Plasma membrane Ca2+ transporting ATPase 1 | P11505 | 139 | 3 | 4 |
| Ras GTPase-activating protein SynGAP | Q9QUH6 | 143 | 3 | 3 |
| Tenascin-R precursor (Restrictin) | Q92752* | 150 | 2 | 3 |
| Clathrin heavy chain 1 | P11442 | 191 | 9 | 13 |
| Microtubule-associated protein 2 | P15146 | 199 | 21 | 46 |
| Microtubule-associated protein 1B | P15205 | 270 | 8 | 13 |
| Spectrin β chain, brain 2 | Q9QWN8 | 271 | 5 | 7 |
| Non-erythroid spectrin β | Q6XD99 | 274 | 29 | 47 |
| Spectrin α chain, brain | P16086 | 285 | 60 | 109 |
| Microtubule-associated protein 1A | G3V7U2 | 300 | 15 | 27 |
| Protein bassoon | O88778 | 418 | 2 | 4 |
| Ankyrin-2 | Q01484* | 437 | 5 | 5 |
| Dynein heavy chain, cytosolic | P38650 | 532 | 8 | 9 |
Predicted protein based on sequence homology from Homo sapiens
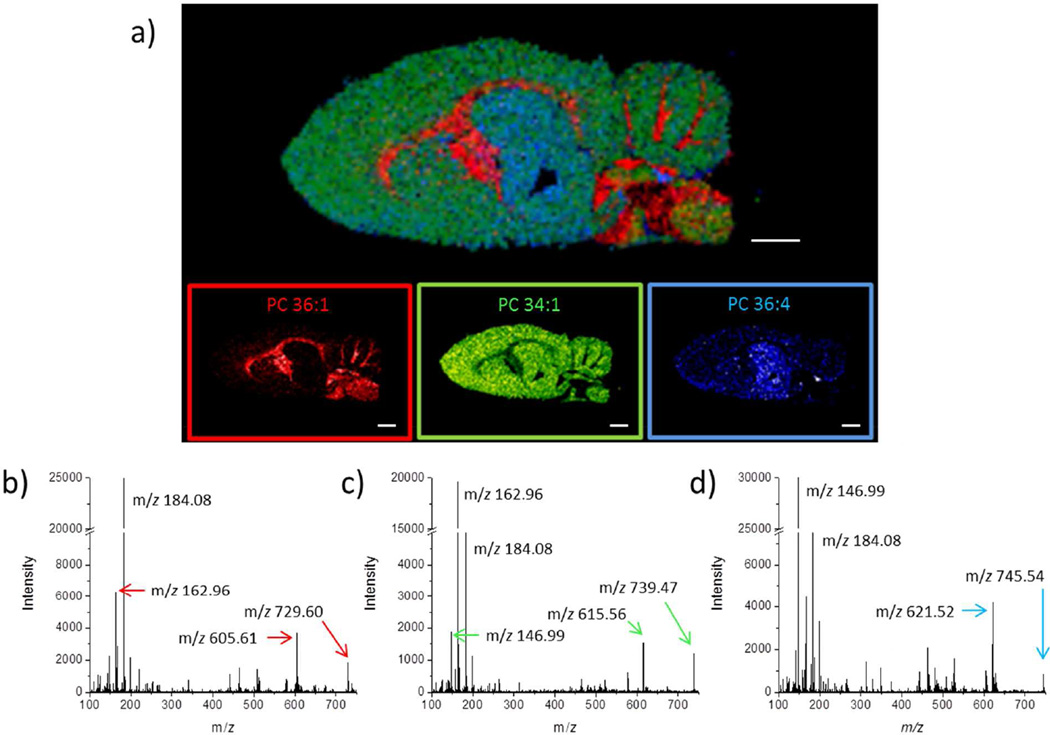
The hydrogels can also be used for a sequential analysis workflow in conjunction with a previously imaged section of tissue. In this case, the digestion and extraction occur after the initial IMS and on-tissue MS/MS, but before histological staining (Figure 3). A 9 µm thick section of rat brain was imaged with the traveling wave ion mobility spectrometry (TWIMS) separation. The use of TWIMS in MALDI IMS experiments separates MALDI matrix ions from the targeted biological ions.26,27 Shown in Figure 3a is an overlaid ion image of three identified lipid species assigned to phosphatidylcholines (PC) at m/z 788.62, 798.54 and 804.55, relating to PC 36:1 (red), PC 34:1 (green) and PC 36:4 (blue), respectively. These species were identified based on accurate mass and MS/MS (Figure 3b–d) of neighboring pixels after the initial full scan IMS experiment, and confirmation that these ions fell into the lipid trend line in the ion mobility dimension of separation (Supporting information Figures S1, S2 and S3 and Appendix on MS/MS identification).
