Abstract
The UPR (unfolded protein response) pathway is comprised of three signalling cascades mediated by the ER (endoplasmic reticulum) stress sensor proteins PERK [PKR (double-stranded RNA-activated protein kinase)-like ER kinase], IRE1 (inositol-requiring kinase 1) and ATF6 (activating transcription factor 6). The present study shows that ASNS (asparagine synthetase) transcription activity was up-regulated in HepG2 cells treated with the UPR activators thapsigargin and tunicamycin. ChIP (chromatin immunoprecipitation) analysis demonstrated that during ER stress, ATF4, ATF3 and C/EBPβ (CCAAT/enhancer-binding protein β) bind to the ASNS proximal promoter region that includes the genomic sequences NSRE (nutrient-sensing response element)-1 and NSRE-2, previously implicated by mutagenesis in UPR activation. Consistent with increased ASNS transcription, ChIP analysis also demonstrated that UPR signalling resulted in enhanced recruitment of general transcription factors, including RNA Pol II (polymerase II), to the ASNS promoter. The ASNS gene is also activated by the AAR (amino acid response) pathway following amino acid deprivation of tissue or cells. Immunoblot analysis of HepG2 cells demonstrated that simultaneous activation of the AAR and UPR pathways did not further increase the ASNS or ATF4 protein abundance when compared to triggering either pathway alone. In addition, siRNA (small interfering RNA)-mediated knockdown of XBP1 (X-box binding protein 1), ATF6α or ATF6β expression did not affect ASNS transcription, whereas siRNA against ATF4 suppressed ASNS transcription during UPR activation. Collectively, these results indicate that the PERK/p-eIF2α (phosphorylated eukaryotic initiation factor 2α)/ATF4 signalling cascade is the only arm of the UPR that is responsible for ASNS transcriptional induction during ER stress. Consequently, the ASNS NSRE-1 and NSRE-2 elements, in addition to ERSE (ER stress response element)-I, ERSE-II and the mUPRE (mammalian UPR element), function as mammalian UPR responsive sequences.
Keywords: activating transcription factor 3 (ATF3), activating transcription factor 4 (ATF4), asparagine synthetase (ASNS), CCAAT/enhancer-binding protein (C/EBP), endoplasmic reticulum stress, nutrient sensing, unfolded protein response (UPR)
INTRODUCTION
ER (endoplasmic reticulum) stress represents an imbalance between ER protein load and ER processing capacity [1]. Cellular stresses that lead to perturbation in calcium homoeostasis, abnormal protein glycosylation, glucose deprivation or expression of mutant membrane and secretory proteins lead to the accumulation of unfolded proteins in the ER lumen [2]. As a consequence, three signal transduction cascades are triggered that are collectively referred to as the UPR (unfolded protein response). These signalling pathways include both translational and transcriptional control mechanisms that reduce protein synthesis, increase the ER folding capacity by up-regulating the transcription of chaperones, activate ERAD (ER-associated protein degradation), and ultimately, provoke cell death [1, 3–5]. Upon accumulation of malfolded proteins, the ER lumenal chaperone BiP (immunoglobulin heavy-chain-binding protein), also known as GRP78 (glucose-regulated protein 78), dissociates from the lumenal domains of three ER resident transmembrane sensors and binds to the unfolded proteins. The ER-stress sensors, IRE1 (inositol-requiring kinase 1), PERK [PKR (double-stranded RNA-activated protein kinase)-like ER kinase] and ATF6 (activating transcription factor 6) are all activated after dissociation from BiP/GRP78 through either dimerization and trans-autophosphorylation (IRE1 and PERK) or translocation to the Golgi complex followed by proteolysis to a functional form (ATF6) [1]. Active IRE1 is an endonuclease that contributes to the splicing of the XBP1 (X-box binding protein 1) precursor mRNA, which then permits synthesis of XBP1 protein [6]. IRE1/XBP-1 and ATF6 mediate transcriptional regulation of chaperones that assist folding in the ER as well as protein degradation mechanisms [7], metabolism [8] and apoptosis [9, 10]. XBP1 or ATF6 binds to genomic elements, called ERSEs (ER stress response elements) (ERSE-I,CCAAT-N9-CCACGor ERSE-II,ATTGG-N1-CCACG) in the presence of a constitutively bound NF-Y (nuclear factor Y), which is a CCAAT/binding factor [11, 12]. XBP1 also binds to another UPR-mediating genomic element called the mUPRE (mammalian UPR element, TGACGTGG/A) [6].
Translational control during UPR activation is exercised through the PERK/eIF2α (eukaryotic initiation factor 2α)/ATF4 branch. ER stress activates PERK, an eIF2α kinase that phosphorylates eIF2α at Ser51 [13]. An increase of p-eIF2α (phosphorylated eIF2α) leads to a transient suppression of general translation, but increased translation of the mRNA for the transcription factor ATF4 as a consequence of a ribosome scanning mechanism and two short upstream opening reading frames in the ATF4 mRNA [14, 15]. Repression of general translation by p-eIF2α alleviates the protein load of the ER, while the increased translation of ATF4 leads to transcriptional induction of ATF4-responsive genes involved in the ER stress response [16, 17].
In contrast to ER stress, the cellular stress of protein limitation or amino acid limitation leads to the activation of a signalling cascade called the AAR (amino acid response) pathway. A limiting amount of an amino acid that is essential for that particular cell type leads to the accumulation of uncharged tRNA that binds to and activates a ribosome-associated protein called GCN2 (general control non-derepressible 2) [18]. GCN2 is a eIF2α kinase that phosphorylates Ser51 [19, 20]. As described above for PERK, phosphorylation of eIF2α at Ser51 leads to increased ATF4 synthesis and downstream transcriptional activation. Among the many ATF4 target genes is ASNS (asparagine synthetase), for which transcription is increased in response to activation of both the UPR and AAR pathways. Mutagenesis and transient transfection of the ASNS promoter region has suggested that the two cis-acting elements, NSRE (nutrient-sensing response element)-1 and NSRE-2 mediate the transcriptional induction by both AAR and UPR pathway activation [21]. NSRE-1 and NSRE-2 function together as an enhancer element and are referred to as the NSRU (nutrient sensing response unit) [22]. The NSRE-1 sequence 5′-TGATGAAAC-3′ is a C/EBP (CCAAT/enhancer-binding protein)–ATF composite site [22, 23], and these sequences have been shown to bind heterodimers of the C/EBP and ATF bZIP subfamilies, including C/EBPβ and ATF4 [24–26]. Electrophoresis mobility shift analysis documented that ATF4 binds to the ASNS NSRE-1 site in vitro [25]. However, those previous studies did not document ATF4 binding to the NSRE-1 site in vivo, nor were the ATF6 and XBP1 arms of the UPR tested for their potential role in ASNS regulation. The question of whether the IRE1/XBP1 and ATF6 arms of the UPR impact ASNS transcription is an important one because the ATF half-site sequence of the C/EBP-ATF site (TGATG) has similarity to the 5′ portion of the mUPRE (TGACG).
The present study characterizes the transcriptional control of the ASNS gene during UPR activation in human HepG2 hepatoma cells. Extensive ChIP (chromatin immunoprecipitation) analysis of the C/EBP-ATF site in the ASNS proximal promoter during UPR activation revealed the time-dependent binding of the transcription factors ATF4, ATF3 and C/EBPβ, providing further evidence that the NSRU genomic element mediates the transcriptional response during both the AAR and UPR. Data obtained from the concomitant activation of the AAR and UPR indicated that these two pathways share a common step in the regulation of the ASNS gene. The siRNA (small interfering RNA)-mediated knockdown of XBP1, ATF6α and ATF6β expression documented that these UPR arms do not have a critical contribution to the ASNS induction following ER stress. Short interference RNA against ATF4 decreased ASNS transcriptional induction during UPR. Collectively, the results demonstrate that the PERK-mediated arm of the UPR is the sole signalling cascade responsible for ASNS transcriptional induction and that, along with the ERSE and mUPRE sequences, the NSRU is an ER stress-responsive genomic element.
