Abstract
T-cell receptors recognize peptides presented by the major histocompatibility complex (MHC) on the surface of antigen-presenting cells (APC). The ability of the T-cell receptor (TCR) to recognize more than one peptide-MHC structure defines cross-reactivity. Cross-reactivity is a documented phenomenon of the immune system whose importance is still under investigation. There are a number of rational arguments for cross-reactivity. These include the discrepancy between the theoretical high number of pathogen-derived peptides and the lower diversity of the T-cell repertoire, the need for recognition of escape variants, and the intrinsic low affinity of this receptor–ligand pair. However, quantifying the phenomenon has been difficult, and its immunological importance remains unknown. In this review, we examined the cases for and against an important role for cross reactivity. We argue that it may be an essential feature of the immune system from the point of view of biological robustness.
Keywords: T cell, T-cell receptor, T-cell repertoire, cross-reactivity
I. INTRODUCTION
T-cell cross-reactivity is a phenomenon of the immune system defined as the recognition of two or more peptide-MHC complexes (pMHCs) by the TCR. The recognition is based on the ability of the TCR to bind sufficiently well to initiate a cellular response. There is an expectation of T-cell cross-reactivity based on the large number of pathogen epitopes that may be encountered and on the probability that low affinity interactions may favor multiple pMHC complexes with sufficient affinity to bind a particular TCR. Yet, despite a number of publications about cross-reactivity of T cells, the overall number of documented cases of cross-reactions is low. This review describes cross-reactivity of T cells and its possible importance in immune systems, with an emphasis on the structural aspects of the recognition of pMHC by TCR. We discuss the basis and the necessity of cross-reactivity from a systemic point of view, with a particular emphasis on survival in humans after thymic involution. We start with some necessary background.
A. T-Cell Recognition of pMHC
The recognition by T cells of antigenic peptides bound to class I or class II MHC initiates an adaptive immune response. Recognition occurs through the TCR, a heterodimeric cell-surface receptor composed of an α- and a β-chain. Each chain is generated by a somatic rearrangement process occurring during thymic maturation in which one of a number of variable (V) segments rearranges next to one of a number of joining (J) segments (α-chain) or a diversity (D) and joining segment (β-chain). Recognition specificity is a function of the variable regions, which contain three highly diverse loops, termed complementarity-determining regions (CDRs) 1, 2, and 3, that make direct contact with the ligand. The CDR1 and CDR2 loops are encoded within the V gene segment, whereas CDR3 loops are generated during rearrangement and span the VJ or VDJ junctions. CDR3 loops define the TCR because the joining process is imprecise, with insertion of non-templated nucleotides (N nucleotides) in the junction site, as well as 3′- and 5′-nucleotide deletion from segments participating in the rearrangement. Thus, the resulting CDR3 has a unique nucleotide sequence that is specific to that particular T cell and all its progeny; hence, the receptors have a clonotypic nature of (reviewed in Davis and Bjorkman1). For this reason, CDR3 sequences have been the main focus in studies aimed at unveiling the mechanisms responsible for the specific recognition by one T cell of multiple pMHC complexes. CDR1 and CDR2 sequences are the focus of investigations of germline TCR-MHC bias, which is discussed below as part of thymic selection (see III.A. Thymic Selection and Cross-reactivity).
The αβ TCR can recognize two major polymorphic classes of MHC molecules. MHC class I (MHCI) is expressed on most cell types and presents internally generated peptides to CD8-expressing T cells. MHC class II (MHCII) is expressed on the surface of professional APCs of the immune system, such as dendritic cells, macrophages, and B cells. These APCs present peptides generated from exogenous sources to CD4 T cells whose function is the production of cytokines and in some cases cytotoxicity. Whereas both MHCI and MHCII have relatively similar structures, MHCI-bound peptides are anchored at their ends and are generally only nine amino acids long. Longer peptides maintain their end anchor points and bulge in the middle. MHCII-bound peptides tend to be more exposed and are of different lengths because the ends of the peptide binding groove are not closed.
B. Generation of TCR Diversity and Repertoire Restriction
The gene structure and rearrangement process leads to the generation of diversity of the TCR. Combinatorial diversity represents the factoring of the number of possible V-J joins with assumptions as to their probability based on their distance apart. To this is added the junctional diversity generated by addition and removal of nucleotides. Finally, pairing of the chains can increase the diversity. During thymic maturation, properly rearranged β-chains are first selected (β-selection), and prior to α-chain rearrangement the thymocytes undergo extensive division (reviewed in Starr et al.2), allowing for cells with the same β-chain but different α-chains. The theoretical diversity based on both combinatorial and junctional processes has been estimated to be approximately 1018 in humans and 1015 in mice. However, individual repertoire diversity is much lower; it is estimated to be less than 108 in humans3,4 and 106 in mice.5 These lower levels in the diversity of the TCR repertoire is in part a result of the negative selections that occur during T-cell development in the thymus. The process of negative selection eliminates T cells having too high an affinity for self-MHC or self-pMHC, preventing autoimmune reactions. This central tolerance results in a repertoire capable of recognizing self-MHC in a complex with foreign peptides but avoiding self-peptide recognition. However, the elimination of T cells during negative selection creates “holes” in the T-cell repertoire,6 which can lead to increased susceptibility or emergence of escape variants that are similar to self-peptides. The lower levels of diversity are also dictated by the carrying capacity of the organism because there is limited space for T cells in the periphery.
C. Peripheral Selection and Memory
After exiting the thymus, a naïve T cell has a limited lifetime that can be extended on the basis of its participation in an immune response (effector function) with a portion of the cells surviving as memory cells. Because the effector stage includes cell division, the clonotypic lineage is preserved even if only a small percent of the effector cells in the lineage are set aside as memory. Memory cells respond more rapidly and robustly upon a second exposure, ensuring that subsequent exposures have less effect (morbidity). As might be expected, an antigen-experienced (memory) T cell does not have the same signaling requirements as a naïve cell.7–10
Initial participation in a peripheral response is mediated by the level of inflammation and the resulting signals from innate cells. The inflammatory state may regulate the activation of threshold of the T cells entering the node or spleen, and this threshold setting may allow for less avid T cells to participate. On the other hand regulatory T cells may increase the threshold.11 The ability to reset thresholds can play a role in cross-reactivity if it allows T cells with lower-avidity TCR to respond to a particular peptide.
II. MOLECULAR BASIS OF T-CELL CROSS-REACTIVITY
A. Structural Mechanisms
There have been a number of mechanisms proposed for the structural basis of TCR cross-reactive recognition of pMHC. A number of these have been summarized in a recent review by Yin & Mariuzza.12 TCR:pMHC interactions are driven by the relatively flat nature of the complex interface, low number of H-bonds, the frequent lack of strong (ionic) interactions, and reliance on van der Waals (VDW) interactions. The overall affinities are relatively weak, especially in comparison to immunoglobulin-antigen (Ig:Ag) interactions.13 Peptide-loaded MHC have to be multimerized to provide a mechanism for identifying specific T cells, and even multimers often miss biologically relevant interactions for CD8 cells and have a poor history of utilization for CD4 cells. The mechanics of cross-reactivity involves changes in CDR loop conformation, altered TCR docking on the pMHC, flexible changes in pMHC, and structural degeneracy.
1. Conformational Plasticity of CDR Loops
A number of studies have observed structural rearrangements occurring during the TCR:pMHC interaction, mostly from comparison of free vs. bound TCR structures or of the same TCR with two different pMHC.14–23 The range of motion is between 0.3 and 11.4 Å overall, averaging 2.6 ± 1.5 Å across loops. In general, CDR3 loops undergo the largest shifts.20–23 Interestingly, the loops do not fold during the engagement with the ligand. Rather, the shifts are “rigid-body” like, followed by occasional remodeling.
Three structures are resolved for the BM3.3 TCR bound to different pMHC complexes: VSV8:H-2Kb, pBM1:H-2Kb, and pBM8:H-2Kbm8.24 The bound peptides are considerably different in their chemistry. The BM3.3 binds the three ligands in the same orientation. The CDR3α varies significantly via loop remodeling and hinge bending between the three complexes. The conformation of the CDR3β is maintained between pBM1 and VSV8 structures, but is shifted by 4.4 Å in the pBM8 structure due to a large wrinkle in the center of the loop. Among the germline loops, only the CDR2α shifts by approximately 2 Å. The affinity of the BM3.3 for pBM1 is greater than for pBM8, probably for the different reorganization of CDR3β and displacement of CDR2α, but also for the different interactions present at the interface.
Of special interest is the analysis of the A6 TCR, which has been resolved in complex with five different pHLA-A2 ligands: the wild-type Tax peptide and four single-substituted variants.25,26 The mutations applied to three of the four peptides dramatically alter the binding interface. The CDR3β undergoes the most significant variation among structures, shifting by an amount that correlates with the modification in the binding plane. Also in the case of the A6 TCR, the greater the conformational change in CDR3β, the weaker the affinity. However, discriminating between the effect of the interactions established at the interface and the effect of loop conformational rearrangements on the differences in affinity is not straightforward.
