Abstract
It is textbook knowledge that inheritance of traits is governed by genetics, and that the epigenetic modifications an organism acquires are reset between generations. Recently, however, transgenerational epigenetic inheritance has emerged as a rapidly growing field, providing evidence suggesting that some epigenetic changes may result in persistent phenotypes across generations. Here, we survey some of the most recent examples of transgenerational epigenetic inheritance in animals, ranging from C. elegans to humans, and describe approaches and limitations to studying this phenomenon. We also review the current body of evidence implicating chromatin modifications and RNA molecules in mechanisms underlying this unconventional mode of inheritance and discuss its evolutionary implications.
Keywords: Transgenerational, epigenetic inheritance, chromatin, non-coding RNA, aging, fertility, metabolism, stress stimuli
Unconventional modes of inheritance
Heredity is overwhelmingly acknowledged to be governed by the laws of Gregor Mendel, with genes as the primary templates of inherited information. However, an increasing number of exceptions to the rules of genetic inheritance have been reported, suggesting that additional layers of information may also be transmitted. In the 1920s, inheritance of mating behavior in toads was reported by Paul Kammerer1, although this study remains controversial2. In the 1940s, Conrad Waddington coined the term ‘epigenetic’3 and described inheritance of wing patterns in response to heat shock in Drosophila4. Shortly after, Alexander Brink discovered an unconventional mode of inheritance for pigment biosynthesis in plants and termed it ‘paramutation’5. The discovery of parental imprinting in mammals in the 1980s provided the first indication that epigenetic modifications, such as DNA methylation, were not entirely erased between generations and could underlie transgenerational epigenetic inheritance6–11. It was also around this time that several cases of transgenerational epigenetic inheritance were reported in mice, both for exogenous transgenes and endogenous alleles affecting coat color12,13. Since these breakthrough reports, many more cases of non-genetic inheritance have been reported in species ranging from worms to mammals, suggesting that this phenomenon is more widespread than originally thought. In parallel, a growing body of work has revealed that exposure of parents to environmental stimuli could affect the phenotype of several generations of descendants, suggesting that exposure to environmental stimuli may also lead to changes in epigenetic modifications that are passed on to descendants via the gametes. Importantly, the field has recently made significant progress in garnering mechanistic insight into the processes underlying transgenerational epigenetic inheritance, by implicating specific chromatin modifications and non-coding RNA molecules.
In this review, we survey the most recent examples of epigenetic inheritance induced by genetic and environmental perturbations in animals (epigenetic inheritance in plants has been reviewed elsewhere14,15). This review also discusses the current state of progress in elucidating molecular mechanisms of this unconventional mode of inheritance, primarily focusing on chromatin and non-coding RNAs, as the role of DNA methylation has been extensively reviewed elsewhere16–21. We mostly focus on instances of non-genetic inheritance that persist for several generations (but that are not permanent), because persistence over multiple generations reduces confounding factors such as maternal effects or direct exposure of the gametes to toxins. We also discuss these important limitations to our understanding of the mechanisms underlying transgenerational epigenetic inheritance. Finally, we reflect on how this previously underappreciated mode of inheritance may affect evolutionary studies.
Epigenetic inheritance induced by alterations of the parental genome
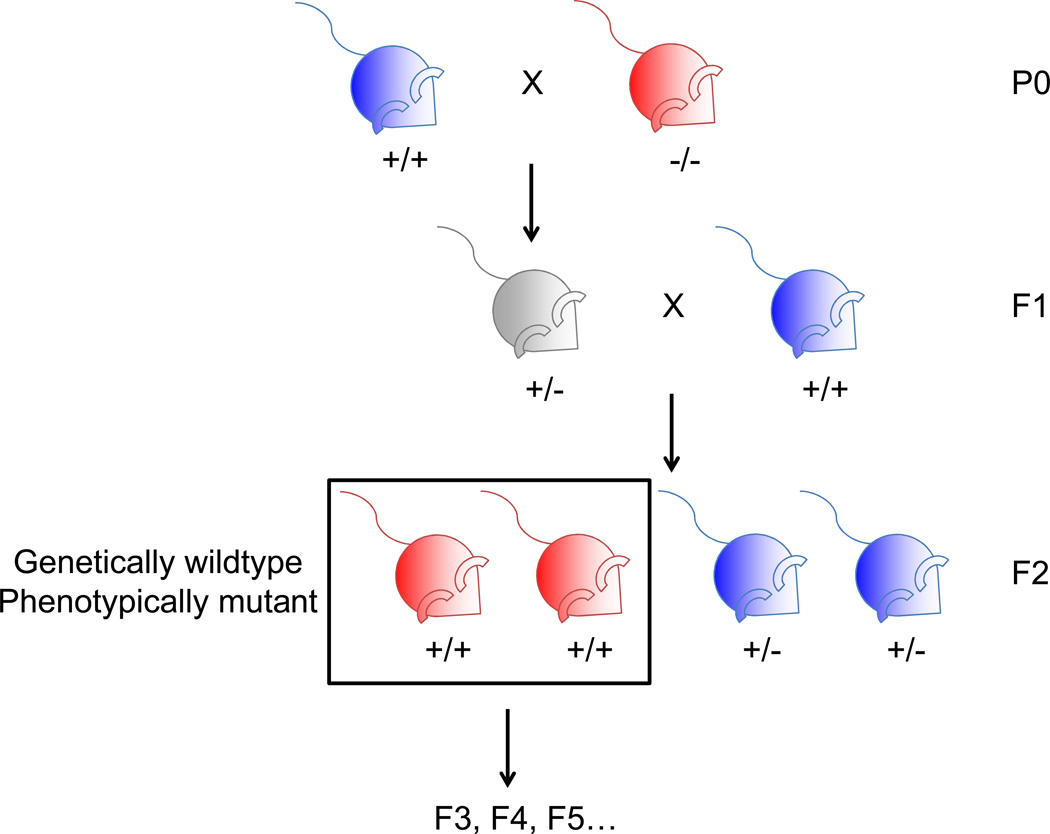
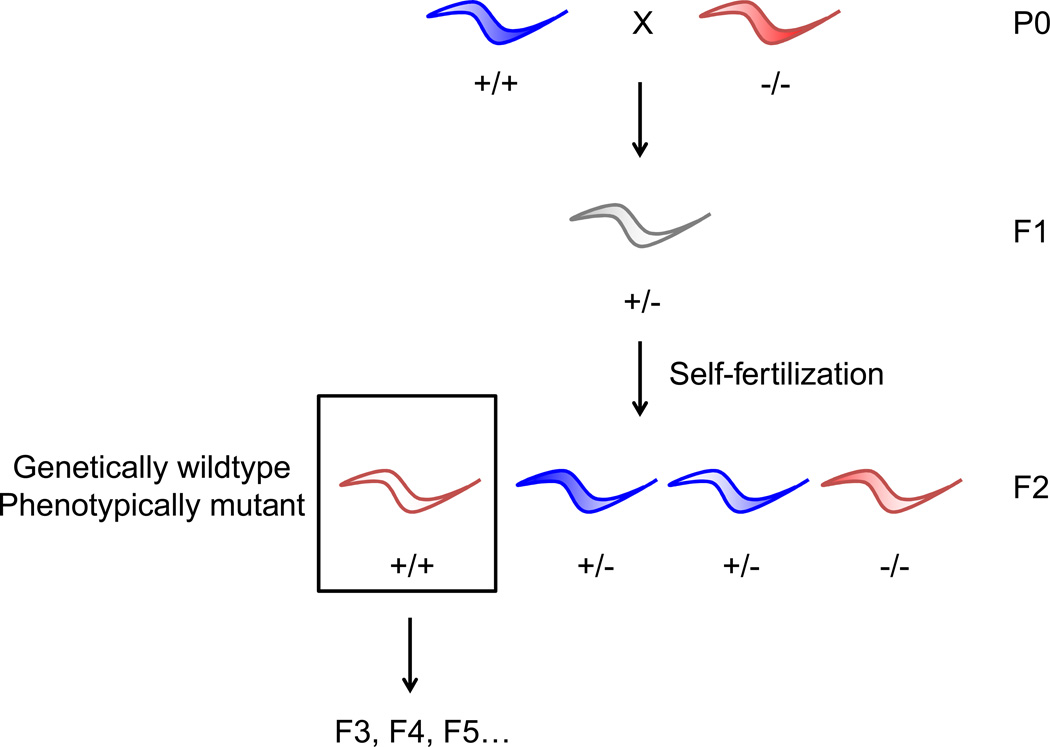
Studying wild type descendants of ancestors with mutations in specific genomic loci has revealed that in some cases, phenotypes characteristic of the mutant ancestors are still present in wild type descendants (Fig. 1). Alterations in the genome, for example transcriptional activation of a chromosomal element and retrotransposon insertion, were shown to lead to transgenerational epigenetic inheritance of Drosophila eye color22 and mouse coat color12, respectively.