MATERIALS AND METHODS
Cell culture
HepG2 human hepatoma cells were grown in T-175 flasks and for experiments plated in either 60 mm or 150 mm culture dishes. Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The medium used was modified Eagle’s MEM (minimal essential medium; pH 7.4) (Mediatech, Herndon, VA, U.S.A.), supplemented with 1×non-essential amino acids, 2 mM glutamine, 100 mg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 mg/ml amphotericin B and 10% (v/v) FBS (fetal bovine serum). To ensure that the cells were in a basal or ‘fed’ status before the stress treatment, fresh MEM and serum was provided to the cells 12 h before initiating drug treatment. For experiments involving histidine deprivation (MEM−His), 10% dialysed FBS was added to the histidine-deficient MEM medium (Invitrogen). To induce the UPR pathway, cells were incubated for a specific time period in MEM containing 300 nM Tg (thapsigargin) (MEM+Tg) or 5µg/ml Tu (tunicamycin) (MEM+Tu).
Immunoblot analysis
Total cell extracts were prepared for immunoblot analysis from HepG2 cells incubated in MEM only, MEM−His or MEM+Tg for 0–24 h. Protein content was quantified by a Lowry assay and 30 µg of protein was separated on a pre-cast Criterion Tris/HCl polyacrylamide gel (Bio-Rad). After electrotransfer to a Bio-Rad nitrocellulose membrane, the membrane was stained with Fast Green to check for equal loading and then incubated with 10% blocking solution [10% (w/v) Carnation non-fat dried skimmed milk and Tris-buffered saline/Tween (30 mM Tris base (pH 7.6), 200 mM NaCl and 0.1% Tween-20)] for 1 h at room temperature (21 °C) with mixing. Immunoblotting was performed using a primary antibody in 10% blocking solution for 2 h at room temperature (21°C) with mixing. The membrane was washed five times for 5 min in 5% blocking solution on a shaker and then incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD, U.S.A.) at a 1:20000 dilution in 5% blocking solution for 1 h at room temperature with mixing. The membrane was then washed five times for 5min in 5% blocking solution and twice for 5 min in TBS/T (30 mM Tris base, pH 7.6, 200 mM NaCl and 0.1% Tween-20). The bound secondary antibody was detected using an Enhanced Chemiluminescence kit (Amersham Biosciences) and exposing the membrane to Biomax® MR film (Kodak). ATF4 antibody [CREB (cAMP-response-element-binding protein)-2, catalogue number sc-200], was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). A monoclonal antibody against ASNS was prepared as described [27]. To provide a demonstration of equal loading beyond the Fast Green staining, membranes were re-probed with a 1:5000 dilution of an antibody specific for actin (Sigma Chemical Co., St. Louis, MO, U.S.A.). The ATF4 quantification was performed using ImageQuant TL v2005 (Amersham Biosciences).
Transcriptional activity and mRNA determination
After treatment of HepG2 cells with Tg or Tu, total RNA was isolated using the Qiagen RNeasy kit (Qiagen). Processing of each sample included a DNase I treatment before final elution to eliminate any DNA contamination. To measure the transcriptional activity, primers derived from the ASNS intron 12 and exon 13 junction were used to measure the short-lived unspliced hnRNA (heterogeneous nuclear RNA) by qRT–PCR (quantitative reverse transcriptase–PCR), as described previously [28]. Reactions without reverse transcriptase were performed as a negative control to rule out amplification from any residual genomic DNA and these tests were always negative. The ASNS transcription activity primers for amplification were sense, 5′-CCTGCCATTTTAAGCCATTTTGC-3′ and anti-sense, 5′-TGGGCTGCATTTGCCATCATT-3′, and the primers for amplification of ASNS exon 7 (coding region) were sense, 5′-GCAGCTGAAAGAAGCCCAAGT-3′ and anti-sense, 5′-TGTCTTCCATGCCAATTGCA-3′. The ATF4 mRNA primers were sense, 5′-TGAAGGAGTTCGACTTGGATGCC-3′ and anti-sense, 5′-CAGAAGGTCATCTGGCATGGTTTC-3′. The reaction mixtures were incubated at 48 °C for 30 min followed by 95°C for 10 min to activate the Taq polymerase and amplification of 35 cycles of 95°C for 15 s and 63°C for 60 s. After PCR, melting curves were acquired by stepwise increase of the temperature from 55° to 95°C to ensure that a single product was amplified in the reaction. The housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a negative control for the drug-induced stress and as an indicator of the variation for the qRT–PCR analysis. The primers used to measure relative steady state mRNA levels for GAPDH were sense, 5′-TTGGTATCGTGGAAGGACTC-3′ and anti-sense, 5′-ACAGTCTTCTGGGTGGCAGT-3′. The ASNS hnRNA, ASNS mRNA and GAPDH mRNA values were determined relative to an RNA standard curve and are represented as fold-change relative to the zero time point (T=0). The PCR reactions were performed in duplicate for each sample, and samples were collected from at least three independent experiments. The data are expressed as means ± S.E.M.
ChIP
ChIP analysis was performed according to our previously published protocol [28]. To monitor binding to the NSRU enhancer site [21], the ASNS proximal promoter primers were sense, 5′-TGGTTGGTCCTCGCAGGCAT-3′ and anti-sense, 5′-CGCTTATACCGACCTGGCTCCT-3′. In addition to the anti-ATF4 mentioned above, antibodies for ChIP were purchased from Santa Cruz Biotechnology as follows: ATF3, sc-188; C/EBPβ, sc-150; RNA Pol II (polymerase II), sc-899; TFIID (transcription factor IIB) [TBP (TATA binding protein)], sc-204; TFIIB, sc-274; TFIIE-α, sc-237 and normal rabbit IgG, sc-2027.