2. Altered TCR:pMHC Docking Geometry
All the TCRs assume a docking orientation that is diagonal with respect to the peptide backbone direction of the pMHC. In general, the TCR variable domains bind roughly at a 35° angle across the complex major axis in complexes involving MHCI and at a 50° angle in the case of MHCII. An exception to the former is class I HLA-A2 in complex with the xenoreactive AHIII 12.2 TCR, which shows an angle of 67°. The Vα domain is centered over the α helical portion of the MHCI α1 domain or of the MHCIIβ domain. The Vβ domain mainly contacts the α–helix of the MHCI α2 domain or of the MHCIIβ domain. In some structures, the TCR axis appears to be nearly orthogonal with respect to the pMHC main axis.27 Thus, it appears that TCR docking, although being conserved in terms of overall geometry, features a certain degree of variability. It is conceivable that this very variability may allow the same TCR to engage different pMHC ligands. The 2C TCR binding to different pMHC is an example of such changes.28
3. Flexibility of the Peptide and MHC
As the TCR engages the pMHC complex, conformational shifts in both the peptide and the MHC have been shown to take place along with the structural rearrangements of the TCR itself. These shifts in the ligand are usually small in magnitude and have been interpreted as “induced-fit” type conformational rearrangement with little or no contribution to the final binding. However, a recent work from Baker and colleagues has shown evidence that peptide-dependent “tuning” of molecular motion distributed throughout the TCR binding surface of the pMHC can contribute to TCR recognition and facilitate cross-reactivity.29
Indeed, in this study the human αβ TCR A6 recognizes the HTLV-1 Tax peptide presented by HLA-A2, as well as the Saccharomyces cerevisiae peptide Tel1p bound to the same MHC. Cross-reactivity between Tax and Tel1p is not unexpected given the similarities in the peptides. However, the interface formed by A6 with Tel1p-HLA-A2 is substantially different from the interface formed with Tax-HLA-A2, although the Tel1p-HLA-A2 complex is an ideal structural mimic of the Tax ligand. Strikingly, the conformational differences involve not only the peptide and the TCR CDR3 β loop but also the HLA-A2 α2 helix.
4. Structural Degeneracy
This mechanism has parallels to the “hydrophobic interactions” used to describe some forms of non-covalent binding. These are not necessarily strong interactions and can easily slip. Some TCR:pMHC interactions are predominantly driven by VDW interactions, and usually the complexes have low affinity. However, complexes whose formation seems to rely on structural degeneracy have been the subject of mutational analyses that generate more tightly binding complexes. The 3A6 TCR, which recognizes myelin basic protein (MBP) 89–101 peptide bound to HLA-DR2, is an example of this mechanism.30 The crystal structure was composed of four asymmetric units, none of which showed identical interfaces. Interactions between TCR and peptide are mainly restricted to VDW contacts, with limited juxtaposition of hydrophobic surfaces. The paucity of interactions between 3A6 and MBP offers ample opportunity for optimizing the TCR:pMHC interface through variations of the peptide. Indeed, combinatorial libraries identified peptides with multiple substitutions at TCR contact positions that stimulate 3A6 T cells far more efficiently than MBP itself.31
B. Molecular Mimicry
Molecular mimicry has been considered a cross-reactive mechanism. It originally defined a situation in which a pathogen expresses an epitope that shares antigenic structures with host tissue-derived protein or peptide,32 and was originally used for both B- and T-cell responses. In the context of a T-cell response, pathogen-derived peptides when presented by MHC may activate potentially self-reactive T cells. As a consequence, tolerance/ignorance is broken, and the pathogen-specific immune response cross-reacts with host-derived epitopes, which can cause tissue damage and disease. Molecular mimicry per se is not a structural definition but rather a functional description. The mimicry may result from one or a combination of the mechanisms described above. However, there can be very restricted mechanistic version of molecular mimicry in which the two epitopes have identical structures. The differences may lie in buried side chains that do not affect the overall structure. Here, we use the term in its original context.
A well-studied example of molecular mimicry is cross-reactivity between an Epstein-Barr virus (EBV) epitope and MBP.33
Molecular mimicry has been extended to alloreactive responses that can be important in transplant settings. One of the first reported examples of this was the 2C TCR, which recognized a self-peptide presented by H2-Kb as well as the foreign QL9 peptide bound to the H2-Ld alloantigen.28 A recent publication34 showed that the LC13 TCR, which recognizes a viral peptide presented by self-HLA-B*0801, and also recognizes B44 allotypes (HLA-B*4402 and HLA-B*4405), bound to two different allopeptides.
C. Thermodynamic Analysis of the TCR:pMHC Interaction and Cross-Reactivity
Thermodynamic analysis of the various TCR:pMHC complexes has enhanced our understanding of the determinants of TCR binding and their involvement in cross-reactivity. For example, the possibility that the TCR CDR loops are flexible and undergo conformational rearrangements as they interact with the pMHC ligand was initially hypothesized on the basis of kinetic analysis35 and the subsequent crystallographic studies.14,15 However, a confirmation of the flexibility of the CDRs in the unbound state was found on the basis of the thermodynamic analysis of the human JM2236,37 and the murine F537 systems, which measured in both cases a negative entropic contribution to the free energy decrease of binding to their respective pMHC ligand.
These initial observations were corroborated by another work focused on the 2B4 TCR interacting with the MCC/I-Ek complex.38 Indeed, the loss of entropy and the restraining of conformational mobility observed in this latter system were interpreted as the evidence that the TCR CDR loops sample multiple conformations, allowing a TCR repertoire with limited variety to interact with a much larger repertoire of pMHC complexes.
This thermodynamic model of TCR recognition would be challenged just few years later, with the discovery of a number of entropically favored TCR:pMHC complexes.16,39–41 Because it was apparent that the TCR/pMHC interaction is not characterized by a specific structure, the question as to whether thermodynamic data could provide insight into the mechanisms of T-cell activation was raised.
By comparing the data of the interactions for which a thermodynamic study has been carried out, it appears that the values of ΔG are very similar, whereas the values of enthalpic and entropic contributions span a wide range. Moreover, the changes in entropy and enthalpy follow a linear trend, suggesting the likelihood of the presence of a compensatory mechanism between the entropic and enthalpic contributions.42 On this basis, it was argued that as far as enthalpy and entropy are concerned, it does not matter how you form the TCR:pMHC complex, just that you do. A similar conclusion obviously leaves room to that permissiveness of recognition that is at the basis of cross-reactivity.
III. IMMUNOLOGY OF T CELL CROSS-REACTIVITY
A. Thymic Selection and Cross-Reactivity
Thymic maturation has a positive selection phase mediated by selection on self-MHC loaded with self-peptides. This selection on MHC-loaded with self-peptide could formally make any T cell peripherally selected in response to a pathogen inherently cross-reactive. However, cross-reactivity in thymic selection has had an MHC-focus.43 The MHC-focus at this stage is thought to be predominantly mediated by CDR1 and CDR2 interactions with the MHC.44 Thus, recognition of self-pMHC during positive selection is usually ignored when thinking about cross-reactivity, even though the peptide is part of the complex being recognized. If the role of the peptide is to assure surface expression of the MHC and not to interfere with the recognition of the MHC, then positive selection does not formally meet the definition of cross-reactivity.
It has been suggested that in normal humans and WT mice, different thymocytes encounter different numbers of self-pMHCs; thus, the level of cross-reactivity of the selected T cells can vary.45
B. Cross-Reactivity in Peripheral Selection and Autoimmunity
Peripheral selection can involve recently matured naïve T cells in cases of the relatively young (antigen inexperienced); it can involve a mix of naïve and previously experienced T cells; or, in older individuals, it can involve primarily experienced T cells. An experienced cell is in the process of developing a memory phenotype. If it participates in a second response, owing to sufficient avidity with the new epitope, it will be cross-reactive. It is very likely that the probability of such an occurrence is driven not only by avidity but by other signals that set the response threshold.
In the case of a pathogen that is inducing a large inflammatory response, the cellular response thresholds is undoubtedly set lower to encourage a T-cell response. While the ability to set response thresholds has survival benefit, the unintended consequence could be cross-reactivity on self. Typically, this is minor and can be dealt with by peripheral tolerance mechanisms. However, some examples, such as reactive arthritis, can lead to autoimmune disease at non-negligible frequencies.46,47 In the case of most autoimmune diseases, an infection is thought to be the trigger,48–50 and cross-reactivity could play an important role.
1. Examples of Cross-Reactivity between Pathogens
There are a number of examples of cross-reactivity between both related and unrelated pathogens.
Examples of cross-reactivity between related pathogens include different serotypes of the Dengue virus that differ in MHCI presenting epitopes,51,52 different strains of the influenza A virus,53–57 hepatitis C virus (HCV) escape variants,58 and human immunodeficiency virus (HIV) common variant epitopes from different clades.59,60 Cross-reactivity can also be observed between different epitopes derived from the same virus. These examples include EBV61 and HIV.62 A majority of these cross-reactive peptides have structural similarity and differ in one or two amino acids.
Cross-reactivity has also been examined between a known peptide antigen and either peptide variants (or mutant viruses) generated by amino acid substitutions,63–67 or by scanning combinatorial peptide libraries to find structurally and/or biochemically similar peptides.6,68–72
Cross-reactive peptides from unrelated pathogens often demonstrate a lower level of structural homology. Examples of such cross-reactivity include HIV and human cytomegalovirus (CMV),73 HIV and Influenza A,74 Influenza A and EBV,75,76 Influenza A and HCV,77–79 and HCV and human herpes virus (HHV1).80
Scanning combinatorial peptide libraries has also been used to identify structurally and/or biochemically unrelated peptides.6,68–72
All these studies demonstrate “true” cross-reactivity, when a particular clone is tested against a set of peptides and recognize more than one. Polyclonal cytotoxic T-cell lines (CTLs) generated against the peptide in question that can recognize different peptide(s) also show true cross-reactivity for that subset that recognizes both the original and the different peptide(s). In the case when multiple peptides are recognized, it is not known whether the same TCR (clone or clonotype) recognizes two or more peptides, or different subset of established CTL able to recognize at least one additional peptide.
Activation of CD8 T cells specific for EBV, CMV, and Influenza A has been observed during HIV infection.81 Cross-reactivity might explain these observations, but other explanations could include possible bystander activation of pre-existing memory T cells specific for other epitopes.
2. Cross-reactivity in Autoimmunity
As a general rule, most autoimmune disorders are thought to result from cross-reactivity based on an initial reaction to a pathogen. This idea gave rise to the term molecular mimicry. It is known that pathogenic peptides can activate autoreactive T cells due to structural similarity between pathogenic and self-peptides and cause autoimmune diseases30,34,72,82–88 or accelerate a previously initiated autoimmune process.89
When considering cross-reactivity, the most-studied examples of autoimmune diseases that might be triggered by pathogens include multiple sclerosis (MS) and diabetes mellitus. MS immunopathology is contributed by recognition of MBP by autoreactive TCRs.90 Thus, the autoimmune 3A6 CD4 clone recognizes the N-terminal portion of MBP in complex with MHCII molecule HLA-DR2a.30 Using a panel of superagonists with substitution at a TCR contact position, it had been shown that 3A6 recognizes the antigen through the structural degeneracy mechanism. Type 1 diabetes is characterized by killing of pancreatic B cells by autoreactive CTLs that target preproinsulin signal peptide-derived epitopes.91 The 1E6 CD8 T-cell clone that recognizes one of the preproinsulin-derived peptides can also recognize up to 106 different peptides from a decamer peptide combinatorial library.72 This high level of cross-reactivity could involve most of the mechanisms described in section II.A. Thus, the TCRs involved have inherent cross-reactive potential; however, the pathogen epitope that may drive the initial response has not been identified.