Figure 1. Transgenerational epigenetic inheritance induced by genetic modification in ancestors.
A) Crossing scheme used in rodent studies to generate genetically wild type progeny from a mutant ancestor. Color of mouse denotes phenotype (blue are wild type and red are mutant). Note that in many instances presented in the review, the phenotype of the F1 heterozygotes is unknown.
B)Crossing scheme used in worm studies to generate genetically wild type progeny from a mutant ancestor. The F1 generation arises from mating between a hermaphrodite and male. The F2 generation arises from the self-fertilization of F1 hermaphrodites. Color of worm denotes phenotype (blue are wild type and red are mutant). Note that in many instances presented here, the phenotype of the F1 heterozygotes is unknown.
Transgenerational epigenetic inheritance of color and size in mice
Recently, additional examples of transgenerational epigenetic inheritance induced by alterations of the parental genome were described in mice, implicating RNA in this phenomenon23–25. In the first study, mice with an insertion of LacZ into the Kit gene gave rise to genetically wild-type offspring that still exhibited the tail and paw color phenotype characteristic of Kit mutants for at least two generations23. Genetically wild type descendants of ancestors that had the Kit mutant phenotype showed an altered pattern of Kit RNA expression, with RNA molecules of shorter size in brain and testis23. Microinjection of RNA from heterozygous mutant animals into one-cell embryos was sufficient to recapitulate the mutant phenotype in the following generation23. Collectively, these data suggest that disruption of a genomic locus in parents (in this case by LacZ insertion) may lead to abnormal RNA production in sperm, which may be transmitted in a transgenerational manner for at least two generations. However, the exact mechanisms underlying this epigenetic memory are still unclear.
The importance of RNA in epigenetic inheritance in mice was bolstered by two examples of transgenerational inheritance of cardiac hypertrophy and organismal growth24,25. Injection of fertilized eggs with RNAs and micro RNAs (miRNAs) targeting Cdk9, a regulator of cardiac growth, resulted in cardiac hypertrophy in the next generations24. Furthermore, injection of miR-124, a miRNA normally expressed in the brain and involved in central nervous system development, into fertilized eggs results in progeny that exhibit a 30% larger than normal body size25. This “giant” phenotype persisted until the F2 generation, after which the body size of the descendants reverted back to normal. These studies suggest that RNA-mediated epigenetic phenomena may underlie the inheritance of select phenotypes. Why some RNAs have a potential for transgenerational effects whereas others do not is still unclear. Non-genetic inheritance of traits such as organ and animal size may have crucial implications for the etiology of human hypertrophic cardiac myopathy and obesity.
Transgenerational epigenetic inheritance of fertility and longevity in C. elegans
Examples of transgenerational epigenetic inheritance induced by alterations of the parental genome were also recently described in the nematode C. elegans. C. elegans is a powerful model system for the study of epigenetic memory given its very rapid generation time and amenability for controlling many environmental and genetic variables. Transgenerational epigenetic inheritance of sterility and longevity have been reported, and involve similar histone modifications26,27. First, mutants of spr-5, the worm homolog of the LSD1/KDM1 histone 3 lysine 4 dimethyl (H3K4me2) demethylase, exhibit progressive decreased brood size beginning in the first generation and progressive progeny sterility starting around generation twenty26. The fertility of severely sterile generations was rescued by introducing one wild type copy of spr-5 for one generation26, indicating that this is sufficient to induce epigenetic resetting. Interestingly, accumulation of H3K4me2 and misregulation of spermatogenesis genes is observed in the primordial germ cells (PGC) of spr-5 mutants26. Indeed, it has been recently shown that H3K4me2 actually increases throughout the entire germline of spr-5 mutants28. Together, these results suggest that failure to reset H3K4me2 marks at select germline genes in the PGCs may result in progressive transgenerational sterility.
More recently, the first example of transgenerational epigenetic inheritance of longevity was described27. Genetically wild type descendants from ancestors mutant for members of the COMPASS H3K4 methylation complex display increased lifespan for up to three generations. The COMPASS complex is composed of ASH-2, WDR-5 and SET-2 in worms. This complex is conserved across species, and catalyzes the trimethylation of lysine 4 of histone H3 (H3K4me3)29, a chromatin modification commonly associated with actively transcribed genes30. Deficiency in COMPASS complex members extends lifespan in a manner that depends on the presence of a functional germline31. Genetically wild-type descendants from a cross between wild type males and hermaphrodites with mutations in the wdr-5 or set-2 genes were long-lived for up to three generations compared to descendants from pure wild type ancestors27. The long lifespan of the descendants is dependent on the H3K4me3 demethylase RBR-2 and requires a functioning germline27, which suggests that longevity of wild-type descendants from a cross between mutant ancestors and wild type worms is unlikely to be due to an extraneous mutation present in the initial strain. Transgenerational inheritance of longevity appeared to be relatively specific to members of the COMPASS complex, as manipulation of other longevity-promoting pathways such as the insulin signaling and mitochondrial pathways, or other chromatin regulators, did not show a transgenerational inheritance of long life27. These findings show that manipulation of specific chromatin modifiers in parents can have lasting effects on complex traits of offspring. It is interesting to note that both cases of transgenerational inheritance26,27 involve the regulation of H3K4 methylation in the germline, suggesting that this histone mark may be particularly important for the mechanism of epigenetic memory.
Together, these data describe instances where initial perturbation of an ancestor’s genome can result in transgenerational epigenetic inheritance of a variety of traits. These observations also raise the intriguing possibility that environmental stimuli that perturb RNA or chromatin states could also affect phenotypes of descendants for multiple generations. However, the prevalence and specificity of this epigenetic mode of inheritance is still unknown. Furthermore, the exact sequence and nature of events by which initial mutations in chromatin modifiers in ancestors lead to relatively persistent phenotype changes in wild-type descendants is not known, and will be further discussed below, in the mechanism section.