siRNA transfection
The human XBP1, ATF6α and ATF6β (CREBL1) siGENOME SMARTpool (Dharmacon M-009552-02-0005; M-009917-00-0005; M-008805-00-0005), human ATF4 ON-TARGETplus SMARTpool (L-005125-00), siControl Non-Targeting siRNA (D-001210-02), and DharmaFECT 4 transfection reagent were purchased from Dharmacon (Lafayette, CO, U.S.A.). HepG2 cells were seeded in 6-well plates at a density of 4 × 105 cells per well in MEM and grown for 16 h. Transfection was performed according to Dharmacon’s instructions using 3 µl of DharmaFECT-4 plus 100 nM per well final siRNA concentration. After treatment of HepG2 cells with transfection reagent for 24 h, cells were incubated in fresh MEM for a second 24 h period. The medium was then removed and replaced with control MEM or MEM + Tg. Total RNA was isolated after 8 h and analysed by qRT–PCR. The mRNA primers were as follows: for ATF6α (sense, 5′-GGAACAGGATTCCAGGAGAATGAACCCTAGTG-3′; anti-sense, 5′-GATGTGTCCTGTGCCTCTTTAGCAGAAAATCC-3′), for ATF6β (sense, 5′-CTGAAGCGGCAGCAGCGAATGATCAAG-3′; anti-sense, 5′-CGAGCCTCCAGTCCCTGCAGATACTCTTTC-3′) and for XBP1 (sense, 5′-CAGAGTAGCAGCTCAGACTGCCAGAGATCG-3′; anti-sense, 5′-GCTGTTCCAGCTCACTCATTCGAGCC-3′)
RESULTS
ASNS transcriptional activity and steady state mRNA levels are up-regulated during ER stress
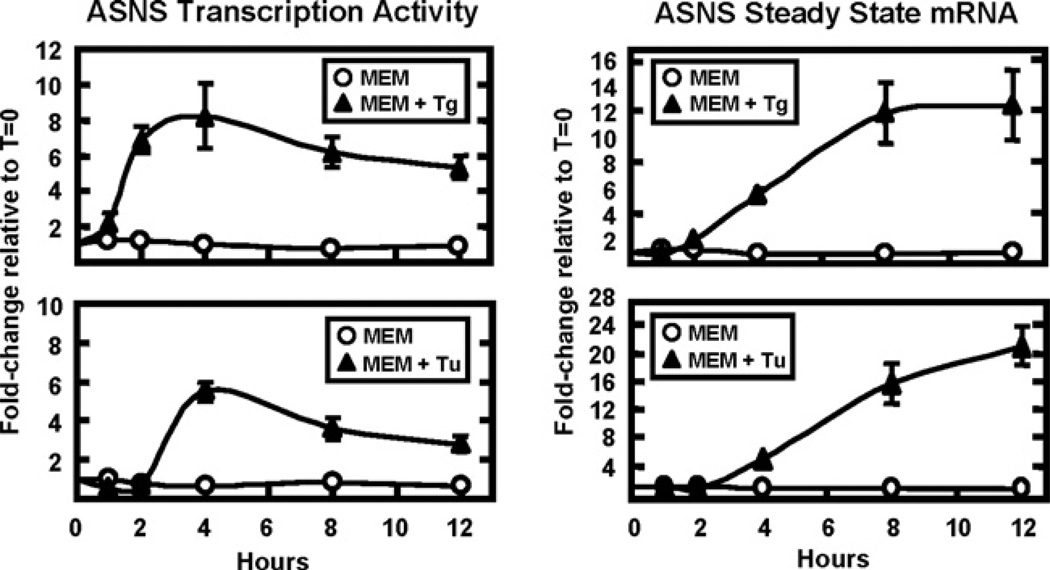
The response of the ASNS gene during UPR activation was investigated by analysing the transcription activity and steady state mRNA levels after treatment of HepG2 cells with the ER calcium ATPase blocker Tg or the N-glycosylation inhibitor Tu (Figure 1), both known and widely used UPR activators [3]. ASNS transcription activity during Tg treatment of HepG2 cells was analysed by specific primers that amplify the ASNS intron 12–exon 13 junction to measure the short-lived hnRNA levels [28]. In response to Tg treatment, the results revealed an increase in ASNS transcription activity starting within 1 h, peaking between 2 to 4 h, and then declining slightly at 8 h and 12 h (Figure 1). Consistent with the transcription activity, steady state ASNS mRNA levels lagged behind, reaching a plateau at 8–12 h. Similar results were obtained using Tu to initiate ER stress, although the time course was slightly delayed relative to Tg, presumably because of the difference in the mechanisms by which they trigger ER stress.
Figure 1. ASNS transcription activity and steady state mRNA are induced during Tg- or Tu-triggered ER stress.
Cultured HepG2 cells, treated for 0–12 h with control medium (MEM) or MEM containing 300 nM Tg (MEM+Tg, upper panels) or 5 µg/ml Tu (MEM+Tu, lower panels), were used to collect total RNA. Specific primers that amplify across the intron 12 – exon 13 junction of the ASNS gene were used to measure the hnRNA (ASNS transcription activity) (left panels), whereas primers that amplify a segment of ASNS exon 7 were used to assay the steady state mRNA levels (right panels). The data were collected from three independent experiments, the qRT–PCR for each sample was performed in duplicate, and the values shown represent the means ± S.E.M. and are plotted as the fold-change relative to T=0.
Transcription factor recruitment to the ASNS promoter during the UPR
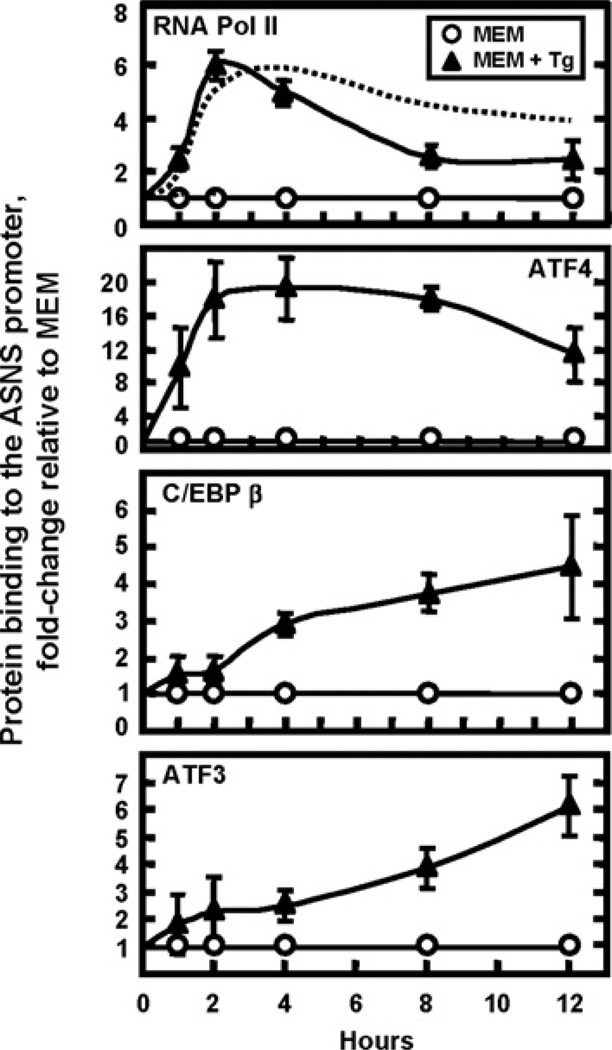
During amino acid limitation, ASNS transcription is controlled by ATF4, ATF3 and C/EBPβ by a self-regulating mechanism involving an initial period of activation by ATF4 and, subsequently, suppression of transcription by ATF3 and C/EBPβ [28]. Whether these same transcription factors mediate the ASNS transcription control during ER stress has not yet been established. Given that transcriptional activation of UPR target genes is mediated by three signalling cascades PERK/eIF2α/ATF4, ATF6 and IRE1/XBP1, investigating which of them contributes to the control of the ASNS gene represents an important question. ChIP analysis of the ASNS promoter, shown in Figure 2, revealed a time course of binding for RNA Pol II and ATF4 that paralleled the ASNS transcription activity (Figure 1 and dashed line in top panel of Figure 2). Upon UPR activation by Tg, RNA Pol II binding increased within 1 h, peaked at about 2 h, and then declined but remained elevated even at 12 h. The binding of ATF4 also occurred within 1 h of Tg treatment, peaked between 2 and 4 h, and slowly decreased at later time points (Figure 2). Following amino acid deprivation, ATF4 induction of C/EBPβ and ATF3 expression leads to a feedback mechanism by which these two factors are subsequently recruited to the NSRU and suppress the ATF4-enhanced ASNS transcription [28]. To determine if a similar mechanism was operational during the UPR, ChIP analysis of C/EBPβ and ATF3 was performed after Tg treatment (Figure 2). Delayed relative to the RNA Pol II and ATF4 binding, increased C/EBPβ binding began at 4 h and steadily increased at 8 h and 12 h. Likewise, relatively little ATF3 binding occurred at the early time points of 1–4 h, but increased recruitment was observed at 8 h and 12 h, at a time when the transcription activity was declining (Figure 1).