This link between peripheral selection and autoimmunity defined the phenomenon of molecular mimicry. However, it is likely that all the mechanisms described in section II.A could play some role in this phenomenon.
C. Frequency of T-Cell Cross-Reactivity
The number of peptides that can be presented by self-MHC is estimated to be greater than 1015;72,92 at the same time, the number of T-cell clonotypes in humans is less than108 3,4 and even less in mice.5 Thus, it has been hypothesized that for effective immune response, T cells need to recognize more than 106 epitopes (peptides).93 Therefore, one would expect a large number of examples of cross-reactive T cells. The question has been how to estimate the rate of this phenomenon.
1. Examples of Cross-Reactivity Measurements
A majority of the reported examples mentioned above include cross-reactivity between two or more peptides. 63,67,70,73,74,77,78 Estimates of cross-reactivity of known clones can be up to 106 different peptides.6,72 At the same time, several reports have indicated an extremely low frequency of cross-reactions between unrelated peptides.71,94,95 Ishizuka and colleagues71 examined approximately 30,000 TCR-pMHC interactions using 18 murine or human CD8 T-cell clones and polyclonal CD8 T cells raised to viral and autoantigens, and they identified only one single cross-reaction. Oseroff et al.94 screened vaccinia virus (VV) for CTL epitopes. They tested peripheral blood mononuclear cells (PBMCs) from healthy donors before and after vaccination. Of 91 peptide pools tested, they identified four positive peptide pool responses in postimmunization PBMCs and none in preimmunization PBMCs. Similarly, a low frequency of cross-reactive responses to CMV-derived peptides was detected in healthy seronegative donors, while seropositive subjects demonstrated CMV-specific T-cell responses.95 Thus, despite a number of reported examples of cross-reactivity, the frequency of observations does not meet the higher estimations.
2. Why is Extensive Cross-Reactivity not Observed on a Regular Basis?
There are several possible answers to this question. The approaches for detection of cross-reactivity are different. Testing ex vivo PBMC for cross-reactivity with unknown peptides might yield a low number of observations or no observations at all due to a very low frequency of cross-reactive T cells and limitations of the methods of detection, such as tetramer staining, ELISPOT, intracellular cytokine staining, spectratype analysis, etc. Expansion of antigen-specific T cells in vitro in short-term cell culture with a peptide or mutant virus allows an increase in the number of these T cells but might not be equivalent to an in situ environment. In vivo infection generates a complex immune response whose intensity can be related to the damage caused by infection. Using an in vitro system simplifies response but might change observed level of cross-reactivity.
Cross-reactive T cells can have a low affinity/avidity to most peptides. It has been reported that cross-reactivity of T cells can be CD8 dependent.78,96 Wooldridge et al.96 used a panel of HLA-A2 molecules exhibiting a range pMHC:CD8 binding affinities and showed that the stronger the pMHC:CD8 interaction, the higher the number of peptides recognized. However, analogous to the earlier discussion of positive selection driven by MHC, if the MHC:CD8 reaction drives the response irrespective of peptide, then we do not consider this true cross-reactivity. It should be pointed out that a TCR:pMHCI:CD8 or TCR:pMHCII:CD4 interaction is complicated, and examples in which one of the component interactions is the main binding energy source could represent an extreme event in the spectrum.
We would like to propose that discrepancies might arise from the systems analyzed. PBMCs from humans represent a rich history of exposures to pathogens and vaccines, yielding a complex T-cell repertoire. This complexity could increase the chance for deriving a T-cell clone that will respond to previously unencountered or self-peptides. Our own studies into the allowable recognition of similar peptides has taken advantage of the complex memory response to influenza.67 In the laboratory mouse, with its ease of manipulation and controlled genetics, the problem is that no peripheral T-cell system of equal complexity has been set up to properly ask the same questions.
D. Cross-Reactivity and Signaling Thresholds
The requirement of adjuvants to break peripheral tolerance in most murine epitope-driven autoimmune models indicates the need for additional signals in the T-cell recognition process. In terms of cross-reactivity, by resetting signaling thresholds, the inflammatory context is most likely very important to overcome situations where epitopes are recognized with lower affinity. Changes promoting higher avidity could also overcome lower-affinity interactions. It has been shown recently that Tregs may suppress expansion of low-avidity antigen-specific CD8 T cells by inhibiting the production of chemokine ligands CCL2/3/4/5 (CC chemokine ligands 2/3/4/5) that stabilize the interaction between antigen-presenting dendritic cells and low-avidity T cells.11 Insight into the signaling threshold process can be gained by examining the effect on signaling of identified cross-reactive epitopes.
Altered peptides that are partial agonists can induce a signal of a different strength than the original epitope. It has been shown that weak ligands alter TCR downstream signaling events such as CDR3 zeta chain phosphorylation and activity of Zap70 kinase,97,98 phosphorylation of ERK ½99 and other kinases,100 and triggering recruitment of SHP-1 with following inactivation of Lck kinase.99,101 Escape variants can suppress CD8 T-cell response by activation of SHP-1.102
The concept of tunable thresholds has been put forth as an explanation for general immune responses.103,104 Immunization of mice with modified myelin proteolipid protein, PLP139-151, where tryptophan (W) at position144 was substituted for glutamine (Q) (W144Q), and following expansion of PLP-specific T cells with the suboptimal cross-reactive self-antigen PLP139-151, lowers the activation threshold to the immunizing peptide, causing heteroclitic response as measured by cytokines production.105 Changes in activation threshold by cross-reactive self-peptides can have implications in autoimmunity.
A certain level of signal threshold is required for polyfunctional effector response. It has been shown that phosphorylation of ERK ½ correlates with degranulation and tumor necrosis factor α (TNF-α) and interferon γ (IFN-γ) production;52 at the same time, production of MIP-1β (CCL4) can be upregulated in response to short-lived epitope-specific TCR-mediated signal in the absence of detectable ERK1/2 phosphorylation. It has been hypothesized that MIP-1 β production may stimulate production of TNF-α and IFN-γ by activated T cells specific for other viral epitopes and that such an increase of these cytokines level may contribute to immunopathology.52
Reports have been published describing computational models that may be used to study different aspects of T-cell activation and T-cell antigen discrimination.106–108 However, which accessory signals might compensate for lower inherent affinity and allow an immune response under sub-optimal conditions has not yet been well elucidated.
E. Heterologous Immunity
The phenomenon of heterologous immunity is well known and has been widely studied. A number of reviews have been dedicated to heterologous immunity in both murine models and humans.109,110 Heterologous immunity is defined as an “immunity that can develop to one pathogen after exposure to non-identical pathogens”.110 Heterologous immunity can be the result of cross-reactivity, a shared epitope, and/or bystander activation of T cells.76,81 If the cross-reactivity is a result of specific recognition epitopes from the different pathogens by the TCR, bystander activation usually is non-specific and can be caused by TCR-independent activation by released cytokines111 or by low-affinity recognition of self-pMHC.88
Heterologous immunity has been well described in mice. Immunity to lymphocytic choriomeningitis virus (LCMV) has been shown to provide partial protection after challenge with either Pichinde virus (PV), VV, or murine cytomegalovirus (MCMV).112–115 This immunity was reciprocal for LCMV and PV, MCMV and PV, and LCMV and MCMV viruses as measured by virus titer level in at least one of the following organs: spleen, liver, and fat pads. Immunity to LCMV, PV, MCMV, or Influenza A provides some level of protection against VV, but immunity to VV did not protect against infections with any of these viruses. Immunity to Influenza A protected against VV but not LCMV or MCMV.116 Mice immune to Influenza A demonstrated an increased viral titer in lungs and signs of lung immunopathology when compared with control naïve mice after challenge with MCMV or LCMV. This immunopathology in part can be explained by activation of cross-reactive T cells by low-avidity cross-reactive epitopes that results in altered cytokine levels and contributes to the observed immunopathology as discussed in the previous paragraph. One study has reported the protective role of Bacillus Calmette–Guérin (BCG) vaccination against a subsequent VV challenge; however, cross-reactive epitopes were not identified.117
The role of cross-reactivity and identification of cross-reactive epitopes has been determined in two cases: LCMV and PV,113 and LCMV and VV.61,118 LCMV NP205-212 and PV NP205-212 peptides demonstrate a high degree of amino acid similarity and differ only by two amino acids at positions 5 and 8 that were identified as anchor residues.119 Crystal structures of corresponding pMHC complexes indicate conformational similarities between peptides;119 nevertheless, the BV composition of responding TCR repertoires was different, with a dominance of VB16 in responses to LCMV and codominance of VB16 and VB5.1 in responses to PV infection.120
Challenge of LCMV immune mice with VV reactivates a subset of LCMV-specific memory T cells that can recognize VV-derived epitopes and protect against a lethal dose of VV.114 Identification of VV-specific epitopes that mediate protection against VV infection and cross-react with known LCMV epitopes revealed a subdominant VV-a11r198-205 peptide.118 An intracellular cytokine staining assay indicated that VV-specific T cell lines from LCMV immune mice stimulated with VV-a11r198-205 peptide recognized up to three LCMV-derived epitopes: NP205-212, GP118-125, and GP34-41.61 The VV-a11r198-205 peptide shared only 3–4 amino acid sequence similarities with any of the LCMV-derived peptides. The lack of a detailed clonotype analysis left open the question of the extent of cross-reactivity (one, two, or three additional peptides recognized). We assume that the observed cross-reactivity is due to the structural degeneracy mechanism.