Epigenetic inheritance induced by metabolic changes in parents
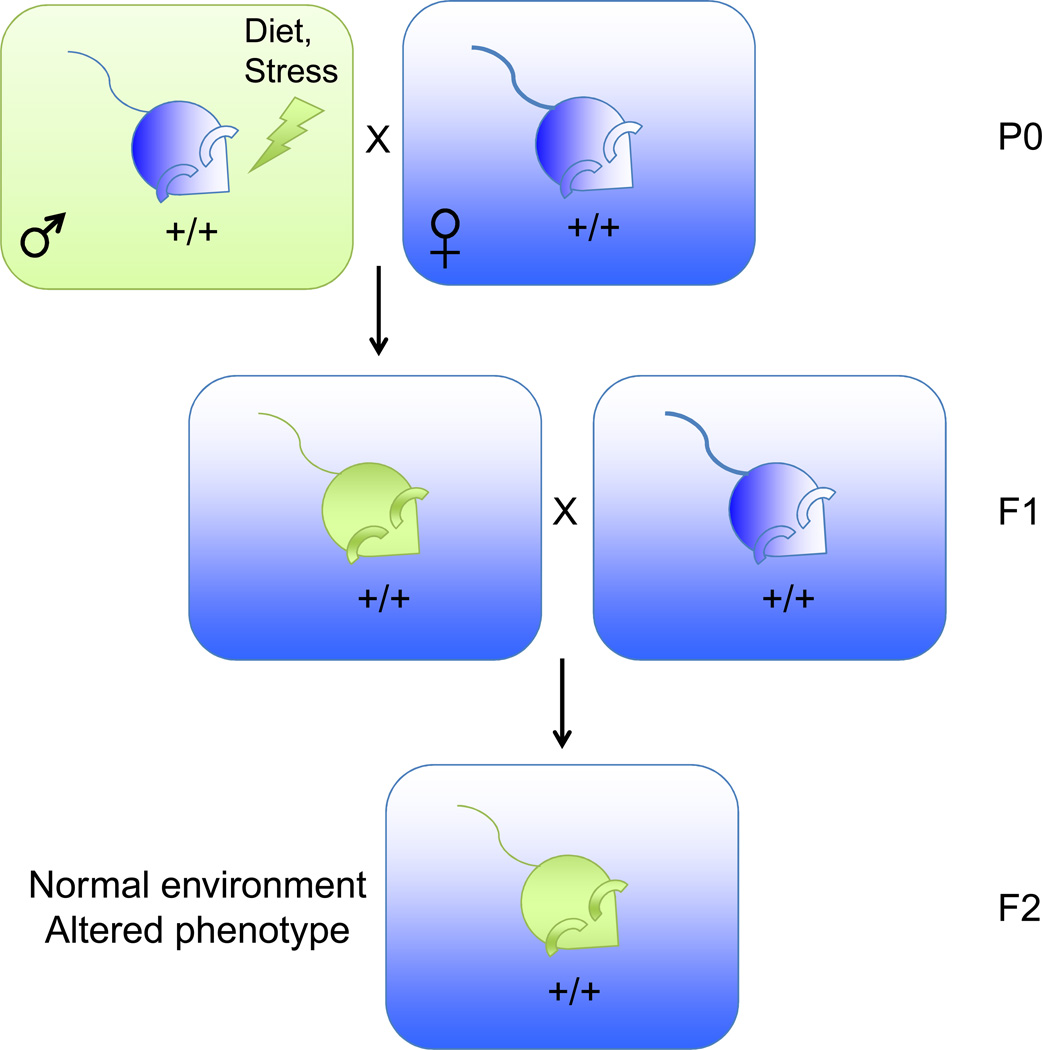
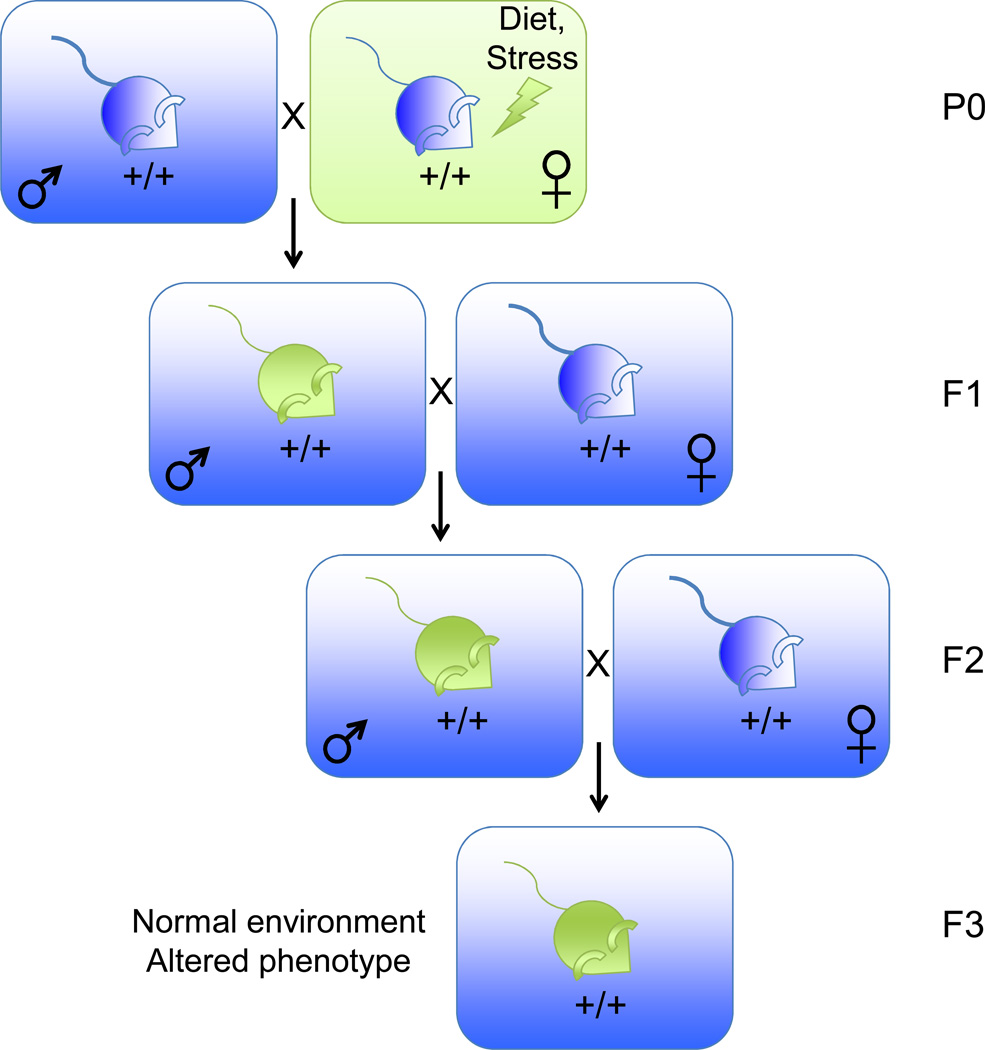
Interestingly, epigenetic inheritance can also be induced by environmental changes in parents. The past few years have seen a flurry of reports on the inheritance of acquired metabolic phenotypes resulting from over- or under- nutrition in parents. To avoid potential confounds of in utero and altered maternal caretaking, studies have investigated transgenerational inheritance of metabolic physiology through the paternal lineage (Fig. 2A) or the maternal lineage (Fig. 2B). In such schemes, the gametes of the P0 father or the F1 mother are directly exposed to the environmental stimulus, which may alter the genome in ways that could be transmitted to the F1 or F2 offspring, respectively. Thus, studying the F2 generation from males or F3 generation from females provides a way to distinguish direct exposure from epigenetic inheritance (Fig. 2).
Figure 2. Transgenerational epigenetic inheritance induced by environmental manipulations in ancestors.
A) Strategy used to generate progeny from a paternal ancestor subject to an environmental perturbation. This scheme minimizes maternal effects in organisms that require significant maternal care during development. Blue: normal phenotype. Green: altered phenotype resulting from a perturbation to the ancestral environment.
B)Scheme used to generate progeny from a maternal ancestor subject to an environmental perturbation. This scheme allows for the study of transgenerational effects induced by an initial change in maternal environment, but minimizes cryptic maternal effects. Blue: normal phenotype. Green: altered phenotype resulting from a perturbation to the ancestral environment.