Figure 3.
a) Image overlay of three PC lipid species analyzed using ion mobility MALDI IMS: PC 36:1 (red), PC 34:1 (green), PC 36:4 (blue). Scale bars are 2 mm. On-tissue MS/MS spectra for the precursor b) protonated ion of PC 36:1 at m/z 788.62, c) potassiated ion of PC 34:1 at m/z 798.54 and d) sodiated ion PC 36:4 at m/z 804.55 were obtained prior to on-tissue hydrogel digestion.
After the lipid IMS and MS/MS experiments were completed, the tissue was not visibly damaged since the laser energy used (energy level 275, approximately 46 % power) was sufficiently high enough to obtain signal, but below the threshold for ablation of the tissue (Supporting Information Figure S4). Before proceeding to the in-situ digestion, the tissue was washed two times for 30 sec (70 % ethanol and 100 % ethanol, respectively) to remove the residual DHB matrix, salts and lipids on the tissue surface. A hydrogel disc (diameter = 1.5 mm, Vtot = 8 µL containing 10 µg trypsin and 0.1 % Triton X-100 in 100 mM NH4HCO3) was placed on the cerebellum region of the tissue and incubated for 6 hours at 50 °C. For comparison, another imaged rat brain tissue section was washed and a 1.5 mm (diameter) region of cerebellum was removed for an in-solution digestion (Vsol = 250 µL) with the same amount of trypsin and Triton X-100. A third imaged tissue section was treated the same as the previous two, but a hydrogel without trypsin was placed on the cerebellum as a control. After 6 hours of incubation, the control gel was placed in a 250 µL solution with 10 µg trypsin and 0.1 % Triton X-100 in 100 mM NH4HCO3 for 6 hours at 50 °C. All samples were dried and reconstituted to identical volumes (V = 50 µL), and the samples were analyzed via LC-MS/MS.
The on-tissue digestion using the hydrogel protocol identified 96 proteins compared to 147 for the in-solution tissue digestion and 22 proteins for the control hydrogel solution digestion. The identified high molecular weight (> 100 kDa) proteins again reveals many membrane (e.g. hexokinase-1, several Na+/K+ transporting ATPases, neural cell adhesion molecule 1, etc.) and cytoskeletal proteins (e.g. microtubule-associated proteins, neurofilament heavy polypeptide, spectrin, etc) identified using the homogenization and hydrogel digestions (Table 2). However, the digestion efficiencies of the two approaches differed. Based on the number of unique peptides identified and the number of assigned spectra, the in-solution homogenization digestion was a more extensive, albeit destructive, digestion procedure compared to the current iteration of the hydrogel digestion. However, given that the same amount of trypsin was used in both methods, the difference may be attributed to the limited volume of the hydrogel used on-tissue (8 µL) that was more than 30× smaller compared to the solution volume of the homogenization (250 µL), and the requirement for proteins and peptides to migrate into the hydrogel during the digestion incubation. The accessibility of proteins for digestion was greater in the homogenization procedure since trypsin mobility in solution is greater than in the hydrogel network, and peptides are released into solution rather than required to diffuse from tissue into the hydrogels. Future iterations of on-tissue hydrogels can be produced to reduce the polymer network density by using alternative hydrogel compositions and creating gels with alternative geometries to increase the total volume-to-surface area ratio of the structure.
Table 2.
