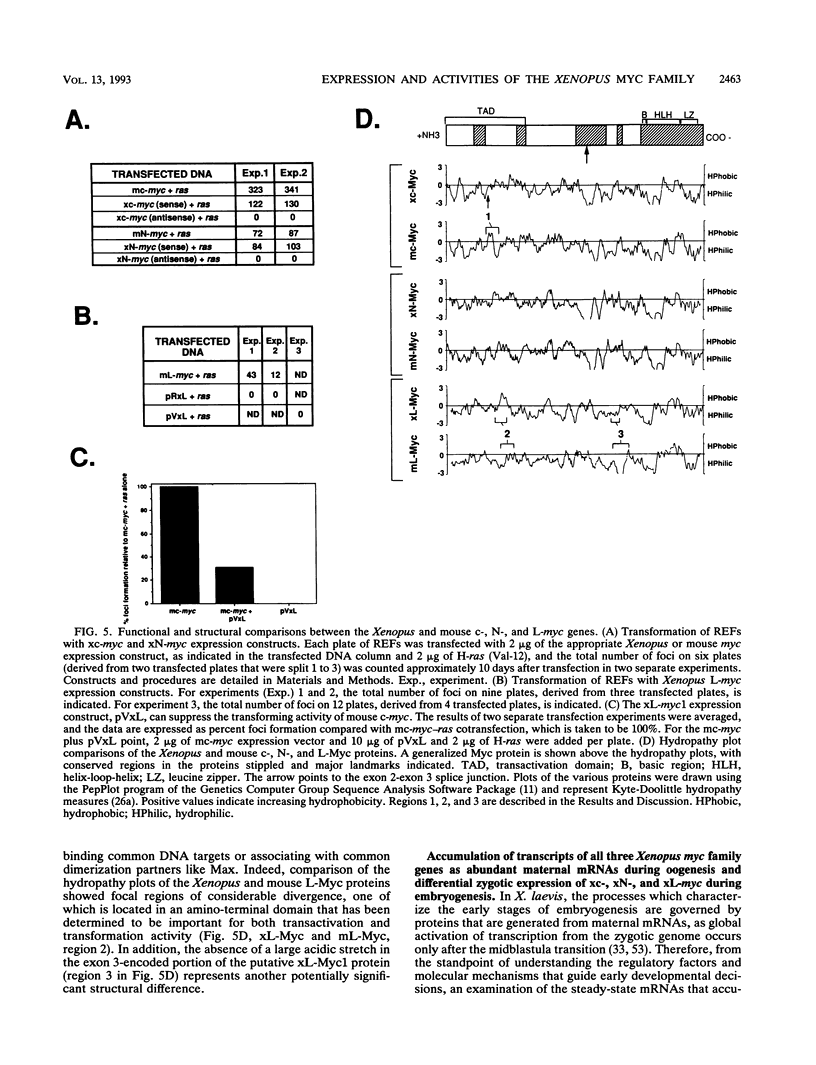
Abstract
A polymerase chain reaction-based cloning strategy allowed for the isolation of two distinct Xenopus L-myc genes, as well as previously isolated xc- and xN-myc genes, thus demonstrating that these three well-defined members of the mammalian myc gene family are present in lower vertebrates as well. Comparison of the Xenopus and mammalian Myc families revealed a high degree of structural relatedness at the gene and protein levels; this homology was consistent with the ability of the xc-myc1 and xN-myc1 genes to function as oncogenes in primary mammalian cells. In contrast, the xL-myc1 gene was found to be incapable of transforming rat embryo fibroblast cells, and this inactivity may relate to localized but significant differences in its putative transactivation domain. Analysis of xc-, xN-, and xL-myc gene expression demonstrated that (i) all three genes were highly expressed during oogenesis and their transcripts accumulated as abundant maternal mRNAs, (ii) each gene exhibited a distinctive pattern of expression during embryogenesis and in adult tissues, and (iii) the xL-myc1 and xL-myc2 genes were coordinately expressed in the maternal and zygotic genomes. The markedly high expression of the Xenopus myc gene family in differentiated tissues, such as the central nervous system and kidney, contrasts sharply with the low levels observed in mammalian adult tissues. These differences may reflect unique functions of the Myc family proteins in processes specific to amphibians, such as tissue regeneration.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson P., Goldfarb M. P., Weinberg R. A. A defined subgenomic fragment of in vitro synthesized Moloney sarcoma virus DNA can induce cell transformation upon transfection. Cell. 1979 Jan;16(1):63–75. doi: 10.1016/0092-8674(79)90188-0. [DOI] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Birrer M. J., Segal S., DeGreve J. S., Kaye F., Sausville E. A., Minna J. D. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol Cell Biol. 1988 Jun;8(6):2668–2673. doi: 10.1128/mcb.8.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbee C. A., Baker M. A., Wilson A. C., Haji-Azimi I., Fischberg M. Albumin phylogeny for clawed frogs (Xenopus). Science. 1977 Feb 25;195(4280):785–787. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Cabada M. O., Darnbrough C., Ford P. J., Turner P. C. Differential accumulation of two size classes of poly(A) associated with messenger RNA during oogenesis in Xenopus laevis. Dev Biol. 1977 Jun;57(2):427–439. doi: 10.1016/0012-1606(77)90227-5. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Hatton K. S., Tesfaye A., Yancopoulos G. D., Alt F. W. The human myc gene family: structure and activity of L-myc and an L-myc pseudogene. Genes Dev. 1987 Dec;1(10):1311–1326. doi: 10.1101/gad.1.10.1311. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs K. M., Martin G. R., Bishop J. M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989 Jun;3(6):860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Dumont J. N. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972 Feb;136(2):153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Fasano O., Taparowsky E., Fiddes J., Wigler M., Goldfarb M. Sequence and structure of the coding region of the human H-ras-1 gene from T24 bladder carcinoma cells. J Mol Appl Genet. 1983;2(2):173–180. [PubMed] [Google Scholar]
- Frykberg L., Graf T., Vennström B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1(4):415–422. [PubMed] [Google Scholar]
- Godeau F., Persson H., Gray H. E., Pardee A. B. C-myc expression is dissociated from DNA synthesis and cell division in Xenopus oocyte and early embryonic development. EMBO J. 1986 Dec 20;5(13):3571–3577. doi: 10.1002/j.1460-2075.1986.tb04684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden L., Schafer U., Rosbash M. Accumulation of individual pA+ RNAs during oogenesis of Xenopus laevis. Cell. 1980 Dec;22(3):835–844. doi: 10.1016/0092-8674(80)90560-7. [DOI] [PubMed] [Google Scholar]
- Grady E. F., Schwab M., Rosenau W. Expression of N-myc and c-src during the development of fetal human brain. Cancer Res. 1987 Jun 1;47(11):2931–2936. [PubMed] [Google Scholar]