Overfeeding
Exposure to a chronic high fat diet in rat fathers results in impaired insulin metabolism and pancreatic cell gene expression in female F1 offspring32. Female offspring from fathers fed a high fat diet mated with mothers fed a control diet exhibit an increase in blood glucose in the glucose tolerance test and a decrease in insulin secretion compared to offspring with both parents fed a control diet32. Microarray analysis of islet cells from female offspring of fathers fed a high fat diet uncovered genes involved in a range of pathways, including insulin and glucose metabolism as well as MAP kinase and Wnt signaling32. The greatest difference observed was an increase in expression of the gene encoding interleukin 13, Il13ra2, a cytokine involved in Jak-Stat signaling32. It will be interesting to understand the extent to which the dysfunctional β cell phenotype is mediated by differential expression of Il13ra2. It will also be important to test whether this phenotype can persist for more than one generation in order to distinguish direct exposure from bona fide transgenerational inheritance. Interestingly, another study reported that overfeeding of male mice, via culling of litter size, resulted in altered insulin and glucose metabolism of two subsequent generations of male offspring33. The observation that F2 male offspring of overfed fathers also exhibited altered metabolic phenotypes supports the notion that this phenomenon is mediated via an epigenetic mechanism rather than permanent damage to the genome induced by changes in metabolism.
The observation that metabolic traits can be inherited in a transgenerational epigenetic manner was further bolstered by studies of three generations of offspring from mothers fed a high fat diet34,35. Both the F1 and F2 generations inherited an increase in body length and a decrease in insulin sensitivity, through both maternal and paternal lineages34. Studying the inheritance of phenotypes to the F3 generation in this type of scheme is crucial to minimize the impacts of in utero exposure to environmental stimuli35. Only F3 females, but not males, inherited the increase in body length, and this was passed on only via the paternal lineage. These observations raise the possibility that imprinted genes are involved in this inheritance of body size in parents. Consistent with this possibility, paternally expressed genes showed a greater change in the magnitude of expression versus maternally expressed genes35.
It will be important to explore further the mechanisms of inheritance of overfeeding-induced phenotypes. One hypothesis is that the contents of sperm and seminal fluid (e.g., chromatin, RNA, metabolites) from fathers are influenced by diet and these changes are sufficient to be transferred to descendants, in some cases over several generations. Such changes may be mediated by relatively stable, yet reversible, molecular entities, and perhaps via amplification loops.
Dietary restriction
Dietary restriction has also been found to result in heritability of various metabolic phenotypes. For example, offspring of male parents fed a low-protein diet exhibit differential metabolic gene expression and changes in lipid metabolism36. Male mice fed a low protein diet and subsequently mated to naïve females gave rise to offspring of both sexes that exhibited increased expression of genes involved in fat and cholesterol biosynthesis as well as genes important for DNA replication36, corresponding to altered lipid metabolism and a hyperproliferative state, respectively. Metabolic analysis of the liver showed that offspring from males on a low protein diet had lowered hepatic cholesterol and other changes in lipid metabolism36. Whether these diet-induced phenotypes can be inherited for more than one generation is unknown. Importantly, this study investigated DNA methylation and histone modifications in sperm of males subjected to a low protein diet and showed that the epigenome of sperm can be altered by paternal diet36. However, which of these (or other) changes are causative of the transgenerational inheritance of metabolic phenotype remains an open question.
Human cohorts of food scarcity
The potential impact of ancestral diet on metabolic phenotypes of descendants has started to be examined in humans via two retrospective cohort studies, the Dutch Hunger Winter Families Study in the western Netherlands and the Overkalix cohort in Sweden. The Dutch Hunger Winter famine occurred at the end of World War II from October 1944 to May 1945, when official rations, consisting of mostly bread and potatoes, fell as low as 500 kcal/day. One study found that female, but not male, offspring from mothers exposed to famine during pregnancy displayed increased total cholesterol, increased triglycerides, and increased LDL cholesterol, compared to unexposed same sex siblings37. Interestingly, second-generation offspring exhibited higher neonatal adiposity and were subject to metabolic disease 1.8 times more frequently than unexposed controls38. Together, these studies suggest transgenerational effects of prenatal famine exposure on birth weight and metabolic disease rates. It would be important to study the F3 generation to assess more thoroughly transgenerational inheritance and to determine the propensity of these cohorts to develop age-related diseases, such as cardiovascular disease, cancer, diabetes, and hypertension.
The other cohort of humans in which differences in metabolism were studied transgenerationally is the Overkalix cohort, a three-generation group of families from northern Sweden in the late 1800s to the early 1900s who experienced a fluctuating food supply. Studies in this cohort revealed that low food intake during a critical period in early adolescence of grandparents correlated with an increase in survival of granddaughters, but not grandsons39. These results suggest that early adolescence is an important period for the response to the nutrient environment in ways that potentially affect future generations. This period could correspond to a critical window in gamete maturation in which metabolic environment may be an important determinant of gamete quality. However, a confound of the Overkalix cohort studies is that the nature of the environmental trigger of these survival phenotypes remains unclear, as the available information is on global food availability, not specific nutritional intake of parents.
Epigenetic inheritance of phenotypes induced by environmental stress
In Drosophila, heat shock and osmotic stress induce phenotypes, including wing changes and the disruption of heterochromatin, that are transgenerationally heritable for multiple generations4,40. The epigenetic inheritance of heat shock-induced chromatin disruption occurs via dATF-2 (drosophila activation transcription factor-2), a transcription factor that functions in heterochromatin nucleation41. Cellular stress induces the phosphorylation of dATF-2 and thus its release from chromatin, resulting in disrupted heterochromatin40. Stress-induction over several consecutive generations caused inheritance of a defective heterochromatin state over multiple generations of descendants, but the chromatin state eventually returned to normal. Thus, while the epigenome may be significantly changed upon environmental intervention, the resulting chromatin state retains the capacity to be reset once the insult is no longer present. This reversal also indicates that this stress induces bona fide epigenetic changes, rather than genetic mutations. This study also suggests that transgenerational epigenetic inheritance may be more evident upon a persistent, rather than temporary, environmental stimulus.
Epigenetic inheritance of RNAs derived from a viral sequence
A recent study in C. elegans reported transgenerational inheritance of small RNAs derived from a virus42. The authors generated a worm transgenic line expressing the Flock House virus, which results in the production of small-interfering RNAs derived from the virus (viRNAs). These viRNAs serve to silence the viral genome. Interestingly, the silencing effect of viRNAs is transmitted in an epigenetic manner to several generations of descendants42. The viRNAs themselves are transmitted in a manner that is independent of the template that generated them42. What is particularly intriguing is that both short-term (~3 generations) and long-term (>50 generations) silencing occurs, but only long-term silencing requires the RNA-dependent RNA polymerase rrf-1. It will be interesting to understand further the mechanisms underlying the difference between worms that lose inherited silencing of viRNAs early on and worms that exhibit stable silencing of viRNAs. One could speculate that in an organism with a very short life cycle, such as the worm, the epigenetic inheritance of a response to pathogens may have beneficial effects for the offspring.
Epigenetic inheritance of behaviors
Depressive-like behaviors
Exposure to prenatal psychological stress in mice appears to impact F2 generation offspring in an epigenetic manner43. F2 males from fathers that were exposed to prenatal stress exhibited a brain gene expression profile that was more similar to that of control females than control males (termed ‘dysmasculinization’)43. In particular, three miRNAs targeting β-glycan, a member of the TGFβ family known to regulate release of gonadal hormones, were significantly reduced in the brains of F2 males from fathers that were exposed to prenatal stress43. In addition, these males had decreased anogenital distance and testis weight, which is another characteristic of dysmaculinization43. The observation that prenatal stress of fathers results in dysmasculinization of male offspring indicates potential epigenetic transfer of neurodevelopmental phenotypes. It will be important to test if any of the observed phenotypes can be passed on to descendants via in vitro fertilization (IVF), which would test if the contents of sperm are sufficient to induce transgenerational inheritance, thereby implicating an epigenetic mode of transfer.