Figure 2. Profile of transcription factor binding at the ASNS proximal promoter during UPR activation.
ChIP analysis was performed on HepG2 cells treated for 0–12 h with control medium (MEM) or MEM containing Tg (MEM+Tg) to induce ER stress. Antibodies against RNA Pol II, ATF4, ATF3 and C/EBPβ were used for the immunoprecipitation step and primers specific for the ASNS promoter region were used for qRT–PCR amplification. Each PCR reaction was run in duplicate and the data were obtained from at least three independent experiments. The data are shown as fold-change relative to the MEM control and represent the means ± S.E.M. The dashed line in the top panel represents the ASNS transcription activity for the Tg-treated cells in Figure 1.
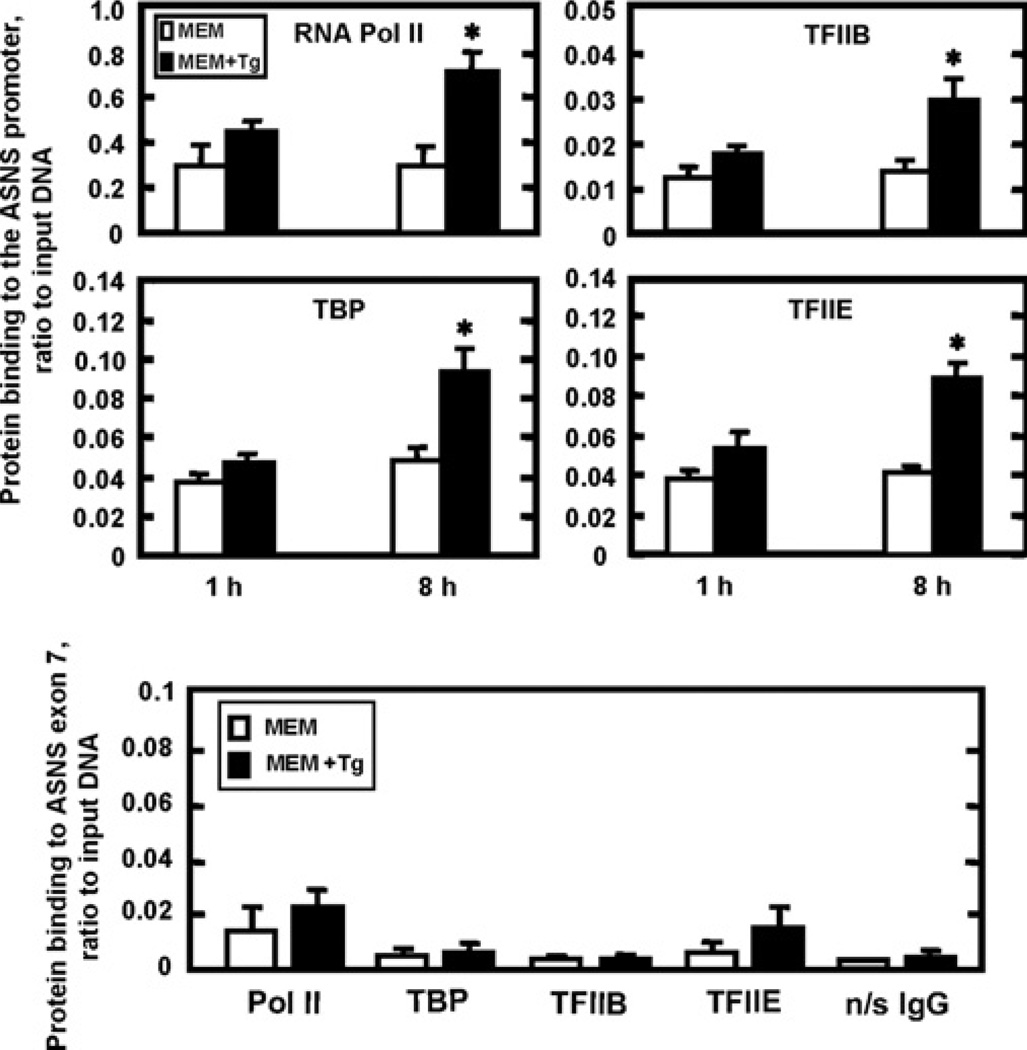
The preinitiation complex is comprised of GTFs (general transcription factors) that are responsible for the recruitment and correct positioning of RNA Pol II to the promoter [29]. Assembly of the preinitiation complex at the ASNS promoter during ER stress was investigated by ChIP analysis of representative GTFs in Tg-treated HepG2 cells (Figure 3). The results at 1 h showed enhanced recruitment for each of the factors tested, RNA Pol II, TBP, TFIIB and TFIIE, and the abundance was even greater at 8 h, consistent with the increased transcription activity (Figure 1). The background binding for the TBP, TFIIB and TFIIE was established by measuring association of these factors with the coding region (exon 7) of the ASNS gene (Figure 3).
Figure 3. ASNS promoter occupancy by general transcription factors is increased during ER stress.
HepG2 cells treated for 1 h or 8 h with control medium (MEM) or ME Mcontaining Tg(MEM+Tg) were analysed by ChIP for binding of RNA Pol II, TBP (TFIID), TFIIB and TFIIE to the ASNS promoter (upper panels). As a negative control (bottom panel), the same 8 h samples were used to amplify a downstream region of the ASNS gene (exon 7) to illustrate the background binding. The qRT–PCR reactions for each experiment were performed in duplicate and the data shown, as the ratio to DNA input, were collected from three independent experiments. The values represent the means ± S.E.M.; *P <0.05.