- Gurdon J. B., Wickens M. P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- Gusse M., Ghysdael J., Evan G., Soussi T., Méchali M. Translocation of a store of maternal cytoplasmic c-myc protein into nuclei during early development. Mol Cell Biol. 1989 Dec;9(12):5395–5403. doi: 10.1128/mcb.9.12.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraudie J., Hourdry J., Vriz S., Singer M., Méchali M. Enhanced c-myc gene expression during forelimb regenerative outgrowth in the young Xenopus laevis. Proc Natl Acad Sci U S A. 1990 May;87(10):3797–3801. doi: 10.1073/pnas.87.10.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Jakobovits A., Schwab M., Bishop J. M., Martin G. R. Expression of N-myc in teratocarcinoma stem cells and mouse embryos. Nature. 1985 Nov 14;318(6042):188–191. doi: 10.1038/318188a0. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. W., Roberts J. M., Eisenman R. N. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol Cell Biol. 1986 Dec;6(12):4499–4508. doi: 10.1128/mcb.6.12.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mugrauer G., Alt F. W., Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol. 1988 Oct;107(4):1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G., Ekblom P. Contrasting expression patterns of three members of the myc family of protooncogenes in the developing and adult mouse kidney. J Cell Biol. 1991 Jan;112(1):13–25. doi: 10.1083/jcb.112.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Morgenbesser S. D., DePinho R. A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992 Aug;6(8):1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982 Oct;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982 Oct;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Expression of c-myc proto-oncogene during the early development of Xenopus laevis. Oncogene Res. 1987 Jul;1(2):179–191. [PubMed] [Google Scholar]
- Pfeifer-Ohlsson S., Rydnert J., Goustin A. S., Larsson E., Betsholtz C., Ohlsson R. Cell-type-specific pattern of myc protooncogene expression in developing human embryos. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5050–5054. doi: 10.1073/pnas.82.15.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Principaud E., Spohr G. Xenopus laevis c-myc I and II genes: molecular structure and developmental expression. Nucleic Acids Res. 1991 Jun 11;19(11):3081–3088. doi: 10.1093/nar/19.11.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid P., Schulz W. A., Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989 Jan 13;243(4888):226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- Schwab M., Varmus H. E., Bishop J. M. Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature. 1985 Jul 11;316(6024):160–162. doi: 10.1038/316160a0. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds G., Hartshorn A., Kennewell A., O'Mara M. A., Bruskin A., Bishop J. M. Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene. 1989 Mar;4(3):285–294. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. V., Gusse M., Evan G. I., Dathan N., Mechali M. Xenopus myc proto-oncogene during development: expression as a stable maternal mRNA uncoupled from cell division. EMBO J. 1986 Dec 20;5(13):3563–3570. doi: 10.1002/j.1460-2075.1986.tb04683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R., Schreiber-Agus N., Morgenbesser S. D., DePinho R. A. Myc and Max: a putative transcriptional complex in search of a cellular target. Curr Opin Cell Biol. 1992 Jun;4(3):468–474. doi: 10.1016/0955-0674(92)90013-3. [DOI] [PubMed] [Google Scholar]
- Vize P. D., Vaughan A., Krieg P. Expression of the N-myc proto-oncogene during the early development of Xenopus laevis. Development. 1990 Nov;110(3):885–896. doi: 10.1242/dev.110.3.885. [DOI] [PubMed] [Google Scholar]
- Vriz S., Taylor M., Méchali M. Differential expression of two Xenopus c-myc proto-oncogenes during development. EMBO J. 1989 Dec 20;8(13):4091–4097. doi: 10.1002/j.1460-2075.1989.tb08593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., Nisen P. D., Tesfaye A., Kohl N. E., Goldfarb M. P., Alt F. W. N-myc can cooperate with ras to transform normal cells in culture. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5455–5459. doi: 10.1073/pnas.82.16.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda G. K., Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992 Apr;8(4):124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]