Studies examining how postnatal exposure to stress in parents affects the behavior and physiology of subsequent generations have reported heritable effects of chronic, unpredictable maternal separation and unpredictable maternal stress (MSUS) on mouse behavior44,45. Depressive-like behaviors were observed in two generations of offspring from mice exposed to MSUS during the first two weeks after birth44. Males, but not females, exposed to MSUS exhibited more depressive-like behaviors. They also consumed less sucrose, a model of anhedonia, or lack of enjoyment in normally rewarding tasks44. Similarly, maternal separation results in heritable increased risk-taking behavior, which is exacerbated when mothers are also subjected to unpredictable maternal stress45. Some behavioral changes were inherited via the male germline through multiple generations44, suggesting an epigenetic mode of inheritance. Consistently, the sperm of males subject to MSUS showed a significant increase in DNA methylation of the chromatin regulator methyl CpG binding protein 2 (MeCP2), along with a decrease in DNA methylation of the stress hormone receptor corticotropin releasing factor receptor 2 (CRFR2)44. It will be important to test the F3 generation to determine if the observed inheritance is truly transgenerational and not due to multigenerational exposure to stressful stimuli.
These findings suggest that unpredictable postnatal stress can lead to the inheritance of increased depressive-like behaviors, impaired social interactions, and increased risky or reckless behaviors. The presence in descendants of less anxious behaviors, decreased anhedonia, and decreased fear of aggressor-specific odors, however, points to the possibility that early stress could blunt subsequent normal responses to stress. Thus, it remains unclear whether these inherited behaviors play a beneficial or detrimental role in the animals’ life.
It is important to note that several confounding factors might affect the interpretation of behavioral experiments (Box 1). For example, mothers may not invest as much energy in raising offspring from males they perceive as deficient, a confound to epigenetic inheritance called ‘maternal provisioning’46. Using IVF with sperm from experimental fathers should help rule out such confounds. For example, offspring of socially defeated males mated with control females displayed increase depressive and anxiety-like behaviors47. However, when IVF with sperm from socially-defeated males was conducted, the only significantly altered behavioral phenotype observed in F1 offspring was a decreased latency to immobility in the forced swim test, indicating that very few or very subtle changes are passed on to offspring via the sperm epigenome alone.
BOX 1: Caveats underlying studies of transgenerational epigenetic inheritance.
Although many of the studies discussed in this review describe compelling and well-controlled examples of transgenerational epigenetic inheritance of acquired traits, it is important to consider the potential confounds that exist in the design and interpretation of such experiments.
In studies involving genetic crosses (Fig. 1), there is the possibility that the parents are not completely isogenic, thereby carrying extraneous mutations and potentially creating “hybrid vigor” effects. Using highly backcrossed strains can minimize these effects, but it is virtually impossible to eliminate differences in genetic backgrounds in ancestor strains. Moreover, initial changes in the ancestors’ genomes may affect chromosomal structure and result in defects in meiosis that could in turn affect the genotype or phenotype of subsequent generations. For example, meiotic silencing of gene expression by unpaired DNA has been observed in fungi, worms, and flies83. This potential confound has been ruled out in one study of transgenerational inheritance42, but it remains unclear whether it could affect others.
In cases of transgenerational inheritance induced by nutrient changes, environmental stress, or psychological stress (Fig. 2), it is possible that such stimuli may result in genetic mutations in the germline of parents that are subsequently passed on to offspring. Deciphering whether epigenetic marks are directly inherited or just downstream consequences of another genetic event will be instrumental in providing further insight into the mechanism underlying transgenerational epigenetic effects.
Other more complex confounds also exist. It is also possible that uncontrolled alterations in the microbiome, the organism’s intestinal flora, could underlie some of the observed non-genetic transmission of metabolic and other phenotypes84,85. Cryptic behavioral phenomena, including maternal provisioning, whereby mothers allocate more or less resources to progeny depending on the quality of their mate46, could also be a confound in the interpretation of epigenetic inheritance of behavioral or metabolic phenotypes.
Thus, although great care and experimental rigor is usually taken in studies of transgenerational epigenetic inheritance, it is important to note that confounding genetic and environmental caveats may be difficult to completely eliminate. The use of in vitro fertilization (IVF) could provide an alternative, and perhaps more direct, way of testing transgenerational inheritance. However, IVF itself also increases the risk of aberrant genomic imprinting86 and might bring a set of epigenetic issues of its own. Discovering the specific molecules involved in the mechanism of transgenerational inheritance, including DNA methylation, histone modifications, and non-coding RNAs, should be a fundamental step in identifying which instances of non-genetic inheritance are truly epigenetic.
Olfactory imprinting behavior
Although most studies on behaviors were performed in rodents and were done mostly in response to stressful stimuli, a recent study described the inheritance of an acquired olfactory imprinting behavior in C. elegans48. Olfactory imprinting is a process whereby exposure to an olfactory cue early in development affects the animal’s behavioral response, in this case positive chemotaxis, upon encountering this chemical in adulthood48. In worms, olfactory imprinting requires the presence of food48. Worms that have been imprinted not only exhibit a more robust ability to migrate toward the chemical, but also lay significantly more eggs, raising the possibility that olfactory imprinting may provide a memory of a favorable environment49. Strikingly, inducing imprinting over multiple consecutive generations of worms results in a stable inheritance of the chemotaxis behavior in subsequently naïve descendants, in some cases for over forty generations48. This study suggests that acquired behavioral plasticity can be inherited for many generations when ancestors were exposed to a persistent stimulus. One could speculate that specific mechanisms of transgenerational epigenetic inheritance may have been selected for, possibly to allow organisms with a short life cycle to pass on information to their offspring about previous favorable environments.
Molecular mechanisms of epigenetic inheritance
DNA methylation
DNA methylation can be transmitted across generations, for example in genomic imprinting, and could thus underlie transgenerational inheritance of specific traits16–19,21. Indeed, several studies discussed here have observed heritable changes in DNA methylation at specific loci, for example in response to a high fat diet and maternal and postnatal stress32,34,44. By contrast, other studies have found DNA methylation to be minimally affected, for example after maternal caloric restriction or a paternal low protein diet36,50. It is also noteworthy that worms display transgenerational epigenetic inheritance, yet do not appear to have classical DNA methylation enzymes. Thus, DNA methylation may play a direct mechanistic role in some, but not all, examples of transgenerational epigenetic inheritance. As DNA methylation has been extensively reviewed elsewhere16–19,21, this review will focus on other epigenetic mechanisms.
Histone modifications
Histone modifiers, in particular those affecting H3K4 methylation, have been implicated in transgenerational epigenetic inheritance of sterility and longevity in C. elegans26,27. In flies, environmental stress induces disruption of heterochromatin over several generations40. In mice, levels of trimethylated histone 3 lysine 27 (H3K27me3), a repressive histone mark, are lower at specific loci in the sperm of offspring from males fed a low protein diet36. Furthermore, chromatin remodelers have previously been identified in a dominant screen in mice associated with epigenetic reprogramming51. These observations suggest that histone modifications – and possibly chromatin states – may be central to mechanisms of transgenerational epigenetic inheritance in many organisms.
The exact sequence of events in which a change in histone marks could lead to transgenerational inheritance of traits is not yet known, but potential scenarios are emerging. The initial depletion of a histone mark (for example H3K4me3) may not be entirely replenished at specific loci in the next generations, perhaps due to an amplification loop involving other epigenetic mechanisms, or a blockage by antagonistic marks at these loci. For example, transgenerational inheritance of fertility or longevity due to deficiencies in H3K4me3 regulators such as the COMPASS complex (WDR-5, ASH-2, SET-2) or LSD-1 could be relayed by changes in H3K36me352. Indeed, the H3K36 methyltransferase MES-4 (maternal effect sterile-4) has been shown to be involved in transgenerational maintenance of chromatin states52,53. In early embryos, MES-4 is bound to genes previously expressed in the maternal germline46. MES-4 plays a role in the maintenance of H3K36me3 at germline genes in a manner that is largely independent of their transcriptional status52. Thus, an initial depletion in H3K4me3 might result in the depletion of MES-4 and/or H3K36m3 at germline genes, which may underlie transgenerational inheritance of longevity and fertility in C. elegans, in a transcription-independent manner.