UPR activation does not trigger increased recruitment of Mediator subunits to the ASNS promoter
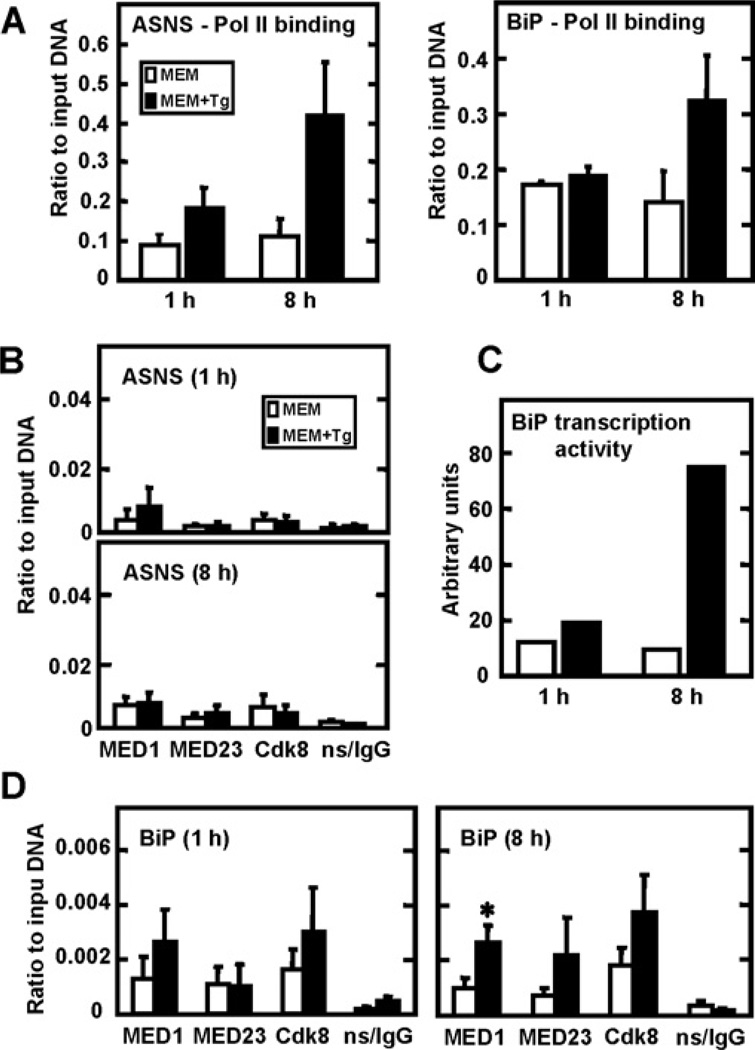
Mediator is large protein complex that has been proposed to be necessary for most, if not all, of Pol II-mediated transcription [30, 31]. However, several studies have shown that in Saccharomyces cerevisiae [32, 33] and Schizosaccharomyces pombe [34], Mediator recruitment does not always correlate with transcription activity or the recruitment of the GTF complex. Fan et al. [32] have suggested that Mediator might be selectively associated with genes that are activated by environmental stress or suboptimal growth conditions. Given that the ChIP data indicated an increase in recruitment of GTFs to the ASNS promoter region following UPR activation, the question of Mediator recruitment was also addressed. The RNA Pol II data (Figure 4A) are shown to demonstrate additional recruitment of the pre-initiation complex to the ASNS promoter during ER stress. ASNS (Figure 1, left hand panels) and BiP/GRP78 (Figure 4, Panel C) transcription activity is presented to demonstrate increased transcription from the promoters of these genes following UPR activation. The ChIP results shown in Figure 4 indicated that, despite the fact that some of the basal values obtained were above the background established by the non-specific IgG antibody, during ER stress there is no enhanced recruitment of the MED1, MED23 and CDK8 (cyclin-dependent kinase 8) Mediator subunits to the ASNS promoter at 1 h or 8 h (Figure 4B). In contrast to the ASNS promoter, ChIP analysis of the BiP/GRP78 promoter revealed a trend toward a greater degree of Mediator association after ER stress, occurring at 1 h for MED1 and CDK8 and at 8 h for all three subunits tested (Figure 4D). However, only the MED1 values at 8 h reached statistical significance.
Figure 4. ASNS transcriptional induction during ER stress does not involve enhanced recruitment of mediator subunits.
HepG2 cells incubated in MEM control medium only (white bars) or MEM+Tg (black bars) for 1 h and 8 h were used to perform ChIP analysis using antibodies specific for RNA Pol II (A), the MED1, MED23 or CDK8 Mediator subunits (B and D), or a non-specific rabbit IgG (ns/IgG), as indicated. During the final qRT–PCR step, primers specific for the ASNS promoter region or BiP/GRP78 were used (A, B and D). The transcriptional activity of BiP/GRP78 (C) was measured using primers that span the exon 2 – intron 2 junction. The qRT–PCR reactions were performed in duplicate for each sample. The ChIP data were collected from at least three independent experiments and the values represent the means ± S.E.M; *P <0.05.
Concurrent activation of the AAR and UPR pathways does not have an additive effect on the induction of ASNS
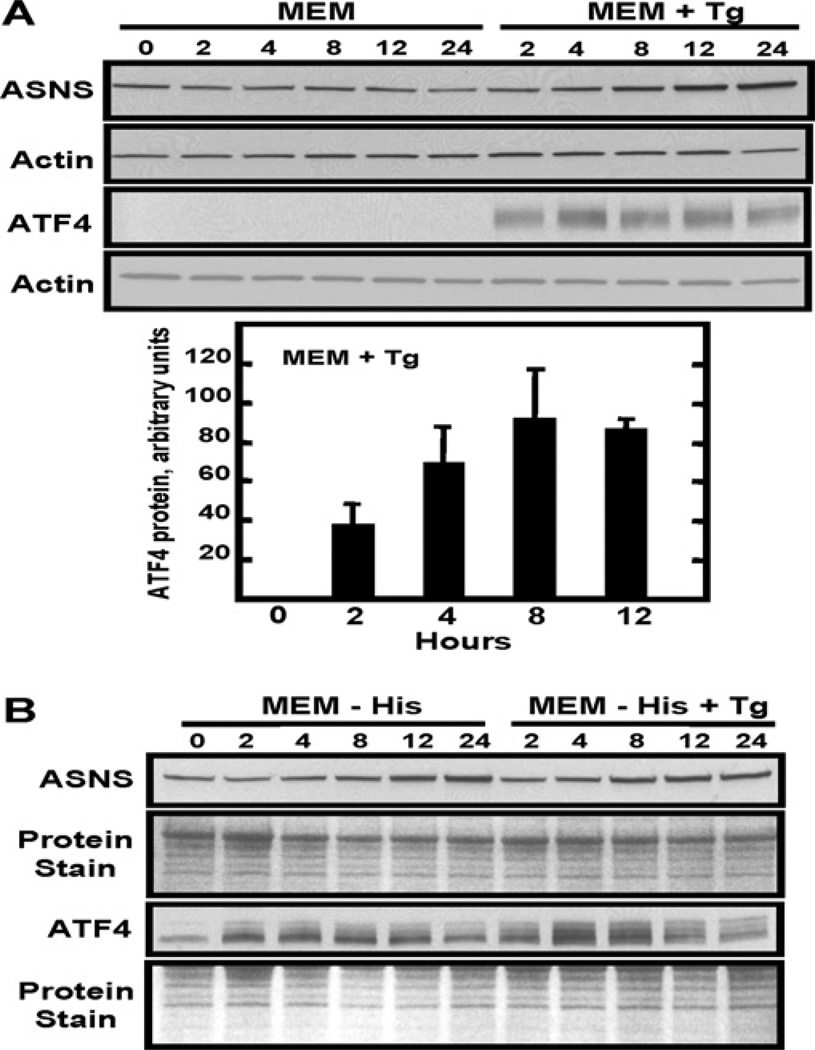
To measure the efficacy of ER stress on ATF4 protein abundance in HepG2 cells, immunoblot analysis was performed after Tg treatment (Figure 5A). Consistent with its prompt translational control and the relatively rapid binding to the ASNS promoter (Figure 2), increased ATF4 protein levels were observed within 2 h (Figure 5A). ATF4 expression levels further increased at 4 h and 8 h and remained relatively high during the later time points (Figure 5A). Depending on the resolution of the gel, ATF4 runs as a smear or a series of bands, which denote potential post-translational modifications of the protein. In contrast, ASNS protein content showed little or no change up to 4 h after which it increased (Figure 5A). This pattern of ASNS protein expression is consistent with the UPR model, which proposes an initial transient period of translational inhibition to lower the ER load, followed by a recovery phase characterized by an increase in protein synthesis to allow for expression of the UPR target proteins [35].
Figure 5. Induction of ASNS expression is not further enhanced by simultaneous activation of the AAR and UPR pathways.
Whole cell lysates were prepared for ASNS and ATF4 immunoblot analysis from HepG2 cells treated for 0–24 h with control medium (MEM) or MEM containing either Tg (A), medium lacking amino acid histidine (MEM−His) or the simultaneous treatment with MEM−His and Tg (B). ATF4 protein quantification during Tg treatment for 0–12 h was performed as described in the Materials and methods section and is presented in (A). As a measure of equal protein loading, the blots were stained with Fast Green or probed with an actin antibody. Antibodies against ATF4 and ASNS protein were used as described in the Materials and methods section. Each blot shown is representative of multiple experiments.