There is no doubt that the phenomenon of heterologous immunity applies to humans. There are examples of immunopathology associated with T-cell cross-reactivity.52,73,75,79
Questions remain about how heterologous immunity shapes the responding repertoire. The history of pathogen exposure in humans is unknown; thus, it is unknown whether the cross-reactive T cells participating in the known responses can also respond to other pathogens.
H. Heteroclitic Immunity and Cross-Reactivity
Heteroclitic immunity is defined by the observation that stimulation with an antigen results in T cells that are even more responsive to another antigen.121 This can be considered the counterbalance to the partial agonist studies described above. A classic example is the pigeon cytochrome C system, in which most T cells raised to this antigen respond even better to moth cytochrome C.122,123 Other examples include identification of heteroclitic peptides in cancer immunotherapy. Modification of self-peptides expressed by malignant cells can result in enhanced immunogenicity of an antigen and generation of CTL recognizing both modified and original self peptides, which can be used in antigen-specific immunotherapy and tumor vaccine development.124-131 The peptide changes result in generation higher stability pMHC complexes, which are linked to generation a more diverse TCR repertoire with range of affinities.132 Heteroclitic peptides can also originate from pathogens. It has been shown that MPHF2 peptide derived from Mycoplasma penetrans HF-2 permease protein stimulate CD4 T cells, exhibiting a higher functional avidity for melanoma antigen MAGE-A6 than MAGE-A6-specific CD4 T cell lines.133 HIV-1 protease 76-84 is considered to be a heteroclitic variant of (IFN-γ)-inducible protein 30 signal peptide.134
Identification of heteroclitic peptides can also be useful in generating highly cross-reactive CTL to prevent escape variants and predict autoimmunity and immunopathology.134–137 Heteroclitic immunity is mostly related to the degeneracy mechanism.
I. Prediction of Cross-Reactivity
Prediction of cross-reactivity is important for vaccine design in two ways. It would be useful to predict protection against a pathogen and escape variants or multiple pathogens with similar epitopes if this is found to increase efficiency. It is also useful to predict possible autoimmune reactions from cross-reactivity on self.
There are a number of practical approaches to predicting cross-reactivity. These include the generation of a panel of mutant peptides with single and/or multiple amino acid substitutions in known epitopes and testing the impact of these substitutions on T-cell effector functions.63–67 Another approach involves scanning combinatorial peptide libraries to find cross-reactive peptides for clones with known antigen specificities6,69,72,96,138–140 as well as specifying antigens for clones of unknown specificities.141 Scanning of a pathogen proteome to reveal unknown epitopes might help in detection of cross-reactive peptides as well.94,95,142
We are currently not in a position to perform ab initio prediction based on structural considerations and mechanisms. A better understanding of the thermodynamics of TCR:pMHC interactions should be able to supply rules that specify the probability of cross-reactivity.
J. Complexity of T-Cell Repertoires and Cross-Reactivity
Peptide antigens can generate oligo- or polyclonal T-cell repertoires. Polyclonality, which would correspond to the abundance of distinct TCR species, can be used as an approximation for diversity. Limited TCR diversity of T-cell responses is associated with CTL escape.143–145 Polyclonal T-cell repertoires are more likely to contain high-avidity T-cell clones that result in better epitope recognition and control of the pathogen.54,146–148 Such repertoires can contain cross-reactive T-cell clones that are able to recognize escape variants and new epitopes. Such functional overlap would avoid loss of immune recognition and maintain responses to the pathogen.
One of the examples of such a diverse repertoire is the CD8 T-cell response to HLA-A2 restricted influenza A–derived M158-66 peptide. The clonotype distribution in the M158-66 repertoire is characterized by an extensive low-frequency tail, which could be indicative of a power law-like distribution.149 To facilitate additional analysis, the repertoire can be graphed on the basis of rank frequencies. In such a graph, the number of clonotypes present once, twice, etc. would be counted and plotted as rank one, two etc. In a power law distribution, a log-log plot of the rank vs. frequency would give a line with a slope that is equal to the exponent of the distribution.150–152 We refer to repertoires with power law-like clonotype distributions as complex. Complex repertoires have also been reported for mouse CD8 T-cell responses to influenza A153 and for Treg populations.154 Recent analyses of ex vivo repertoires of total CD4 or CD8 cells or subsets thereof155,156 show that these repertoires exhibit the power law distribution.
Two questions arise about complex repertoires. The first is how the complexity arises. The second is whether the complexity affects cross-reactivity. A complex repertoire may be the result of a random distribution of TCR affinities for the pMHC. There would be many ways to generate low-affinity interactions but fewer and fewer to generate high-affinity interactions. If affinity is related to proliferation then there would be a few high-affinity, high-frequency clonotypes and many low-frequency, low-affinity clonotypes in the repertoire. It could be expected that cross-reactive clonotypes may be most frequent in the low-affinity subset of the repertoire, as these would have a higher probability of having higher affinity toward a different peptide.
We examined the distribution of cross-reactive clonotypes from the M158-66 repertoire, which can form a fully connected cross-reactive network.67 Plotting the total repertoire on the basis of rank frequency yields a line. Plotting the subset of clonotypes that are cross-reactive also gives a line that extends out to the high-rank clonotypes (Figure 1).
Figure 1.
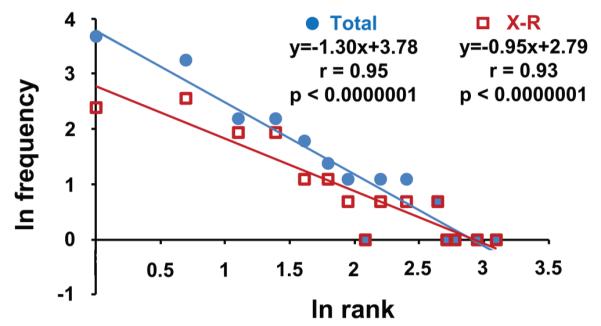
The power law–like distribution of frequencies of overall (Total, blue circles) and cross-reactive (X-R, red squares) clonotypes in M158-66 repertoire. Logn-transformed values are fit to a linear regression line. The equation y=mx+b describes the regression line, where m is a slope and b is the intercept with the y axis. The distribution of cross-reactive clonotypes indicates self-similarity to the overall (total) repertoire.
If cross-reactivity is a side-effect of a TCR affinity toward different pMHC complexes, then cross-reactive clonotypes should be found only in the low-frequency tail of the repertoire (Figure 1, ranks 1–3) as we have suggested above. However, cross-reactive clonotypes are distributed through the all ranks, which indicates power law-like distribution. This distribution indicates a selection of cross-reactive clonotypes that is similar to the total repertoire. Selection is an indication of an important role in the immune response.
When imagining an immune system in which cross-reactivity plays an important role, repertoire complexity is expected. In spite of an early tendency to look for relatively monoclonal results in the peripheral selection process, complex repertoires appear to be the normal phenomenon and cross-reactivity may be playing a role in the selection of such repertoires.
IV. POTENTIAL SIGNIFICANCE OF CROSS-REACTIVITY
A. Cross-Coverage
The potential of cross-coverage has been developed in terms of heterologous immunity. The PV–LCMV experiments of Welsh and colleagues112,113,115 point the way and supply a possible probability argument for cross-reactivity, with a virus that generates many stably recognized epitopes providing better protection against a smaller virus, whereas in the opposite direction the effect is less pronounced. However, the nature of the pathogen itself may dictate the extent to which cross-reactivity may be important, because VV infection provided poor protection against subsequent LCMV or PV challenges.
With human immunology, the system becomes much more complicated due to the large and varied number of pathogens encountered. The concept of heterologous immunity may have to be extended to include different pathogen classes. For mouse models, this would mean virologists collaborating with bacteriologists, mycologists, and parasitologists.
Where cross-reactivity may play an especially important role is in the period after thymic involution. After production of naïve T cells has fallen by 2 to 3 orders of magnitude, good outcomes to exposure to novel pathogens in middle-age may rely heavily on cross-reactivity providing sufficient numbers of T cells that can be involved in the response. The ability to recognize a novel pathogen would be an indicator of a robust system.
B. Robustness
The presence of complex T-cell repertoires poses the question of why such repertoires are maintained. Power law–like distributions have been linked to system robustness. Robustness has been defined as an ability of a system to maintain “some desired characteristics despite fluctuations in the behavior of its component parts or its environment”,157 which in the case of immune response means the ability of a T-cell repertoire to recognize most of the pathogens and escape variants while minimizing the chance of inducing autoimmune reactions.
What does biological, or in this particular case immunological, robustness mean? An acceptable definition is that robustness is the ability of a complex biological system to respond to environmental and genetic perturbations and maintain an ability to evolve.158 Robustness gives the system flexibility against environmental perturbations by allowing changes in structure and components and, thus, maintaining specific functions. By this definition the immune system is the epitome of a robust system, with the innate system responding at a programmed lower level of responses and the adaptive immune system harnessing genetic instability and real-time selection to provide a truly adaptive evolving response. Several mechanisms have been proposed as indicative of robust systems:158 1) system control, 2) “fail-safe” mechanisms, which includes redundancy and diversity, 3) modularity, and 4) decoupling, which buffers high-level functionalities from low-level variation, to which we add adaptation. All of these mechanisms apply to the immune system.
In terms of the contribution of cross-reactivity to this definition of robustness, redundancy (cross-coverage) is obvious. Buffering high-level functionality from low-level variation applies to protection from epitope drift/escape. If we deal with the similar shape-space of pMHC defined by our network of conservative M158-66 substitutions, the 45–58% of cross-reactivity is relatively high. If we generalize these observations, we picture the map of repertoire to epitope:MHC shape (Figure 2, x-y plane) as consisting of many T cells that recognize only the epitope structure (rose cone) with decreasing numbers of cross-reactive (X-R) clonotypes that can probe into the shape–space neighborhood. If a virus mutates the epitope and is still within the X-R shape–space (Figure 2A), we assume that these X-R clonotypes are activated and increase their frequency. Other low-frequency X-R clonotypes that recognize the mutant peptide then increase to the point that they become observable, generating a new polyclonal peak recognizing the new epitope (Figure 2B). Other mutant epitopes may stray into self-shape–space (Figure 2C) that is not recognized by any T cells due to ignorance or central tolerance, resulting in pathogen escape (Figure 2D). Some mutant epitopes may move into the shape–space neighborhood of another epitope for which X-R clonotypes exist (Figure 2E). This possibility would also result in a response to the new epitope (Figure 2F). If the shape–space difference between the original and mutant epitope it sufficiently large, it is unlikely that any of these X-R clonotypes would recognize the original epitope. This model has similarities to epitope spread as first defined for autoimmunity.159,160
FIGURE 2.