Alternatively, the transcriptional status of specific genes may play a role in transgenerational epigenetic inheritance54. Chromatin modifications and histone variants established in the germline can be retained in mature gametes and even in the early embryo, specifically H3K4me2, a mark associated with active transcription, and H3.3, an alternative histone associated with memory of an active transcriptional state55. Heritable epigenetic information appears to be influenced by parental germline transcription54. Active gene expression in adult germ cells in the parent correlated with more robust expression of the same gene in the somatic cells of offspring54. In addition, active transcriptional states were found to be strongly inherited in Dictyostelium, and this phenomenon required H3K4 methylation56. Thus, transgenerational inheritance could be mediated by transcription-dependent mechanisms, in line with the observation that gene expression can also be altered in a transgenerational manner27,54.
It is also possible that depletion of an activating mark at specific loci may allow a repressive mark to expand at these loci. In turn, this repressive mark could itself be inherited, thereby preventing the replenishment of the activating mark over several generations. For example, the repressive mark H3K9me3 has been shown to be involved in transgenerational epigenetic inheritance of genomic loci targeted by non-coding RNAs in C. elegans57,58, and this could mediate the transgenerational inheritance due to changes in H3K4me326,27.
Environmental stimuli that trigger transgenerational phenotypes could also do so by modifying specific chromatin marks in egg and/or sperm. Even though most histones in sperm are replaced by protamines, the remaining histones and DNA still retain extensive epigenetic modifications59,60. In addition, protamines themselves are known to be modified post-translationally61. Protamine modifications could be influenced by the modification of the histones they are replacing, and vice versa, thereby providing a way of transferring epigenetic information at specific loci. Interestingly, genomic loci encoding genes that are crucial for early development, miRNA promoters, and imprinted gene clusters are enriched for specific epigenetic modifications, for example H3K27me3, in human sperm59. In mature human spermatozoa, genes with higher H3K27me3 at their transcriptional start sites are more likely to be repressed during gametogenesis and early embryogenesis60, suggesting that this mark may allow memory of a transcriptional state across generations. Because H3K27me3 at specific loci can be affected by paternal diet36, one could speculate that certain environmentally-induced chromatin modifications may be selectively retained across generations, leading to phenotypic changes in descendants.
Non-coding RNAs
Mounting evidence indicates that a variety of non-coding RNAs, including small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), and miRNAs, are involved in transgenerational epigenetic inheritance in worms, flies, and mice23–25,42,62–65. As non-coding RNAs can also influence epigenetic marks and chromatin states, it is tempting to speculate that these RNAs may also underlie instances of transgenerational inheritance that is initiated by changes in chromatin states.
The RNA interference machinery, chromatin states, and heritability
The RNA interference (RNAi) machinery appears to be important for both maintenance of chromatin state and heritability of non-coding RNAs. In the yeast S. pombe, the RNAi machinery is necessary for initiation and maintenance of centromeric heterochromatin, and double-stranded (ds)RNA-triggered release of RNA polymerase II (Pol II) at the replication fork induces heterochromatin spreading66,67. In C. elegans, several siRNAs are known to be inherited for several generations68–70. The RNAi machinery is necessary for the maintainance of heritable expression of dsRNAs and deposition of the heterochromatic mark histone 3 lysine 9 methylation (H3K9me)57,58. dsRNA accumulation precedes the appearance of the H3K9me3 chromatin mark57,58, and the spread of this repressive mark can extend for several kb around the trigger region targeted by dsRNAs58. The H3K9me3 mark can in turn be maintained for at least two generations in the absence of the initial dsRNA trigger58. Thus, small non-coding RNAs appear to be necessary for the deposition of repressive chromatin modification on genomic regions with sequence homology in C. elegans. An intriguing possibility is that small non-coding RNAs may induce transgenerational changes in selective gene expression by directing deposition of histone modifications in a sequence-specific manner.
The nuclear RNAi pathway appears to be particularly important for the transgenerational inheritance of small RNA and the maintenance of the chromatin state. Indeed, the gene heritable RNAi defective-1 (hrde-1) (also known as wago-9) was recently identified as a gene encoding a nuclear Argonaute involved in transgenerational inheritance of germline RNAi in worms64,65,71. In hrde-1 worm mutants, H3K9me3 at germline target genes is progressively lost over generations, which correlates with overexpression of target genes that are normally repressed in the germline64. Correspondingly, hrde-1 mutants become progressively sterile over generations64. Thus, the nuclear argonaute protein HRDE-1 is required to transmit epigenetic information to future generations in C. elegans. It will be interesting to determine whether nuclear argonaute proteins play a more general role in transgenerational inheritance of traits in different species.
piRNAs
In C. elegans, piRNAs can trigger stable, multi-generational silencing of transgenes62–65. This silencing activity involves imperfect base pairing of transcripts and is independent of the Piwi endonuclease slicer activity62,63. piRNA silencing of transcripts occurs in trans by triggering the secondary siRNA response62–65. Initiation of silencing requires the Piwi argonaute piwi-related gene prg-1, and maintenance of this response requires RNAi-related factors such as the germline-specific nuclear argonaute hrde-1/wago-9, nrde-2, rde-1, rde-3, rde-4, and mut-751,64. Additionally, maintenance of piRNA-induced silencing requires components of chromatin modifiers such as the HP1 ortholog HPL-251,64, and putative H3K9 methyltransferases SET-25 and SET-3264. Consistent with the involvement of H3K9 regulators, the transgene sequences silenced by piRNAs were enriched for H3K9me365. Thus, as was the case for siRNA inheritance57,58, these studies provide support for the interplay between non-coding RNA molecules and chromatin in establishing transgenerational epigenetic inheritance. It is tempting to speculate that this small RNA/chromatin pathway could underlie transgenerational inheritance of other traits that do not involve transgene silencing (for instance those induced by deficiencies in endogenous genes or environmental stresses).
In flies, piRNAs have also been shown to underlie transgenerational inheritance of germline gene silencing72,73. piRNAs can protect the germline against transposable elements (TEs) by triggering heterochromatin in these regions74. In flies, TE silencing is achieved via an amplification loop involving the generation of primary and secondary piRNAs73. Interestingly, the ability to produce secondary – but not primary– piRNAs in fly ovaries can be inherited in a transgenerational epigenetic manner72. Recently, clusters of transgenes have been found to induce strong trans-silencing effects on homologous clusters of transgenes and convert them into strong silencers themselves75. This transgenerational silencing of transgenes lasts for about 50 generations and is reminiscent of the plant ‘paramutation’ phenomenon14,15. Interestingly, this epigenetic phenomenon involves piRNAs that are maternally inherited through the cytoplasm75. Together, these results suggest that piRNAs are also involved in transgenerational inheritance in flies. As many different classes of piRNAs have been observed in different stages of mouse spermatogenesis76, piRNAs might play a role in epigenetic inheritance in mammals as well.
miRNAs
Injection of specific miRNAs into fertilized eggs in mice results in transgenerational inheritance of cardiac hypertrophy and large body size24,25. Furthermore, prenatal exposure to stress in male mice is associated with reduction of three miRNAs that target regulation of gonadal hormone release43. These results suggest that miRNAs may contribute to transgenerational inheritance of some traits. However, changes in miRNA expression do not correlate with opposing changes in the expression of potential RNA targets for these miRNAs in livers from offspring of fathers fed a low protein diet36. Thus, whether miRNAs are key mediators of epigenetic memory, or merely are susceptible to the heritable effect, remains to be determined.