The AAR pathway and the PERK-mediated arm of the UPR both lead to an increase in ATF4 translation, whereas the XBP1 and ATF6 arms of the UPR activate transcription independent of ATF4 action [3]. To determine if either of the latter two UPR arms influences ASNS protein expression, the simultaneous activation of the AAR and UPR pathways in HepG2 cells was investigated. The concurrent AAR and UPR activation in HepG2 cells revealed an increase in both ATF4 and ASNS protein abundance that was similar to that after induction of either pathway alone (Figure 5B). This observation provides initial, but not conclusive evidence for the interpretation that the IRE1/XBP1 and ATF6 branches of the UPR do not participate in the induction of ASNS transcription. Furthermore, the lack of an additive effect by these two pathways is consistent with the known convergence of the UPR PERK arm and the AAR at the common step of eIF2α phosphorylation and subsequent increased ATF4 translation.
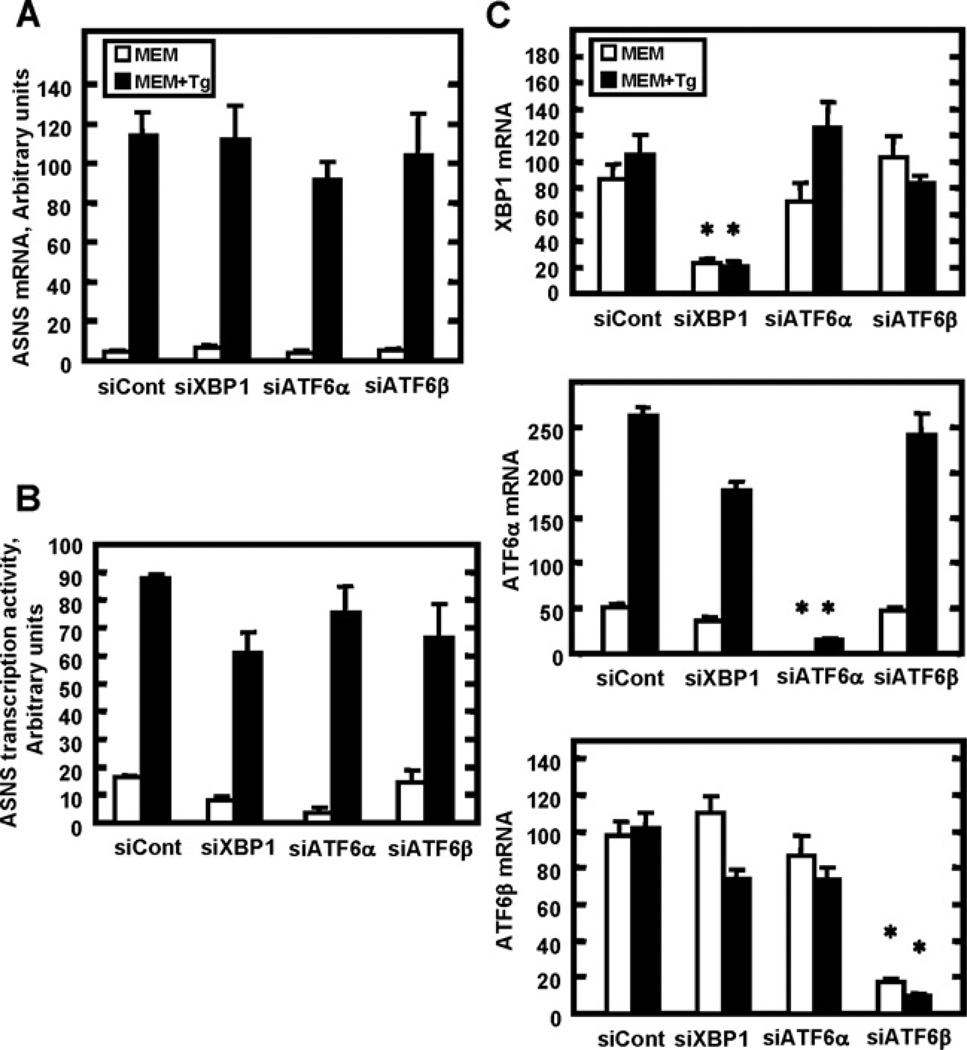
siRNAs against UPR effectors XBP1, ATF6α and ATF6β have minimal effect on ASNS transcription
To explore further whether or not the IRE1/XBP1 and ATF6 branches of the UPR participate in the induction of the ASNS gene by ER stress, an siRNA strategy was used to knockdown expression of XBP1, ATF6α and ATF6β (Figure 6). Despite significant reductions of XBP1, ATF6α or ATF6β expression (Figure 6C), there was no significant effect on the induction by ER stress of either ASNS steady state mRNA (Figure 6A) or ASNS transcription activity (Figure 6B). As an aside, it is noteworthy that ER stress of the HepG2 cells induced the ATF6α mRNA levels, whereas it did not affect those for XBP1 and ATF6β.
Figure 6. Knockdown of XBP1, ATF6α or ATF6β expression by siRNA does not affect induction of ASNS transcription during ER stress.
siRNA transfections with a non-targeting siRNA (siCont) or against XBP1, ATF6α or ATF6β were performed in HepG2 cells. After the first 24 h incubation in complete MEM, the medium was changed to fresh control MEM or MEM+Tg medium for 8 h to induce ER stress. Total RNA was isolated and ASNS steady state mRNA content (A), ASNS transcription activity (B) or XBP1, ATF6α and ATF6β mRNA (C) were assayed. The qRT–PCR reactions were performed in duplicate for each sample and samples were collected from three independent experiments. Values are expressed as means ± S.E.M.; *P <0.05 relative to the respective siControl value.
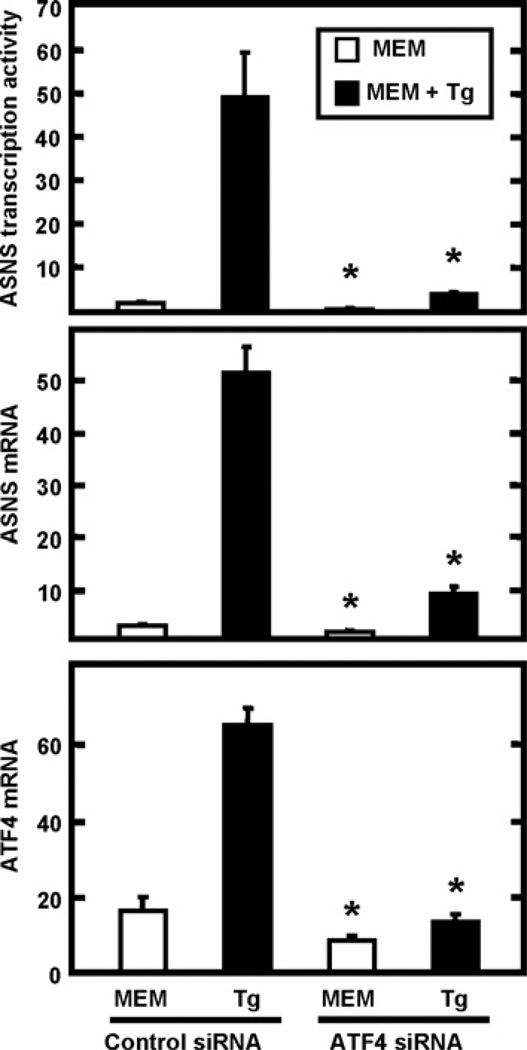
ASNS transcription up-regulation during ER stress is suppressed by the action of siRNA against ATF4
To address directly the role of the PERK/eIF2α/ATF4 arm of the UPR in the ASNS transcriptional up-regulation, an siRNA transfection against ATF4 was performed. The data presented in Figure 7 showed that ASNS expression (transcription activity and mRNA levels) was significantly decreased following knockdown of ATF4. These results confirmed that ATF4 action is required for the ASNS transcriptional induction during UPR activation. These data are consistent with the report of Cui et al. [36] who showed that siATF4 suppressed ASNS induction during ER stress generated by glucose deprivation. The knockdown effect of the siRNA transfection on ATF4 expression levels is presented in the bottom panel of Figure 7.