Comparison of the high molecular weight (>100 kDa) proteins identified with an in-solution homogenization protocol and an on-tissue hydrogel-mediated protein digestion on a previously imaged rat brain cerebellum.
| Protein Name | UniProt Accession Number |
Molecular Weight (kDa) |
In-solution Homogenization |
On-tissue Hydrogel |
||
|---|---|---|---|---|---|---|
| # Unique Peptides |
# Assigned Spectra |
# Unique Peptides |
# Assigned Spectra |
|||
| Microtubule-associated protein 6 | Q63560 | 100 | 6 | 6 | 4 | 4 |
| Hexokinase-1 | P05708 | 102 | 3 | 3 | 3 | 4 |
| Na+/K+ transporting ATPase subunit α 3 | P06687 | 112 | 18 | 24 | 8 | 9 |
| Na+/K+ transporting ATPase subunit α 2 | P06686 | 112 | 7 | 8 | 2 | 2 |
| Na+/K+ transporting ATPase subunit α 1 | P06685 | 113 | 6 | 7 | 2 | 2 |
| Contactin-1 | Q63198 | 113 | 3 | 3 | 3 | 4 |
| Neurofilament heavy polypeptide | F1LRZ7 | 114 | 7 | 9 | 2 | 2 |
| Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2 | P11507 | 115 | 4 | 4 | 2 | 2 |
| Neural cell adhesion molecule 1 | F1LUV9 | 119 | 5 | 6 | 3 | 3 |
| Plasma membrane Ca2+ transporting ATPase 2 | P11506 | 137 | 7 | 9 | 7 | 7 |
| Clathrin heavy chain 1 | D4AD25 | 192 | 14 | 16 | 2 | 2 |
| Microtubule-associated protein 2 | P15146 | 202 | 6 | 6 | 2 | 2 |
| Microtubule-associated protein 1 light chain LC1 | F1LRL9 | 270 | 5 | 5 | 3 | 3 |
| Spectrin β 2, isoform CRA_a | G3V6S0 | 274 | 20 | 20 | 9 | 10 |
| Spectrin α chain, brain | E9PU22 | 285 | 38 | 41 | 10 | 11 |
| Microtubule-associated protein 1A | P34926 | 300 | 4 | 5 | 2 | 2 |
| Cytoplasmic dynein 1 heavy chain 1 | P38650 | 532 | 6 | 6 | 3 | 3 |
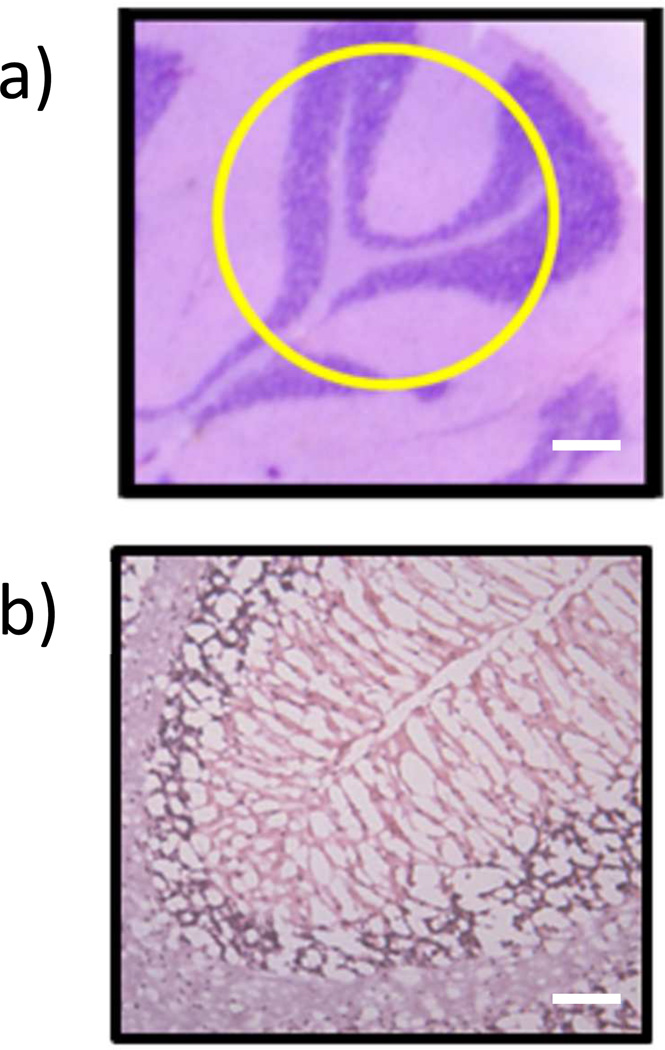
The hydrogel digested tissue sections can be used for additional analysis such as histological staining (hematoxylin and eosin stain, H & E) for evaluation of cells and surrounding tissue regions. The hydrogel-digested tissue was washed, stained and analyzed by microscopy (Figure 4). A serial rat brain section that had not been imaged or treated previously was stained with H & E by the identical procedure for comparison purposes (Figure 4a). It is apparent that there are differences between the two stained tissue sections mainly in the cellular density. There are a lower number of nuclei (stained blue/purple by hematoxylin) present in the imaged and extracted tissue section compared to an untreated section (Figure 4b). The losses are due to a combination of cellular material being extracted into the hydrogel during incubation/digestion, and the extensive washing steps prior to staining which may have dislodged cellular material from the slide surface. The loss of tissue into the hydrogel is a critical insight into the current function of the gels. Future refinements to these materials will focus on enhancing extraction of already imaged cells and cellular material to maximize proteomic coverage and/or minimizing the observed tissue damage while still obtaining useful proteomic information to be used in conjunction with histological examination of the same tissue section. Nonetheless, differential identification of regions (e.g. extent of myelination, white and grey matter regions, location of granular cells) within the tissue was possible in this first study using the protocol for H & E staining. We are currently developing new staining protocols that are amenable to the pretreatment of the tissue sections to prevent any potential damage to the tissue section.