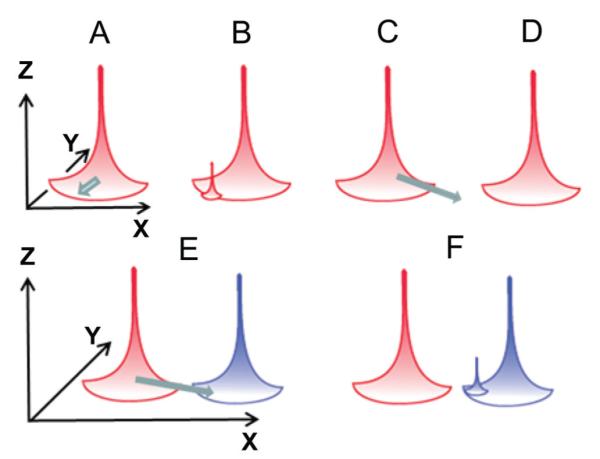
Buffering immune responses against epitope perturbations in middle-aged individuals. Diagram of pMHC-shape–space (x–y plane) showing a mature repertoire against a particular epitope (rose). Height reflects number of T cells, and spread in the x–y plane represents cross-reactivity to adjacent (similar) pMHC-shape. A. The epitope mutates within the cross-reactive (X-R) zone. B. X-R clonotypes expand and are added. C. The epitope moves into a repertoire hole resulting from ignorance or negative selection. D. Epitope escapes detection. E. Epitope mutates into a shape in the X-R neighborhood of a different epitope (blue). F. X-R clonotypes from second repertoire expand to counteract perturbation.
A cross-reactive T cell recognizing a mutated epitope may not respond as readily to presentation of that epitope as it would to the original epitope. However, such a memory cell would be more adapted to responding, and it would probably take fewer semiotic signals161 to involve it fully. Involvement of memory in novel epitopes can be seen in B-cell responses in middle-aged individuals where, in spite of new B-cell production in the marrow, a large portion of circulating Ab secreting cells have undergone previous somatic mutagenesis and tune their receptors to the new antigen.162 Arguing by analogy, memory T cells with potentially cross-reactive TCR, may be more amenable to being repositioned into the responding repertoire of a new pathogen by retuning their response threshold. This effect may be increased if they are selected on more than one epitope prior to the next encounter.
V. CONCLUSIONS
T-cell cross-reactivity based on recognition of more than one peptide by a TCR can be a very important facet of immune responses. Networks of cross-reactive T cells might generate a robust system that can deal with most novel pathogens in the absence of large-scale de novo production of naïve T cells, as is found in human post-thymic involution.
ABBREVIATIONS
- APCs
antigen-presenting cells
- BCG
Bacillus Calmette–Guérin
- CDR
complementarity-determining region
- CMV
cytomegalovirus
- CTL
cytotoxic T-cell line
- EBV
Epstein-Barr virus
- IFN-γ
interferon γ
- Ig:Ag
immunoglobulin-antigen
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HHV1
human herpes virus 1
- HLA
human leukocyte antigen
- LCMV
lymphocytic choriomeningitis virus
- MBP
myelin basic protein
- MCMV
murine cytomegalovirus
- MHC
major histocompatibility complex
- MHCI
MHC class I
- MHCII
MHC class II
- MS
multiple sclerosis
- PBMCs
peripheral blood mononuclear cells
- PLP
myelin proteolipid protein
- pMHC
peptide-MHC complex
- PV
Pichinde virus
- TCR
T-cell receptor
- TNF-α
tumor necrosis factor alpha
- VDW
van der Waals interactions
REFERENCES
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988 Aug 4;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–76. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 3.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999 Oct 29;286(5441):958–61. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 4.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009 Nov 5;114(19):4099–107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J Immunol. 2000 Jun 1;164(11):5782–7. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 6.Frankild S, de Boer RJ, Lund O, Nielsen M, Kesmir C. Amino acid similarity accounts for T cell cross-reactivity and for “holes” in the T cell repertoire. PLoS One. 2008;3(3):e1831. doi: 10.1371/journal.pone.0001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne JA, Butler JL, Cooper MD. Differential activation requirements for virgin and memory T cells. J Immunol. 1988 Nov 15;141(10):3249–57. [PubMed] [Google Scholar]
- 8.Croft M, Bradley LM, Swain SL. Naive versus memory CD4 T cell response to antigen. Memory cells are less dependent on accessory cell costimulation and can respond to many antigen-presenting cell types including resting B cells. J Immunol. 1994 Mar 15;152(6):2675–85. [PubMed] [Google Scholar]
- 9.Tanchot C, Lemonnier FA, Perarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science. 1997 Jun 27;276(5321):2057–62. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 10.Kimachi K, Sugie K, Grey HM. Effector T cells have a lower ligand affinity threshold for activation than naive T cells. Int Immunol. 2003 Jul;15(7):885–92. doi: 10.1093/intimm/dxg087. [DOI] [PubMed] [Google Scholar]
- 11.Pace L, Tempez A, Arnold-Schrauf C, Lemaitre F, Bousso P, Fetler L, Sparwasser T, Amigorena S. Regulatory T cells increase the avidity of primary CD8+ T cell responses and promote memory. Science. 2012 Oct 26;338(6106):532–6. doi: 10.1126/science.1227049. [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, Mariuzza RA. The multiple mechanisms of T cell receptor cross-reactivity. Immunity. 2009 Dec 18;31(6):849–51. doi: 10.1016/j.immuni.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Tsourkas PK, Liu W, Das SC, Pierce SK, Raychaudhuri S. Discrimination of membrane antigen affinity by B cells requires dominance of kinetic proofreading over serial engagement. Cell Mol Immunol. 2012 Jan;9(1):62–74. doi: 10.1038/cmi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998 Feb 20;279(5354):1166–72. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 15.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996 Oct 11;274(5285):209–19. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 16.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007 Apr 6;129(1):135–46. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 17.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000 Mar;12(3):251–61. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- 18.Luz JG, Huang M, Garcia KC, Rudolph MG, Apostolopoulos V, Teyton L, Wilson IA. Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions. J Exp Med. 2002 May 6;195(9):1175–86. doi: 10.1084/jem.20011644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, Burrows SR, McCluskey J, Rossjohn J. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007 Mar;8(3):268–76. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 20.Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Turner RV, Damirjian M, Wojnarowicz A, Biddison WE, Baker BM. T cell receptor recognition via cooperative conformational plasticity. J Mol Biol. 2006 Oct 13;363(1):228–43. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 21.Hare BJ, Wyss DF, Osburne MS, Kern PS, Reinherz EL, Wagner G. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat Struct Biol. 1999 Jun;6(6):574–81. doi: 10.1038/9359. [DOI] [PubMed] [Google Scholar]
- 22.Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002 Nov;10(11):1521–32. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 23.Reiser JB, Gregoire C, Darnault C, Mosser T, Guimezanes A, Schmitt-Verhulst AM, Fontecilla-Camps JC, Mazza G, Malissen B, Housset D. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002 Mar;16(3):345–54. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 24.Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003 Mar;4(3):241–7. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 25.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999 Jul;11(1):45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 26.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996 Nov 14;384(6605):134–41. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 27.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 28.Speir JA, Garcia KC, Brunmark A, Degano M, Peterson PA, Teyton L, Wilson IA. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 1998 May;8(5):553–62. doi: 10.1016/s1074-7613(00)80560-9. [DOI] [PubMed] [Google Scholar]
- 29.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009 Dec 18;31(6):885–96. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. Embo J. 2005 Sep 7;24(17):2968–79. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmer B, Pinilla C, Gran B, Vergelli M, Ling N, Conlon P, McFarland HF, Houghten R, Martin R. Contribution of individual amino acids within MHC molecule or antigenic peptide to TCR ligand potency. J Immunol. 2000 Jan 15;164(2):861–71. doi: 10.4049/jimmunol.164.2.861. [DOI] [PubMed] [Google Scholar]
- 32.Fujinami RS, Oldstone MB. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: mechanism for autoimmunity. Science. 1985 Nov 29;230(4729):1043–5. doi: 10.1126/science.2414848. [DOI] [PubMed] [Google Scholar]
- 33.Lang HL, Jacobsen H, Ikemizu S, Andersson C, Harlos K, Madsen L, Hjorth P, Sondergaard L, Svejgaard A, Wucherpfennig K, Stuart DI, Bell JI, Jones EY, Fugger L. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol. 2002 Oct;3(10):940–3. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- 34.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MC, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J. T cell allorecognition via molecular mimicry. Immunity. 2009 Dec 18;31(6):897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 35.Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12862–6. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishizuka J, Stewart-Jones GB, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008 Feb;28(2):171–82. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, van der Merwe PA. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 1999 Mar;10(3):357–65. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 38.Boniface JJ, Reich Z, Lyons DS, Davis MM. Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning. Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11446–51. doi: 10.1073/pnas.96.20.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong KM, Baker BM. A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction. Biophys J. 2007 Jul 15;93(2):597–609. doi: 10.1529/biophysj.107.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis-Harrison RL, Armstrong KM, Baker BM. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol. 2005 Feb 18;346(2):533–50. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 41.Miller PJ, Pazy Y, Conti B, Riddle D, Appella E, Collins EJ. Single MHC mutation eliminates enthalpy associated with T cell receptor binding. J Mol Biol. 2007 Oct 19;373(2):315–27. doi: 10.1016/j.jmb.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armstrong KM, Insaidoo FK, Baker BM. Thermodynamics of T-cell receptor-peptide/MHC interactions: progress and opportunities. J Mol Recognit. 2008 Jul-Aug;21(4):275–87. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008 Mar;28(3):324–34. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009 Feb;10(2):143–7. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisen HN, Chakraborty AK. Evolving concepts of specificity in immune reactions. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22373–80. doi: 10.1073/pnas.1012051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kingsley G, Sieper J. Current perspectives in reactive arthritis. Immunol Today. 1993 Aug;14(8):387–91. doi: 10.1016/0167-5699(93)90139-C. [DOI] [PubMed] [Google Scholar]