Figure 7. The effect of ATF4 knockdown on the expression of the ASNS gene.
HepG2 cells were transfected with either control siRNA or ATF4 siRNA. After transfection, cells were incubated in MEM or MEM+Tg for 8 h. Total RNA was isolated and subjected to qRT–PCR analysis of ASNS mRNA and ASNS transcriptional activity or ATF4 mRNA. The values represent the means ± S.E.M. of three independent experiments. *P <0.05, compared with the corresponding control.
DISCUSSION
Although previously published work had established that steady state ASNS mRNA content is induced by ER stress [37], the transcription activity of the endogenous gene, the individual factors responsible and the possible role of the IRE1/XBP1 and ATF6 arms of the UPR had not been investigated fully. Given that all three arms of the UPR are activated in a co-ordinated manner and contribute to the overall transcriptional response to ER stress, firmly establishing which signalling cascade or combination of them regulates the induction of ASNS was necessary. The present study demonstrates that the PERK/p-eIF2α/ATF4 signalling cascade is the only arm of the UPR that is responsible for ASNS transcriptional induction during ER stress of HepG2 human hepatoma cells. Indeed, neither the IRE1/XBP1 nor the ATF6 arms of the UPR appear to influence regulation of the ASNS gene. This interpretation is based on two independent approaches. First, activation of both the AAR and UPR pathways simultaneously was not additive with regard to ASNS induction. Second, knockdown of the IRE1/XBP1 and ATF6 arms of the UPR pathway did not affect the induction of ASNS transcription activity or steady state mRNA, whereas knockdown of ATF4 blocked the ASNS induction. The latter result also indicates that the IRE1/XBP1 and ATF6 arms do not substantially alter ASNS mRNA stability either. The data from the present study, obtained by siRNA treatment of human cells, agree with the observation that ASNS induction is intact in ATF6 and IRE1 knockout mouse embryonic fibroblasts [38, 39]. Furthermore, in vivo ChIP analysis revealed that the same three transcription factors, ATF4, C/EBPβ and ATF3, that have been identified as members of C/EBP-ATF enhanceosome during the ATF4-dependent AAR, are also associated with the ASNS NSRU during UPR activation.
The present results show that following ER stress the increase in ATF4 protein abundance is associated with increased ATF4 binding to the ASNS promoter. ChIP assays demonstrated RNA Pol II recruitment, which paralleled the increased transcription activity and ATF4 binding, consistent with the role of ATF4 as a potent activator of ASNS transcription. In contrast, an increase in C/EBPβ and ATF3 binding was observed at later time points when the ASNS transcription activity was declining. Therefore the NSRU enhancer binding proteins involved and the time course of their recruitment to the ASNS promoter region during ER stress is similar to that of ASNS transcription regulation during amino acid limitation [28]. These observations are consistent with the conclusion that the induction of ASNS by the UPR is purely ATF4 driven and does not involve the IRE1/XBP1 or ATF6 arms.
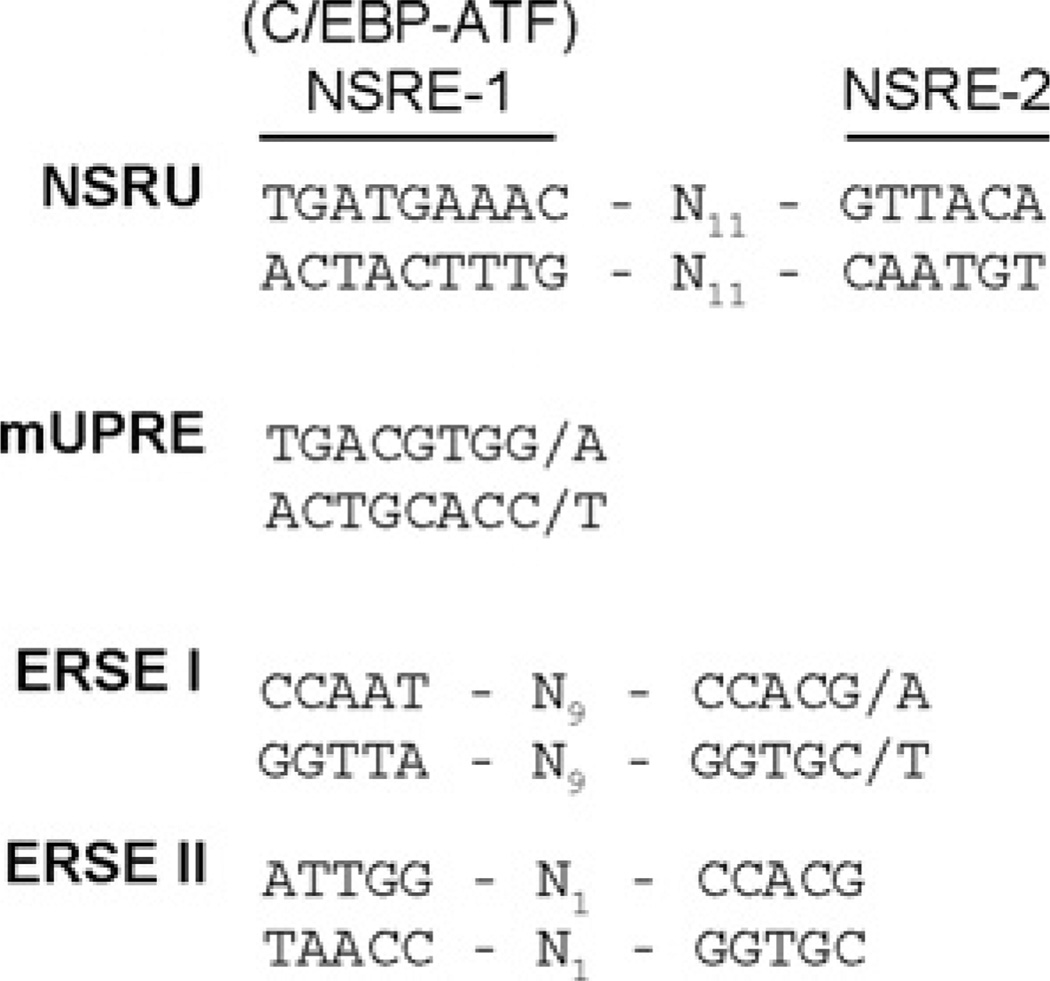
The transcriptional response for most of the ER stress target genes studied to date was shown to be mediated by ERSE-I or ERSE-II or the mUPRE [6], shown in Figure 8. For example, genes such as BiP and CHOP (C/EBP homology protein) contain an ERSE-I, whereas the Herp gene contains both an ERSE-I and an ERSE-II [40]. The mUPRE is composed of a single contiguous sequence (5′-TGACGTGG/A-3′) that binds XBP1 and is present in genes such as EDEM [41], HRD1 [41] and C/EBPβ [42]. In contrast, the NSRU within the ASNS promoter is composed of two sequences (NSRE-1 and NSRE-2) that are separated by a spacer of 11 nucleotides (Figure 8). Single nucleotide mutagenesis of the ASNS promoter demonstrated that both components of the NSRU are required to mediate the transcriptional response to either amino acid limitation or ER stress [21]. This requirement makes the ASNS gene unique thus far, in that no other gene has been discovered that responds to these two stress pathways through the same genomic elements. In contrast to the ASNS gene that requires both NSRE-1 and NSRE-2, the Cat-1 cationic amino acid transporter gene is induced by amino acid limitation through a sequence identical to NSRE-1 without the presence of an NSRE-2 like sequence [43]. Consistent with this lack of an NSRE-2 element, the Cat-1 NSRE-1 site does not mediate this gene’s response to the UPR [43]. Like all other known AAREs (AAR elements) [44], the ASNS NSRE-1 (5′-TGATGAAAC-3′) is a C/EBP–ATF composite site. C/EBP–ATF composite sites are sequences that bind heterodimers of ATF and C/EBP bZIP transcription factors [24–26]. The binding proteins for NSRE-2 (5′-GTTACA-3′) have yet to be determined. Interestingly, although NSRE-2 does not have the ability to mediate the ER stress signal when NSRE-1 is deleted or mutated in the ASNS promoter [21], the NSRE-2 sequence can confer ER stress responsiveness to an otherwise unresponsive AARE [45]. The CHOP gene is induced by both amino acid limitation and ER stress, but the CHOP promoter contains both a C/EBP–ATF composite site that acts as an AARE (nt −301 to −310) and a separate ERSE-I (nt −93 to −75) that mediates the UPR [12, 46]. Ma et al. [9] have presented evidence that the CHOP C/EBP–ATF site and the ERSE are both required for maximal induction during the UPR. Conversely, Jousse et al. [46] showed that a promoter deletion construct retaining the ERSE-I, but lacking the C/EBP-ATF site, exhibited a complete loss of the amino acid response and no reduction of UPR activation. Thus, in this circumstance, the C/EBP-ATF sequence appears to function primarily as an AARE. However, when the ASNS NSRE-2 sequence was placed downstream of the CHOP C/EBP-ATF sequence, the CHOP promoter gained UPR sensitivity [45].