Figure 4.
a) Hematoxylin and eosin (H & E) stain of an untreated rat cerebellum region with outlined (yellow) region of where the hydrogel was placed on the serial digested tissue (scale bar 250 µm) and b) the H & E stain of the serial sectioned, previously imaged (Figure 3) and hydrogel digested rat cerebellum (scale bar 50 µm).
In future studies, hydrogels that are an order-of-magnitude smaller in size (diameters of < 250 µm) will be utilized. Making hydrogels of this size with the paper template method described here will be challenging due to the difficulties involved with manual spotting of the hydrogel solution and laser template printing of features that are useable in the sub-millimeter range. Preliminary attempts to make sub-millimeter gels by hand have achieved diameters down to ~400 µm (Supporting Information Figure S5), however at these sizes it is impractical to manually place and recover the gels for an IMS-directed experiment without damaging the tissue and/or the hydrogel disc. Acoustic deposition of reactants,28 as used with MALDI matrix application, would allow for co-regisitration of previously imaged regions with protein extracted regions similar to a histology-directed approach.5 Additionally, patterning of the hydrogels into ordered micro patterns is a strong possibility to add new functionality and IMS applications to existing hydrogel microarray techniques.29–32
CONCLUSION
Hydrogel mediated on-tissue digestion has been demonstrated to be a simple and inexpensive tool to be added to existing IMS methods for protein identification. The hydrogels used in this study are rapidly formed, and can be shaped to match the size of the particular tissue region in question. This study demonstrates that sequential experiments can be performed on a single tissue section, i.e. IMS, on-tissue MS/MS, protein identification via LC-MS/MS and histology. Moving forward, the polymer composition of the hydrogels will be investigated to improve extraction of intact proteins digested peptides, and also to increase digestion efficiencies. Additionally, the hydrogel methodology presented here could be adapted to a prefabricated IMS target for rapid on-tissue digestions.
Supplementary Material
ACKNOWLEDGEMENTS
This project was supported by grants from the National Center for Research Resources (5P41RR031461-02) and the National Institute of General Medical Sciences (8 P41 GM103391-02) from the National Institutes of Health. We would like to acknowledge the work and insight of Kevin Schey, W. Hayes McDonald and Anna Felix at the Vanderbilt Proteomics core facility concerning future perspective and LC-MS/MS characterization of the samples.