- 47.Kim PS, Klausmeier TL, Orr DP. Reactive arthritis: a review. J Adolesc Health. 2009 Apr;44(4):309–15. doi: 10.1016/j.jadohealth.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Christen U, von Herrath MG. Infections and autoimmunity—good or bad? J Immunol. 2005 Jun 15;174(12):7481–6. doi: 10.4049/jimmunol.174.12.7481. [DOI] [PubMed] [Google Scholar]
- 49.Bach JF. Infections and autoimmune diseases. J Autoimmun. 2005;25(Suppl):74–80. doi: 10.1016/j.jaut.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Rose NR. Autoimmunity, infection and adjuvants. Lupus. 2010 Apr;19(4):354–8. doi: 10.1177/0961203309360670. [DOI] [PubMed] [Google Scholar]
- 51.Friberg H, Bashyam H, Toyosaki-Maeda T, Potts JA, Greenough T, Kalayanarooj S, Gibbons RV, Nisalak A, Srikiatkhachorn A, Green S, Stephens HA, Rothman AL, Mathew A. Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci Rep. 2011;1:51. doi: 10.1038/srep00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friberg H, Burns L, Woda M, Kalayanarooj S, Endy TP, Stephens HA, Green S, Rothman AL, Mathew A. Memory CD8+ T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol Cell Biol. 2010 Jan;89(1):122–9. doi: 10.1038/icb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Townsend AR, Skehel JJ. The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J Exp Med. 1984 Aug 1;160(2):552–63. doi: 10.1084/jem.160.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haanen JB, Wolkers MC, Kruisbeek AM, Schumacher TN. Selective expansion of cross-reactive CD8(+) memory T cells by viral variants. J Exp Med. 1999 Nov 1;190(9):1319–28. doi: 10.1084/jem.190.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008 Feb 1;180(3):1758–68. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cusick MF, Wang S, Eckels DD. In vitro responses to avian influenza H5 by human CD4 T cells. J Immunol. 2009 Nov 15;183(10):6432–41. doi: 10.4049/jimmunol.0901617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT, Richards MJ, Rimmelzwaan GF, Kelso A, Doherty PC, Turner SJ, Rossjohn J, Kedzierska K. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12599–604. doi: 10.1073/pnas.1007270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbani S, Amadei B, Cariani E, Fisicaro P, Orlandini A, Missale G, Ferrari C. The impairment of CD8 responses limits the selection of escape mutations in acute hepatitis C virus infection. J Immunol. 2005 Dec 1;175(11):7519–29. doi: 10.4049/jimmunol.175.11.7519. [DOI] [PubMed] [Google Scholar]
- 59.Lee JK, Stewart-Jones G, Dong T, Harlos K, Di Gleria K, Dorrell L, Douek DC, van der Merwe PA, Jones EY, McMichael AJ. T cell cross-reactivity and conformational changes during TCR engagement. J Exp Med. 2004 Dec 6;200(11):1455–66. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conrad JA, Ramalingam RK, Smith RM, Barnett L, Lorey SL, Wei J, Simons BC, Sadagopal S, Meyer-Olson D, Kalams SA. Dominant clonotypes within HIV-specific T cell responses are programmed death-1high and CD127low and display reduced variant cross-reactivity. J Immunol. 2011 Jun 15;186(12):6871–85. doi: 10.4049/jimmunol.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol. 2010 Mar 15;184(6):2825–38. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shirai M, Kurokohchi K, Pendleton CD, Arichi T, Boyd LF, Takahashi H, Margulies DH, Berzofsky JA. Reciprocal cytotoxic T lymphocyte cross-reactivity interactions between two major epitopes within HIV-1 gp160. J Immunol. 1996 Nov 15;157(10):4399–411. [PubMed] [Google Scholar]
- 63.Gotch F, McMichael A, Rothbard J. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. Use of analogues to orientate the matrix peptide in the HLA-A2 binding site. J Exp Med. 1988 Dec 1;168(6):2045–57. doi: 10.1084/jem.168.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu N, D’Souza C, Cheung H, Lang H, Cheuk E, Chamberlain JW. Highly conserved pattern of recognition of influenza A wild-type and variant CD8+ CTL epitopes in HLA-A2+ humans and transgenic HLA-A2+/H2 class I-deficient mice. Vaccine. 2005 Nov 1;23(45):5231–44. doi: 10.1016/j.vaccine.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 65.Bennett MS, Ng HL, Dagarag M, Ali A, Yang OO. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J Virol. 2007 May;81(10):4973–80. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valkenburg SA, Gras S, Guillonneau C, La Gruta NL, Thomas PG, Purcell AW, Rossjohn J, Doherty PC, Turner SJ, Kedzierska K. Protective efficacy of cross-reactive CD8+ T cells recognising mutant viral epitopes depends on peptide-MHC-I structural interactions and T cell activation threshold. PLoS Pathog. 2010;6(8):e1001039. doi: 10.1371/journal.ppat.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrova GV, Naumova EN, Gorski J. The polyclonal CD8 T cell response to influenza M158-66 generates a fully connected network of cross-reactive clonotypes to structurally related peptides: a paradigm for memory repertoire coverage of novel epitopes or escape mutants. J Immunol. 2011 Jun 1;186(11):6390–7. doi: 10.4049/jimmunol.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubio-Godoy V, Dutoit V, Zhao Y, Simon R, Guillaume P, Houghten R, Romero P, Cerottini JC, Pinilla C, Valmori D. Positional scanning-synthetic peptide library-based analysis of self- and pathogen-derived peptide cross-reactivity with tumor-reactive Melan-A-specific CTL. J Immunol. 2002 Nov 15;169(10):5696–707. doi: 10.4049/jimmunol.169.10.5696. [DOI] [PubMed] [Google Scholar]
- 69.Nino-Vasquez JJ, Allicotti G, Borras E, Wilson DB, Valmori D, Simon R, Martin R, Pinilla C. A powerful combination: the use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol Immunol. 2004 Feb;40(14–15):1063–74. doi: 10.1016/j.molimm.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Kan-Mitchell J, Bajcz M, Schaubert KL, Price DA, Brenchley JM, Asher TE, Douek DC, Ng HL, Yang OO, Rinaldo CR, Jr, Benito JM, Bisikirska B, Hegde R, Marincola FM, Boggiano C, Wilson D, Abrams J, Blondelle SE, Wilson DB. Degeneracy and repertoire of the human HIV-1 Gag p17(77-85) CTL response. J Immunol. 2006 Jun 1;176(11):6690–701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 71.Ishizuka J, Grebe K, Shenderov E, Peters B, Chen Q, Peng Y, Wang L, Dong T, Pasquetto V, Oseroff C, Sidney J, Hickman H, Cerundolo V, Sette A, Bennink JR, McMichael A, Yewdell JW. Quantitating T cell cross-reactivity for unrelated peptide antigens. J Immunol. 2009 Oct 1;183(7):4337–45. doi: 10.4049/jimmunol.0901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wooldridge L, Ekeruche-Makinde J, van den Berg HA, Skowera A, Miles JJ, Tan MP, Dolton G, Clement M, Llewellyn-Lacey S, Price DA, Peakman M, Sewell AK. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem. 2012 Jan 6;287(2):1168–77. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vali B, Tohn R, Cohen MJ, Sakhdari A, Sheth PM, Yue FY, Wong D, Kovacs C, Kaul R, Ostrowski MA. Characterization of cross-reactive CD8+ T-cell recognition of HLA-A2-restricted HIV-Gag (SLYNTVATL) and HCV-NS5b (ALYDVVSKL) epitopes in individuals infected with human immunodeficiency and hepatitis C viruses. J Virol. 2011 Jan;85(1):254–63. doi: 10.1128/JVI.01743-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Acierno PM, Newton DA, Brown EA, Maes LA, Baatz JE, Gattoni-Celli S. Cross-reactivity between HLA-A2-restricted FLU-M1:58-66 and HIV p17 GAG:77-85 epitopes in HIV-infected and uninfected individuals. J Transl Med. 2003 Aug 14;1(1):3. doi: 10.1186/1479-5876-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clute SC, Watkin LB, Cornberg M, Naumov YN, Sullivan JL, Luzuriaga K, Welsh RM, Selin LK. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J Clin Invest. 2005 Dec;115(12):3602–12. doi: 10.1172/JCI25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clute SC, Naumov YN, Watkin LB, Aslan N, Sullivan JL, Thorley-Lawson DA, Luzuriaga K, Welsh RM, Puzone R, Celada F, Selin LK. Broad cross-reactive TCR repertoires recognizing dissimilar Epstein-Barr and influenza A virus epitopes. J Immunol. 2010 Dec 1;185(11):6753–64. doi: 10.4049/jimmunol.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J Virol. 2001 Dec;75(23):11392–400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasprowicz V, Ward SM, Turner A, Grammatikos A, Nolan BE, Lewis-Ximenez L, Sharp C, Woodruff J, Fleming VM, Sims S, Walker BD, Sewell AK, Lauer GM, Klenerman P. Defining the directionality and quality of influenza virusspecific CD8+ T cell cross-reactivity in individuals infected with hepatitis C virus. J Clin Invest. 2008 Mar;118(3):1143–53. doi: 10.1172/JCI33082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urbani S, Amadei B, Fisicaro P, Pilli M, Missale G, Bertoletti A, Ferrari C. Heterologous T cell immunity in severe hepatitis C virus infection. J Exp Med. 2005 Mar 7;201(5):675–80. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kennedy PT, Urbani S, Moses RA, Amadei B, Fisicaro P, Lloyd J, Maini MK, Dusheiko G, Ferrari C, Bertoletti A. The influence of T cell cross-reactivity on HCV-peptide specific human T cell response. Hepatology. 2006 Mar;43(3):602–11. doi: 10.1002/hep.21081. [DOI] [PubMed] [Google Scholar]