Figure 8. Comparison of the NSRU, ERSE-I, ERSE-II and mUPRE sequences.
Collectively, the present observations extend our knowledge of genomic targets of the UPR and demonstrate that the ASNS NSRU genomic element is unique in that, independent of the well-known UPR-associated sequences of ERSE-I, ERSE-II and mUPRE, the NSRU is capable of mediating the transcriptional response to ER stress.
ACKNOWLEDGEMENTS
We thank Elizabeth Dudenhausen and other members of the Kilberg laboratory for technical advice and helpful discussion.
FUNDING
This research was supported by grants to M.S. K. from the Institute of Diabetes, Digestive and Kidney Diseases, the National Institutes of Health [grant numbers DK-59315, DK-52064].
Abbreviations used
- AAR
amino acid response
- AARE
AAR element
- ASNS
asparagine synthetase
- ATF
activating transcription factor
- BiP
immunoglobulin-heavy-chain-binding protein
- CDK
cyclin-dependent kinase
- C/EBP
CCAAT/enhancer-binding protein
- ChIP
chromatin immunoprecipitation
- CHOP
C/EBP homology protein
- CREB
cAMP-response-element-binding protein
- eIF
eukaryotic initiation factor
- ER
endoplasmic reticulum
- ERSE
ER stress element
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GCN2
general control non-derepressible
- GRP78
glucose regulated protein 78
- GTF
general transcription factor
- hnRNA
heterogeneous nuclear RNA
- IRE1
inositol-requiring kinase 1
- MEM
minimal essential medium
- NSRE
nutrient-sensing response element
- NRSU
nutrient sensing response unit
- p-eIF
phosphorylated eIF
- PERK
PKR (double-stranded RNA-activated protein kinase)-like ER kinase
- Pol II
polymerase II
- qRT–PCR
quantitative reverse transcriptase–PCR
- siRNA
small interfering RNA
- TBP
TATA binding protein
- TFII
transcription factor II
- Tg
thapsigargin
- Tu
tunicamycin
- UPR
unfolded protein response
- mUPRE
mammalian UPR element
- XBP1
X-box binding protein 1
REFERENCES
- 1.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 2.van Anken E, Braakman I. Endoplasmic reticulum stress and the making of a professional secretory cell. Crit. Rev. Biochem. Mol. Biol. 2005;40:269–283. doi: 10.1080/10409230500315352. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 4.Guo F, Cavener DR. The GCN2 eIF2 α kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Ron D. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 7.Romisch K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 8.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 10.Pirot P, Ortis F, Cnop M, Ma Y, Hendershot LM, Eizirik DL, Cardozo AK. Transcriptional regulation of the endoplasmic reticulum stress gene chop in pancreatic insulin-producing cells. Diabetes. 2007;56:1069–1077. doi: 10.2337/db06-1253. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 14.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameri K, Harris AL. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid. Redox. Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 18.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 19.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2 α kinase. Eur. J. Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 20.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbosa-Tessmann IP, Chen C, Zhong C, Siu F, Schuster SM, Nick HS, Kilberg MS. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 22.Zhong C, Chen C, Kilberg MS. Characterization of the nutrient sensing response unit in the human asparagine synthetase promoter. Biochem. J. 2003;372:603–609. doi: 10.1042/BJ20030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siu FY, Chen C, Zhong C, Kilberg MS. CCAAT/enhancer-binding protein beta (C/EBPb) is a mediator of the nutrient sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2001;276:48100–48107. doi: 10.1074/jbc.M109533200. [DOI] [PubMed] [Google Scholar]
- 25.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 26.Lopez AB, Wang C, Huang CC, Yaman I, Li Y, Chakravarty K, Johnson PF, Chiang CM, Snider MD, Wek RC, Hatzoglou M. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 2007;402:163–173. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng S, Moraga DA, Van Heeke G, Schuster SM. High-level expression of human asparagine synthetase and production of monoclonal antibodies for enzyme purification. Prot. Exp. Purif. 1992;3:337–346. doi: 10.1016/1046-5928(92)90010-t. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 29.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 30.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J. Biol. Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 32.Fan x, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 33.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 35.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, Higashino F, Hamuro J, Okada F, Kobayashi M, et al. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res. 2007;67:3345–3355. doi: 10.1158/0008-5472.CAN-06-2519. [DOI] [PubMed] [Google Scholar]
- 37.Barbosa-Tessmann IP, Chen C, Zhong C, Schuster SM, Nick HS, Kilberg MS. Activation of the unfolded protein response pathway induces human asparagine synthetase gene expression. J. Biol. Chem. 1999;274:31139–31144. doi: 10.1074/jbc.274.44.31139. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6 α and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 40.Ma Y, Hendershot LM. Herp is dually regulated by both the endoplasmic reticulum stress-specific branch of the unfolded protein response and a branch that is shared with other cellular stress pathways. J. Biol. Chem. 2004;279:13792–13799. doi: 10.1074/jbc.M313724200. [DOI] [PubMed] [Google Scholar]
- 41.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Dudenhausen EE, Pan Y, Zhong C, Kilberg MS. Human CCAAT/enhancer-binding protein β (C/EBP β) gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J. Biol. Chem. 2004;279:27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez J, Lopez AB, Wang C, Mishra R, Zhou L, Yaman I, Snider MD, Hatzolgou M. Transcriptional control of the arginine/lysine transporter, cat-1, by physiological stress. J. Biol. Chem. 2003;278:50000–50009. doi: 10.1074/jbc.M305903200. [DOI] [PubMed] [Google Scholar]
- 44.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: How mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruhat A, Averous J, Carraro V, Zhong C, Reimold AM, Kilberg MS, Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- 46.Jousse C, Bruhat A, Harding HP, Ferrara M, Ron D, Fafournoux P. Amino acid limitation regulates CHOP expression through a specific pathway independent of the unfolded protein response. FEBS Lett. 1999;448:211–216. doi: 10.1016/s0014-5793(99)00373-7. [DOI] [PubMed] [Google Scholar]