REFERENCES
- 1.Caprioli RM, Farmer TB, Gile J. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Castellino S, Groseclose MR, Wagner D. Bioanal. 2011;3:2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 3.Casadonte R, Caprioli RM. Nat. Protoc. 2011;6:1695–1709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zemski Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. Chem. Rev. 2011;111:6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwamborn K, Caprioli RM. Nat. Rev. Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 6.Angel PM, Spraggins JM, Baldwin HS, Caprioli RM. Anal. Chem. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mainini V, Angel PM, Magni F, Caprioli RM. Rap. Commun. Mass Spectrom. 2011;25:199–204. doi: 10.1002/rcm.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chansela P, Goto-Inoue N, Zaima N, Hayasaka T, Sroyraya M, Kornthong N, Engsusophon A, Tamtin M, Chaisri C, Sobhon P, Setou M. PLoS ONE. 2012;7:e33154. doi: 10.1371/journal.pone.0033154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarnold JE, Hamilton BR, Welsh DT, Pool GF, Venter DJ, Carroll AR. Molec. BioSys. 2012;8:2249–2259. doi: 10.1039/c2mb25152c. [DOI] [PubMed] [Google Scholar]
- 10.Grüner B, Hahne H, Mazur PK, Trajkovic-Arsic M, Maier S, Esposito I, Kalideris E, Michalski CW, Kleeff J, Rauser S, Schmid RM, Küster B, Walch A, Siveke JT. PLoS ONE. 2012;7:e39424. doi: 10.1371/journal.pone.0039424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenian I, Spiteller M, Ghaste M, Wirth R, Herz H, Spiteller D. Proc. Nat. Acad. Sci. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, Vinatier D, Wisztorski D, Day R, Fournier I, Salzet M. Mol. Cell. Prot. 2009;8:2023–2033. doi: 10.1074/mcp.R800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardesty WM, Kelley MC, Mi D, Low RL, Caprioli RM. J. Proteom. 2011;74:1002–1014. doi: 10.1016/j.jprot.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. J. Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 15.Yao I, Sugiura Y, Matsumoto M, Setou M. Proteom. 2008;8:3692–3701. doi: 10.1002/pmic.200701121. [DOI] [PubMed] [Google Scholar]
- 16.Luk VN, Fiddes LK, Luk VM, Kumacheva E, Wheeler AR. Proteom. 2012;12:1310–1318. doi: 10.1002/pmic.201100608. [DOI] [PubMed] [Google Scholar]
- 17.Bracher PJ, Gupta M, Mack ET, Whitesides GM. ACS App. Mat. Inter. 2009;1:1807–1812. doi: 10.1021/am900340m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracher PJ, Gupta M, Whitesides GM. Adv. Mat. 2009;21:445–450. doi: 10.1002/adma.200801186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracher PJ, Gupta M, Whitesides GM. Soft Mat. 2010;6:4303–4309. doi: 10.1039/C0SM00031K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates JR, Eng JK, McCormack AL, Schieltz D. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 21.Rahman SMJ, Gonzalez AL, Li M, Seeley EH, Zimmerman LJ, Zhang XJ, Manier LM, Olson SJ, Shah RN, Miller AN, Putnam JB, Miller YE, Franklin WA, Blot WJ, Carbone DP, Shyr Y, Caprioli RM, Massion PP. Can. Res. 2011;71:3009–3017. doi: 10.1158/0008-5472.CAN-10-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu D, Xu F, Sun B, Fu R, He H, Matyjaszewski K. Chem. Rev. 2012;112:3959–4015. doi: 10.1021/cr200440z. [DOI] [PubMed] [Google Scholar]
- 23.Pawar SN, Edgar KJ. Biomat. 2012;33:3279–3305. doi: 10.1016/j.biomaterials.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Aerni HR, Cornett DS, Caprioli RM. Anal. Chem. 2009;81:7490–7495. doi: 10.1021/ac900974j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko G, Musunuri S, Wetterhall M, Bergquist J. J. Prot. Res. 2012;11:2441–2451. doi: 10.1021/pr201169q. [DOI] [PubMed] [Google Scholar]
- 26.McLean JA, Ridenour WB, Caprioli RM. J. Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 27.Djidja MC, Claude E, Snel MF, Scriven P, Francese S, Carolan V, Clench MR. J. Prot. Res. 2009;8:4876–4884. doi: 10.1021/pr900522m. [DOI] [PubMed] [Google Scholar]
- 28.Aerni HR, Cornett DS, Caprioli RM. Anal. Chem. 2005;78:827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 29.Mayer M, Yang J, Gitlin I, Gracias DH, Whitesides GM. Proteom. 2004;4:2366–2376. doi: 10.1002/pmic.200300748. [DOI] [PubMed] [Google Scholar]
- 30.Mohan T, Kargl R, Köstler S, Doliška A, Findenig G, Ribitsch V, Stana-Kleinschek K. ACS App. Mat. Inter. 2012;4:2743–2751. doi: 10.1021/am300375m. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Leulmi RF, Juncker D. Lab Chip. 2011;11:528–534. doi: 10.1039/c0lc00291g. [DOI] [PubMed] [Google Scholar]
- 32.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Proc. Nat. Acad. Sci. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.