- 81.Doisne JM, Urrutia A, Lacabaratz-Porret C, Goujard C, Meyer L, Chaix ML, Sinet M, Venet A. CD8+ T cells specific for EBV, cytomegalovirus, and influenza virus are activated during primary HIV infection. J Immunol. 2004 Aug 15;173(4):2410–8. doi: 10.4049/jimmunol.173.4.2410. [DOI] [PubMed] [Google Scholar]
- 82.Hausmann S, Wucherpfennig KW. Activation of autoreactive T cells by peptides from human pathogens. Curr Opin Immunol. 1997 Dec;9(6):831–8. doi: 10.1016/S0952-7915(97)80186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995 Mar 10;80(5):695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.von Herrath MG, Holz A, Homann D, Oldstone MB. Role of viruses in type I diabetes. Semin Immunol. 1998 Feb;10(1):87–100. doi: 10.1006/smim.1997.0108. [DOI] [PubMed] [Google Scholar]
- 85.von Herrath MG, Oldstone MB. Role of viruses in the loss of tolerance to self-antigens and in autoimmune diseases. Trends Microbiol. 1995 Nov;3(11):424–30. doi: 10.1016/s0966-842x(00)88995-7. [DOI] [PubMed] [Google Scholar]
- 86.Hudrisier D, Riond J, Burlet-Schiltz O, von Herrath MG, Lewicki H, Monsarrat B, Oldstone MB, Gairin JE. Structural and functional identification of major histocompatibility complex class I-restricted self-peptides as naturally occurring molecular mimics of viral antigens. Possible role in CD8+ T cell-mediated, virus-induced autoimmune disease. J Biol Chem. 2001 Jun 1;276(22):19396–403. doi: 10.1074/jbc.M008864200. [DOI] [PubMed] [Google Scholar]
- 87.Harkiolaki M, Holmes SL, Svendsen P, Gregersen JW, Jensen LT, McMahon R, Friese MA, van Boxel G, Etzensperger R, Tzartos JS, Kranc K, Sainsbury S, Harlos K, Mellins ED, Palace J, Esiri MM, van der Merwe PA, Jones EY, Fugger L. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity. 2009 Mar 20;30(3):348–57. doi: 10.1016/j.immuni.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Munz C, Lunemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol. 2009 Apr;9(4):246–58. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christen U, Edelmann KH, McGavern DB, Wolfe T, Coon B, Teague MK, Miller SD, Oldstone MB, von Herrath MG. A viral epitope that mimics a self antigen can accelerate but not initiate autoimmune diabetes. J Clin Invest. 2004 Nov;114(9):1290–8. doi: 10.1172/JCI22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhib-Jalbut S. Pathogenesis of myelin/oligodendrocyte damage in multiple sclerosis. Neurology. 2007 May 29;68(22 Suppl 3):S13–21. doi: 10.1212/01.wnl.0000275228.13012.7b. discussion S43–54. [DOI] [PubMed] [Google Scholar]
- 91.Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, Zaremba A, Rackham C, Allen JS, Tree TI, Zhao M, Dayan CM, Sewell AK, Unger WW, Drijfhout JW, Ossendorp F, Roep BO, Peakman M. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest. 2008 Oct;118(10):3390–402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holler PD, Kranz DM. T cell receptors: affinities, cross-reactivities, and a conformer model. Mol Immunol. 2004 Feb;40(14–15):1027–31. doi: 10.1016/j.molimm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 93.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998 Sep;19(9):395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 94.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13980–5. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005 Sep 5;202(5):673–85. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wooldridge L, Laugel B, Ekeruche J, Clement M, van den Berg HA, Price DA, Sewell AK. CD8 controls T cell cross-reactivity. J Immunol. 2010 Oct 15;185(8):4625–32. doi: 10.4049/jimmunol.1001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sloan-Lancaster J, Shaw AS, Rothbard JB, Allen PM. Partial T cell signaling: altered phospho-zeta and lack of zap70 recruitment in APL-induced T cell anergy. Cell. 1994 Dec 2;79(5):913–22. doi: 10.1016/0092-8674(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 98.Madrenas J, Wange RL, Wang JL, Isakov N, Samelson LE, Germain RN. Zeta phosphorylation without ZAP-70 activation induced by TCR antagonists or partial agonists. Science. 1995 Jan 27;267(5197):515–8. doi: 10.1126/science.7824949. [DOI] [PubMed] [Google Scholar]
- 99.Stefanova I, Hemmer B, Vergelli M, Martin R, Biddison WE, Germain RN. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat Immunol. 2003 Mar;4(3):248–54. doi: 10.1038/ni895. [DOI] [PubMed] [Google Scholar]
- 100.Irie A, Chen YZ, Tsukamoto H, Jotsuka T, Masuda M, Nishimura Y. Unique T cell proliferation associated with PKCmu activation and impaired ZAP-70 phosphorylation in recognition of over-expressed HLA/partially agonistic peptide complexes. Eur J Immunol. 2003 Jun;33(6):1497–507. doi: 10.1002/eji.200323618. [DOI] [PubMed] [Google Scholar]
- 101.Monu N, Frey AB. Suppression of proximal T cell receptor signaling and lytic function in CD8+ tumor-infiltrating T cells. Cancer Res. 2007 Dec 1;67(23):11447–54. doi: 10.1158/0008-5472.CAN-07-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schnell FJ, Alberts-Grill N, Evavold BD. CD8+ T cell responses to a viral escape mutant epitope: active suppression via altered SHP-1 activity. J Immunol. 2009 Feb 15;182(4):1829–35. doi: 10.4049/jimmunol.0801798. [DOI] [PubMed] [Google Scholar]
- 103.Viola A, Salio M, Tuosto L, Linkert S, Acuto O, Lanzavecchia A. Quantitative contribution of CD4 and CD8 to T cell antigen receptor serial triggering. J Exp Med. 1997 Nov 17;186(10):1775–9. doi: 10.1084/jem.186.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Itoh Y, Germain RN. Single cell analysis reveals regulated hierarchical T cell antigen receptor signaling thresholds and intraclonal heterogeneity for individual cytokine responses of CD4+ T cells. J Exp Med. 1997 Aug 29;186(5):757–66. doi: 10.1084/jem.186.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nicholson LB, Anderson AC, Kuchroo VK. Tuning T cell activation threshold and effector function with cross-reactive peptide ligands. Int Immunol. 2000 Feb;12(2):205–13. doi: 10.1093/intimm/12.2.205. [DOI] [PubMed] [Google Scholar]
- 106.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005 Nov;3(11):e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008 Aug 22;321(5892):1081–4. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coward J, Germain RN, Altan-Bonnet G. Perspectives for computer modeling in the study of T cell activation. Cold Spring Harb Perspect Biol. 2010 Jun;2(6):a005538. doi: 10.1101/cshperspect.a005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev. 2006 Jun;211:164–81. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010 May;235(1):244–66. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bangs SC, Baban D, Cattan HJ, Li CK, McMichael AJ, Xu XN. Human CD4+ memory T cells are preferential targets for bystander activation and apoptosis. J Immunol. 2009 Feb 15;182(4):1962–71. doi: 10.4049/jimmunol.0802596. [DOI] [PubMed] [Google Scholar]
- 112.Selin LK, Varga SM, Wong IC, Welsh RM. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med. 1998 Nov 2;188(9):1705–15. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brehm MA, Pinto AK, Daniels KA, Schneck JP, Welsh RM, Selin LK. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat Immunol. 2002 Jul;3(7):627–34. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 114.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol. 2001 Nov;2(11):1067–76. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 115.Kim SK, Brehm MA, Welsh RM, Selin LK. Dynamics of memory T cell proliferation under conditions of heterologous immunity and bystander stimulation. J Immunol. 2002 Jul 1;169(1):90–8. doi: 10.4049/jimmunol.169.1.90. [DOI] [PubMed] [Google Scholar]
- 116.Chen HD, Fraire AE, Joris I, Welsh RM, Selin LK. Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am J Pathol. 2003 Oct;163(4):1341–55. doi: 10.1016/S0002-9440(10)63493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mathurin KS, Martens GW, Kornfeld H, Welsh RM. CD4 T-cell-mediated heterologous immunity between mycobacteria and poxviruses. J Virol. 2009 Apr;83(8):3528–39. doi: 10.1128/JVI.02393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cornberg M, Sheridan BS, Saccoccio FM, Brehm MA, Selin LK. Protection against vaccinia virus challenge by CD8 memory T cells resolved by molecular mimicry. J Virol. 2007 Jan;81(2):934–44. doi: 10.1128/JVI.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen AT, Cornberg M, Gras S, Guillonneau C, Rossjohn J, Trees A, Emonet S, de la Torre JC, Welsh RM, Selin LK. Loss of anti-viral immunity by infection with a virus encoding a cross-reactive pathogenic epitope. PLoS Pathog. 2012 Apr;8(4):e1002633. doi: 10.1371/journal.ppat.1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006 May;116(5):1443–56. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakamura I, Cudkowicz G. Fine specificity of auto- and alloreactive cytotoxic T-lymphocytes: heteroclitic cross-reactions between mutant and original H-2 antigens. Curr Top Microbiol Immunol. 1982;99:51–80. doi: 10.1007/978-3-642-68528-6_2. [DOI] [PubMed] [Google Scholar]