Concluding remarks
Trangenerational epigenetic inheritance has been observed for a series of different phenotypes and can result from a variety of genetic or environmental manipulations in either parent. Many recent studies aimed at elucidating the mechanism of this unconventional mode of inheritance have identified promising candidates for mediating these lasting epigenetic effects, including histone modifications and non-coding RNAs. It is possible that different instances of transgenerational epigenetic inheritance have distinct underlying mechanisms. Alternatively, transgenerational inheritance may be mediated by a few major players involved in selective retention of key epigenomic molecules between generations.
Many important questions remain, including how epigenomic changes at key loci are maintained via the germline, how the duration of maintenance is determined, and how the marks or signals are eventually erased and reset. It would be interesting to explore how changes in the strength or duration of stimuli could affect the number of generations an epigenetically inherited phenotype lasts.
Transgenerational epigenetic inheritance has been observed for an increasing number of traits in several species. Thus, mechanisms of epigenetic inheritance are likely to have been selected for during evolution. The ability to pass on information about one’s environment to descendants could be evolutionarily advantageous. For example, the fact that some viral RNAs can be transmitted through the worm germline could provide viral immunity to several generations of progeny42, which might in turn help protect them against this virus. However, it is also possible that some cases of transgenerational epigenetic inheritance are not selected for, and only represent erroneous transmission of epigenetic marks, which would normally be erased and replenished.
Could transgenerational epigenetic inheritance itself increase an organism’s rate of evolution, or evolvability? Although evolvability is itself a controversial notion77, one could speculate that inheritance of epigenetic marks might affect chromatin states at specific loci, which could in turn make these regions more or less accessible to DNA repair enzymes, and thereby more or less prone to mutations78,79. Whether transgenerational epigenetic inheritance potentially impacts evolvability has never been tested, but model systems such as worms or flies would be particularly well-suited for these studies due to their short generation times.
Although genetic information is clearly the main mediator of inheritance, it is becoming increasingly evident that epigenetic mechanisms can also play a role in transmission of a number of traits. An important next step is to decipher the exact mechanisms of epigenetic inheritance, and identify the molecular entities that are either transmitted or reset between generations. It will be interesting to test whether the mechanisms of transgenerational inheritance are similar or different between organisms with very different generation time, such as C. elegans and mammals. It will also be important to test if other phenotypes, and possibly diseases, can be inherited in epigenetic manner. Indeed, epigenetic memory may fill some gaps in the “missing heritability” that human genome-wide association studies have been unable to reconcile for complex traits, such as longevity, and disorders, such as obesity and schizophrenia80,81.
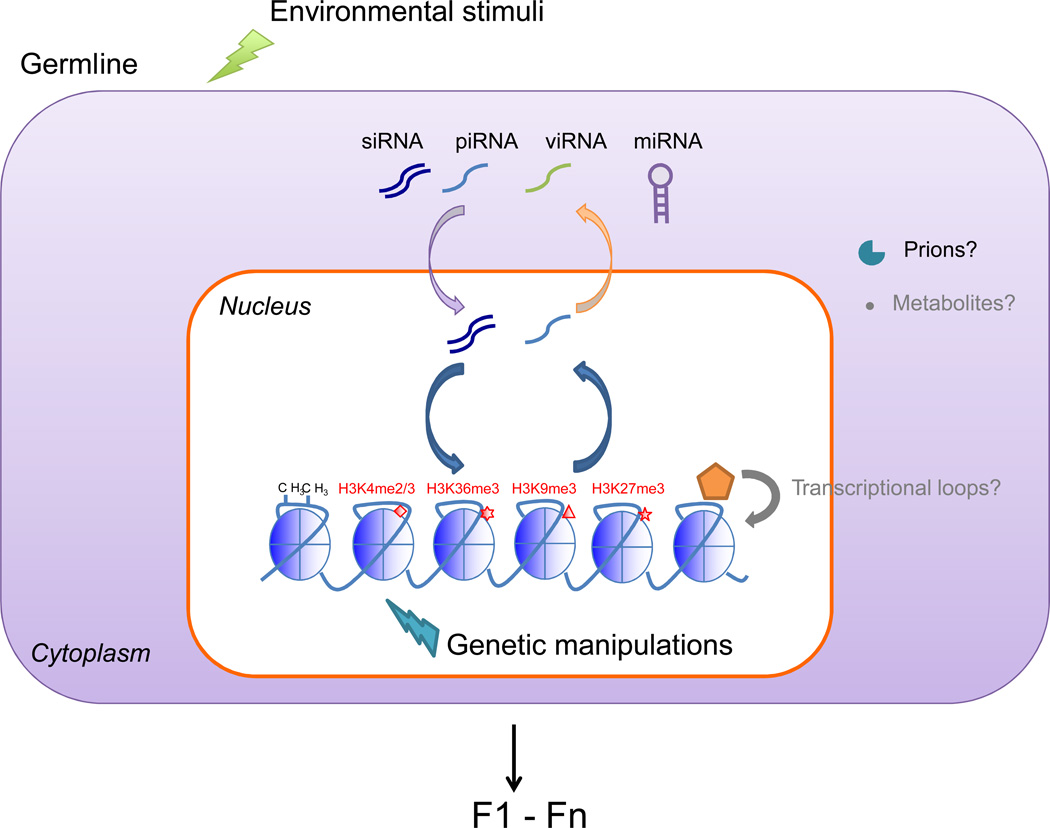
Figure 3. Model of proposed mechanisms of transgenerational epigenetic inheritance.
Alterations to the parental genome (aqua arrow) or environmental stimuli (green arrow) could trigger epigenetic changes in the germline of the organism that could be transmitted to the next generation. Such epigenetic changes could possibly be relayed or amplified in the germline of subsequent generations, and persist for several generations. Eventually, epigenetic changes would be reset to a basal state. So far, the epigenetic mechanisms that have been described include changes to chromatin (histone marks, DNA methylation) and changes to non-coding RNAs, in particular those involving the nuclear RNAi pathway. An amplification loop could be initiated by alteration in chromatin marks (e.g. H3K9me3) at a genomic locus, followed by the generation of non-coding RNAs at this particular locus, which would then be transmitted via the germline, and in turn guide H3K9me3 deposition at that same genomic locus in the germline of the next generation. In grey are additional potential mechanisms that remain to be investigated (Box 2).
Table 1.
Summary of molecular players involved in transgenerational epigenetic inheritance
| Epigenetic molecule | Species | References | ||
|---|---|---|---|---|
| Histone modifications | Active | H3K4me2/3 | Worm | 26, 27, 54 |
| H3K36me3 | Worm | 52, 53 | ||
| Repressive | H3K9me2/3 | Worm, Fly | 40, 58, 65, 57, 71 | |
| H3K27me3 | Mouse, Human | 36, 59, 60 | ||
| DNA methylation | Mouse, Rat | 32, 34, 44 | ||
| Non-coding RNAs | viRNA | Worm | 42 | |
| siRNA | Worm | 57, 58, 68, 69, 70, 71 | ||
| piRNA | Worm, Fly | 65, 62, 63, 64, 72, 75 | ||
| miRNA | Mouse | 23, 24, 25, 36, 43 | ||
| Prions | Yeast | 82 |
BOX 2: Other possible mechanisms for transgenerational inheritance: prions, metabolites, and transcriptional loops.
Although chromatin modifications and non-coding RNAs have received most of the attention for mediating transgenerational inheritance mechanisms, other non-genetic mechanisms could be at play, including prions, metabolites, and transcriptional loops.
Protein-based mechanisms could serve as an alternate means of transmitting adaptive phenotypes across generations87. Many wild yeast strains harbor a variety of proteins that behave as prions and confer adaptive phenotypes to the responses of different yeast strains to a range of environmental stresses such as osmotic, oxidative, pH, ethanol as well as stress from antifungal drugs and DNA damaging agents82. These observations raise the possibility that endogenous prions may be transmitted through meiosis and fertilization and perhaps underlie modes of transgenerational inheritance.
Another potential mechanism for transgenerational epigenetic inheritance that remains to be explored is the possibility of metabolites or other small molecules serving as the transmitted signal. Changes in metabolite levels can affect the activity of various chromatin-modifying enzymes88,89, which could result in altered chromatin states that would then be inherited over generations. Alternatively, small amounts of metabolites present in the cytoplasm of oocytes could be directly transmitted to the zygote, which could either interact with chromatin or affect cellular physiology directly. This model would require the existence of bioenergetic loops that, when triggered, would be capable of changing metabolite profiles for several generations before decaying.
Finally, it is also possible that the mechanisms underlying transgenerational inheritance involve altered transcriptional feedback loops that do not directly require any histone modifications or other epigenetic players90. Such self-propagating gene regulation governed by transcriptional machinery has been observed in the fungus Candida albicans91. In this example, the changes in cellular state occurred stochastically, but it is possible that in other instances, an extracellular signal could activate a transcription factor that would upregulate transcription of its own gene. These feedback loops, once activated, could cause changes in gene expression that persist in the absence of the original activator. For example, an initial genomic mutation in the ancestor could cause a change in the DNA binding affinity of a transcription factor, which could in turn initiate a transcriptional loop. Although most mechanistic work to date has largely focused on chromatin and nucleic acid-based mechanisms, it is important to keep in mind that others may exist and play a role in certain cases of transgenerational inheritance.
Acknowledgements
We would like to thank E. Greer, S. Han, B. Benayoun, and D. Valenzano for critical reading of this manuscript, and G. Sherlock and A. Sidow for advice on potential evolutionary implications. Supported by NIH DP1-AG044848, NIH R01-AG031198, and the Glenn Laboratories for the Biology of Aging at Stanford University. J.P.L. is supported by NIH T32-MH020016 and a NSF Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no financial conflicts of interest.
References
- 1.Kammerer P. The inheritance of acquired characteristics. New York: Boni and Liveright; 1924. [Google Scholar]
- 2.Vargas AO. Did Paul Kammerer discover epigenetic inheritance? A modern look at the controversial midwife toad experiments. J Exp Zool B Mol Dev Evol. 2009;312:667–678. doi: 10.1002/jez.b.21319. [DOI] [PubMed] [Google Scholar]
- 3.Waddington CH. The epigenotype. Endeavor. 1942;1:18–20. [Google Scholar]
- 4.Waddington CH. Genetic Assimilation of an Acquired Character. Evolution. 1953;7:118–126. [Google Scholar]
- 5.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadchouel M, et al. Maternal inhibition of hepatitis B surface antigen gene expression in transgenic mice correlates with de novo methylation. Nature. 1987;329:454–456. doi: 10.1038/329454a0. [DOI] [PubMed] [Google Scholar]
- 7.Swain JL, et al. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- 8.Sapienza C, et al. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987;328:251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- 9.Kearns M, et al. Complex patterns of inheritance of an imprinted murine transgene suggest incomplete germline erasure. Nucleic Acids Res. 2000;28:3301–3309. doi: 10.1093/nar/28.17.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland HG, et al. Reactivation of heritably silenced gene expression in mice. Mamm Genome. 2000;11:347–355. doi: 10.1007/s003350010066. [DOI] [PubMed] [Google Scholar]
- 11.Reik W, et al. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 12.Morgan HD, et al. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 13.Rakyan VK, et al. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 14.Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollick JB. Paramutation and development. Annu Rev Cell Dev Biol. 2010;26:557–579. doi: 10.1146/annurev.cellbio.042308.113400. [DOI] [PubMed] [Google Scholar]
- 16.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 17.Feng S, et al. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: evidence for the impact of adverse environmental effects. Neurobiol Dis. 2010;39:61–65. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol Cell Endocrinol. 2012;354:3–8. doi: 10.1016/j.mce.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reik W, et al. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 21.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 22.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 23.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 24.Wagner KD, et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Grandjean V, et al. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 26.Katz DJ, et al. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nottke AC, et al. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci U S A. 2011;108:12805–12810. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng SF, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 33.Pentinat T, et al. Transgenerational inheritance of glucose intolerance in a mouse model of neonatal overnutrition. Endocrinology. 2010;151:5617–5623. doi: 10.1210/en.2010-0684. [DOI] [PubMed] [Google Scholar]
- 34.Dunn GA, Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150:4999–5009. doi: 10.1210/en.2009-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn GA, Bale TL. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011;152:2228–2236. doi: 10.1210/en.2010-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumey LH, et al. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am J Clin Nutr. 2009;89:1737–1743. doi: 10.3945/ajcn.2008.27038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Painter RC, et al. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. Bjog. 2008;115:1243–1249. doi: 10.1111/j.1471-0528.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 39.Pembrey ME, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 40.Seong KH, et al. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 41.Jia S, et al. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 42.Rechavi O, et al. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31:11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franklin TB, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 45.Weiss IC, et al. Inheritable effect of unpredictable maternal separation on behavioral responses in mice. Front Behav Neurosci. 2011;5:3. doi: 10.3389/fnbeh.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pryke SR, Griffith SC. Genetic incompatibility drives sex allocation and maternal investment in a polymorphic finch. Science. 2009;323:1605–1607. doi: 10.1126/science.1168928. [DOI] [PubMed] [Google Scholar]
- 47.Dietz DM, et al. Paternal transmission of stress-induced pathologies. Biol Psychiatry. 2011;70:408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remy JJ. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr Biol. 2010;20:R877–R878. doi: 10.1016/j.cub.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Remy JJ, Hobert O. An interneuronal chemoreceptor required for olfactory imprinting in C. elegans. Science. 2005;309:787–790. doi: 10.1126/science.1114209. [DOI] [PubMed] [Google Scholar]
- 50.Jimenez-Chillaron JC, et al. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chong S, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39:614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arico JK, et al. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 2011;7:e1001391. doi: 10.1371/journal.pgen.1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 56.Muramoto T, et al. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 57.Burton NO, et al. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 61.Caron C, et al. How to pack the genome for a safe trip. Prog Mol Subcell Biol. 2005;38:65–89. doi: 10.1007/3-540-27310-7_3. [DOI] [PubMed] [Google Scholar]
- 62.Bagijn MP, et al. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee HC, et al. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashe A, et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shirayama M, et al. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 67.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grishok A, et al. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 69.Vastenhouw NL, et al. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 70.Alcazar RM, et al. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grentzinger T, et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–1888. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aravin AA, et al. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 74.Rangan P, et al. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Vanssay A, et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 76.Gan H, et al. piRNA profiling during specific stages of mouse spermatogenesis. Rna. 2011;17:1191–1203. doi: 10.1261/rna.2648411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colegrave N, Collins S. Experimental evolution: experimental evolution and evolvability. Heredity (Edinb) 2008;100:464–470. doi: 10.1038/sj.hdy.6801095. [DOI] [PubMed] [Google Scholar]
- 78.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. Embo J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buard J, et al. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. Embo J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson VR, Nadeau JH. Transgenerational genetic effects. Epigenomics. 2010;2:797–806. doi: 10.2217/epi.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 82.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 84.Rosenberg E, et al. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol. 2009;11:2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 85.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 86.Cortessis VK. Imprinting Errors and IVF. Biennial Review of Infertility. 2009;1:239–246. [Google Scholar]
- 87.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 88.Ladurner AG. Rheostat control of gene expression by metabolites. Mol Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 89.Teperino R, et al. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ptashne M. On the use of the word 'epigenetic'. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 91.Zordan RE, et al. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]