- 122.Solinger AM, Ultee ME, Margoliash E, Schwartz RH. T-lymphocyte response to cytochrome c. I. Demonstration of a T-cell heteroclitic proliferative response and identification of a topographic antigenic determinant on pigeon cytochrome c whose immune recognition requires two complementing major histocompatibility complex-linked immune response genes. J Exp Med. 1979 Oct 1;150(4):830–48. doi: 10.1084/jem.150.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reay PA, Kantor RM, Davis MM. Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93-103) J Immunol. 1994 Apr 15;152(8):3946–57. [PubMed] [Google Scholar]
- 124.Dyall R, Bowne WB, Weber LW, LeMaoult J, Szabo P, Moroi Y, Piskun G, Lewis JJ, Houghton AN, Nikolić-Zugić J. Heteroclitic immunization induces tumor immunity. J Exp Med. 1998 Nov 2;188(9):1553–61. doi: 10.1084/jem.188.9.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tanaka Y, Amos KD, Joo HG, Eberlein TJ, Goedegebuure PS. Modification of the HER2/ NEU-derived tumor antigen GP2 improves induction of GP2-reactive cytotoxic T lymphocytes. Int J Cancer. 2001 Nov;94(4):540–4. doi: 10.1002/ijc.1508. [DOI] [PubMed] [Google Scholar]
- 126.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Topalian SL, Sherry RM, Restifo NP, Wunderlich JR, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, Gritz L, Panicali DL, White DE. Recombinant fowlpox viruses encoding the anchor-modified gp100 melanoma antigen can generate antitumor immune responses in patients with metastatic melanoma. Clin Cancer Res. 2003 Aug 1;9(8):2973–80. [PMC free article] [PubMed] [Google Scholar]
- 127.Gold JS, Ferrone CR, Guevara-Patino JA, Hawkins WG, Dyall R, Engelhorn ME, Wolchok JD, Lewis JJ, Houghton AN. A single heteroclitic epitope determines cancer immunity after xenogeneic DNA immunization against a tumor differentiation antigen. J Immunol. 2003 May 15;170(10):5188–94. doi: 10.4049/jimmunol.170.10.5188. [DOI] [PubMed] [Google Scholar]
- 128.Bae J, Martinson JA, Klingemann HG. Heteroclitic CD33 peptide with enhanced anti-acute myeloid leukemic immunogenicity. Clin Cancer Res. 2004 Oct 15;10(20):7043–52. doi: 10.1158/1078-0432.CCR-04-0322. [DOI] [PubMed] [Google Scholar]
- 129.Bae J, Martinson JA, Klingemann HG. Identification of novel CD33 antigen-specific peptides for the generation of cytotoxic T lymphocytes against acute myeloid leukemia. Cell Immunol. 2004 Jan;227(1):38–50. doi: 10.1016/j.cellimm.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 130.Zirlik KM, Zahrieh D, Neuberg D, Gribben JG. Cytotoxic T cells generated against heteroclitic peptides kill primary tumor cells independent of the binding affinity of the native tumor antigen peptide. Blood. 2006 Dec 1;108(12):3865–70. doi: 10.1182/blood-2006-04-014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwartzentruber DJ, Lawson DH, Richards JM, Conry RM, Miller DM, Treisman J, Gailani F, Riley L, Conlon K, Pockaj B, Kendra KL, White RL, Gonzalez R, Kuzel TM, Curti B, Leming PD, Whitman ED, Balkissoon J, Reintgen DS, Kaufman H, Marincola FM, Merino MJ, Rosenberg SA, Choyke P, Vena D, Hwu P. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med. 2011 Jun 2;364(22):2119–27. doi: 10.1056/NEJMoa1012863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baumgartner CK, Ferrante A, Nagaoka M, Gorski J, Malherbe LP. Peptide-MHC class II complex stability governs CD4 T cell clonal selection. J Immunol. 2010 Jan 15;184(2):573–81. doi: 10.4049/jimmunol.0902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vujanovic L, Mandic M, Olson WC, Kirkwood JM, Storkus WJ. A mycoplasma peptide elicits heteroclitic CD4+ T cell responses against tumor antigen MAGE-A6. Clin Cancer Res. 2007 Nov 15;13(22 Pt 1):6796–806. doi: 10.1158/1078-0432.CCR-07-1909. [DOI] [PubMed] [Google Scholar]
- 134.Mason RD, Bowmer MI, Howley CM, Grant MD. Cross-reactive cytotoxic T lymphocytes against human immunodeficiency virus type 1 protease and gamma interferon-inducible protein 30. J Virol. 2005 May;79(9):5529–36. doi: 10.1128/JVI.79.9.5529-5536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nicholson LB, Waldner H, Carrizosa AM, Sette A, Collins M, Kuchroo VK. Heteroclitic proliferative responses and changes in cytokine profile induced by altered peptides: implications for autoimmunity. Proc Natl Acad Sci U S A. 1998 Jan 6;95(1):264–9. doi: 10.1073/pnas.95.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Butler NS, Theodossis A, Webb AI, Nastovska R, Ramarathinam SH, Dunstone MA, Rossjohn J, Purcell AW, Perlman S. Prevention of cytotoxic T cell escape using a heteroclitic subdominant viral T cell determinant. PLoS Pathog. 2008 Oct;4(10):e1000186. doi: 10.1371/journal.ppat.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cole DK, Edwards ES, Wynn KK, Clement M, Miles JJ, Ladell K, Ekeruche J, Gostick E, Adams KJ, Skowera A, Peakman M, Wooldridge L, Price DA, Sewell AK. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J Immunol. 2010 Aug 15;185(4):2600–10. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.La Rosa C, Krishnan R, Markel S, Schneck JP, Houghten R, Pinilla C, Diamond DJ. Enhanced immune activity of cytotoxic T-lymphocyte epitope analogs derived from positional scanning synthetic combinatorial libraries. Blood. 2001 Mar 15;97(6):1776–86. doi: 10.1182/blood.v97.6.1776. [DOI] [PubMed] [Google Scholar]
- 139.Rubio-Godoy V, Pinilla C, Dutoit V, Borras E, Simon R, Zhao Y, Cerottini JC, Romero P, Houghten R, Valmori D. Toward synthetic combinatorial peptide libraries in positional scanning format (PS-SCL)-based identification of CD8+ Tumor-reactive T-Cell Ligands: a comparative analysis of PS-SCL recognition by a single tumor-reactive CD8+ cytolytic T-lymphocyte clone. Cancer Res. 2002 Apr 1;62(7):2058–63. [PubMed] [Google Scholar]
- 140.Tung CW, Ziehm M, Kamper A, Kohlbacher O, Ho SY. POPISK: T-cell reactivity prediction using support vector machines and string kernels. BMC Bioinformatics. 2011;12:446. doi: 10.1186/1471-2105-12-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rubio-Godoy V, Ayyoub M, Dutoit V, Servis C, Schink A, Rimoldi D, Romero P, Cerottini JC, Simon R, Zhao Y, Houghten RA, Pinilla C, Valmori D. Combinatorial peptide library-based identification of peptide ligands for tumor-reactive cytolytic T lymphocytes of unknown specificity. Eur J Immunol. 2002 Aug;32(8):2292–9. doi: 10.1002/1521-4141(200208)32:8<2292::AID-IMMU2292>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 142.Hemmer B, Gran B, Zhao Y, Marques A, Pascal J, Tzou A, Kondo T, Cortese I, Bielekova B, Straus SE, McFarland HF, Houghten R, Simon R, Pinilla C, Martin R. Identification of candidate T-cell epitopes and molecular mimics in chronic Lyme disease. Nat Med. 1999 Dec;5(12):1375–82. doi: 10.1038/70946. [DOI] [PubMed] [Google Scholar]
- 143.Price GE, Ou R, Jiang H, Huang L, Moskophidis D. Viral escape by selection of cytotoxic T cell-resistant variants in influenza A virus pneumonia. J Exp Med. 2000 Jun 5;191(11):1853–67. doi: 10.1084/jem.191.11.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004 Dec;21(6):793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 145.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004 Aug 2;200(3):307–19. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999 Feb 15;189(4):701–10. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen ZW, Li Y, Zeng X, Kuroda MJ, Schmitz JE, Shen Y, Lai X, Shen L, Letvin NL. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J Immunol. 2001 Apr 1;166(7):4525–33. doi: 10.4049/jimmunol.166.7.4525. [DOI] [PubMed] [Google Scholar]
- 148.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002 Nov 29;298(5599):1797–800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 149.Schroeder MR. Fractals, Chaos, Power Laws: Minutes from an Infinite Paradise. New York: 1991. [Google Scholar]
- 150.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in “real-world” conditions. J Immunol. 1998 Mar 15;160(6):2842–52. [PubMed] [Google Scholar]
- 151.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J Immunol. 2003 Apr 15;170(8):3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 152.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006 Aug 1;177(3):2006–14. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 153.Kedzierska K, Day EB, Pi J, Heard SB, Doherty PC, Turner SJ, Perlman S. Quantification of repertoire diversity of influenza-specific epitopes with predominant public or private TCR usage. J Immunol. 2006 Nov 15;177(10):6705–12. doi: 10.4049/jimmunol.177.10.6705. [DOI] [PubMed] [Google Scholar]
- 154.Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B, Ziegelbauer J, Yassai M, Li SH, Relland LM, Wise PM, Chen A, Zheng YQ, Simpson PM, Gorski J, Salzman NH, Hessner MJ, Chatila TA, Williams CB. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011 Jul 22;35(1):109–22. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010 Sep 1;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr., Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1518–23. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carlson JM, Doyle J. Complexity and robustness. Proc Natl Acad Sci U S A. 2002 Feb 19;99(Suppl 1):2538–45. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kitano H. Biological robustness. Nat Rev Genet. 2004 Nov;5(11):826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 159.Lehmann PV, Forsthuber T, Miller A, Sercarz EE. Spreading of T-cell autoimmunity to cryptic determinants of an autoantigen. Nature. 1992 Jul 9;358(6382):155–7. doi: 10.1038/358155a0. [DOI] [PubMed] [Google Scholar]
- 160.Powell AM, Black MM. Epitope spreading: protection from pathogens, but propagation of autoimmunity? Clin Exp Dermatol. 2001 Jul;26(5):427–33. doi: 10.1046/j.1365-2230.2001.00852.x. [DOI] [PubMed] [Google Scholar]
- 161.Janeway CA., Jr The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992 Jan;13(1):11–6. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 162.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011 Jan 17;208(1):181–93. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]


